We narrowed to 20,061 results for: INO
-
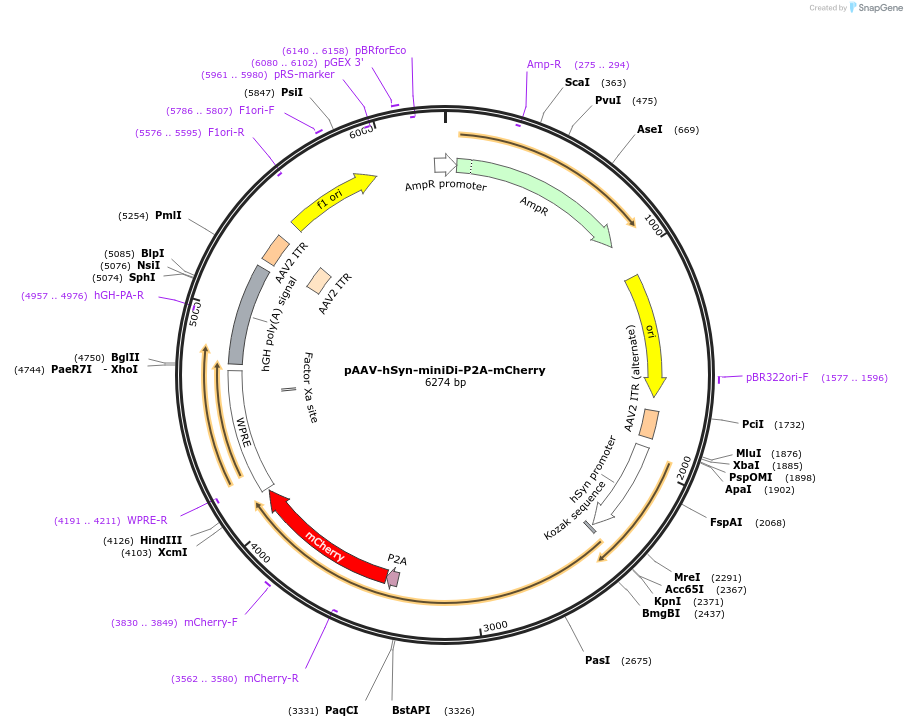
Plasmid#204358PurposeAAV vector for miniDi expression under the control of human synapsin promoterDepositorInsertminiDi-P2A-mCherry
UseAAVTagsmCherryMutationThe third intracellular loop (ICL3) of hM4Di was …PromoterhSynAvailable SinceOct. 22, 2024AvailabilityAcademic Institutions and Nonprofits only -
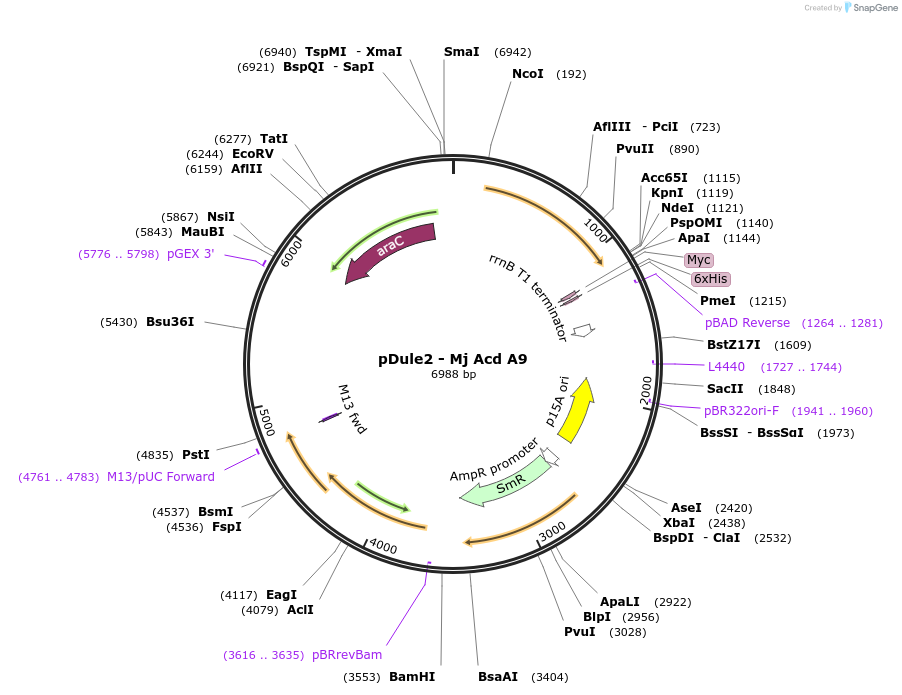
pDule2 - Mj Acd A9
Plasmid#197652PurposeE. coli machinery vector for the genetic encoding of acridone at TAG codons using the M. jannaschii Acd-A9 synthetase/tRNA pairDepositorInsertsMj Acd A9 synthetase
Methanocaldococcus jannaschii tRNA
TagsNoneExpressionBacterialMutationY32A, L65D, F108L, Q109E, D158S, L162TPromoterGlnS and lppAvailable SinceMarch 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
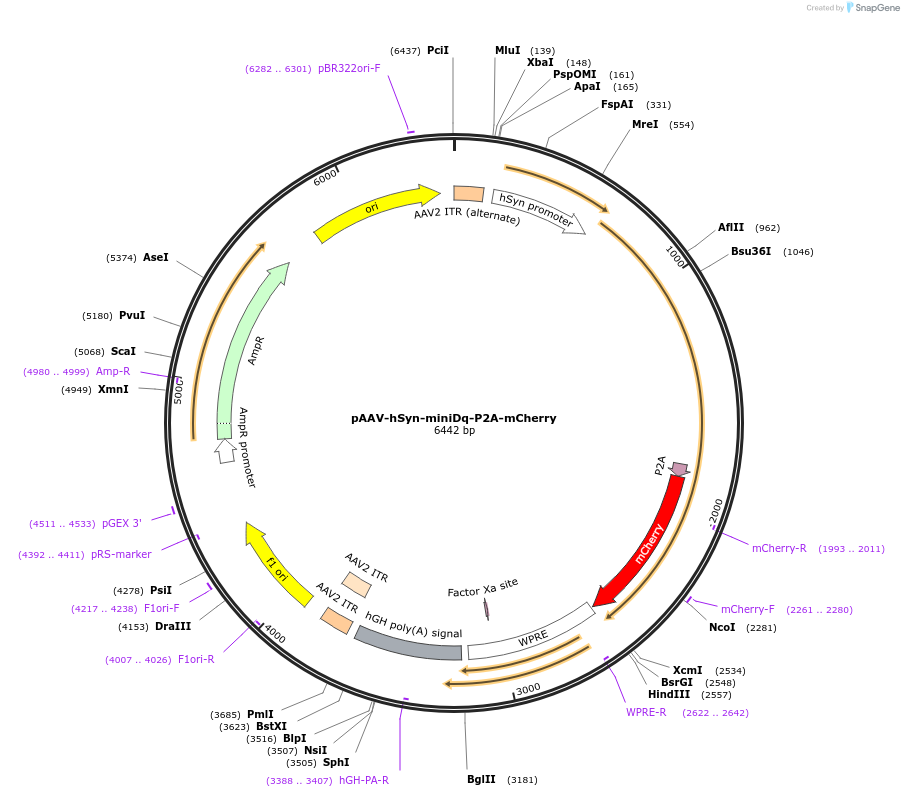
pAAV-hSyn-miniDq-P2A-mCherry
Plasmid#204357PurposeAAV vector for miniDq expression under the control of human synapsin promoterDepositorInsertminiDq-P2A-mCherry
UseAAVTagsmCherryMutationThe third intracellular loop (ICL3) of hM3Dq was …PromoterhSynAvailable SinceOct. 22, 2024AvailabilityAcademic Institutions and Nonprofits only -
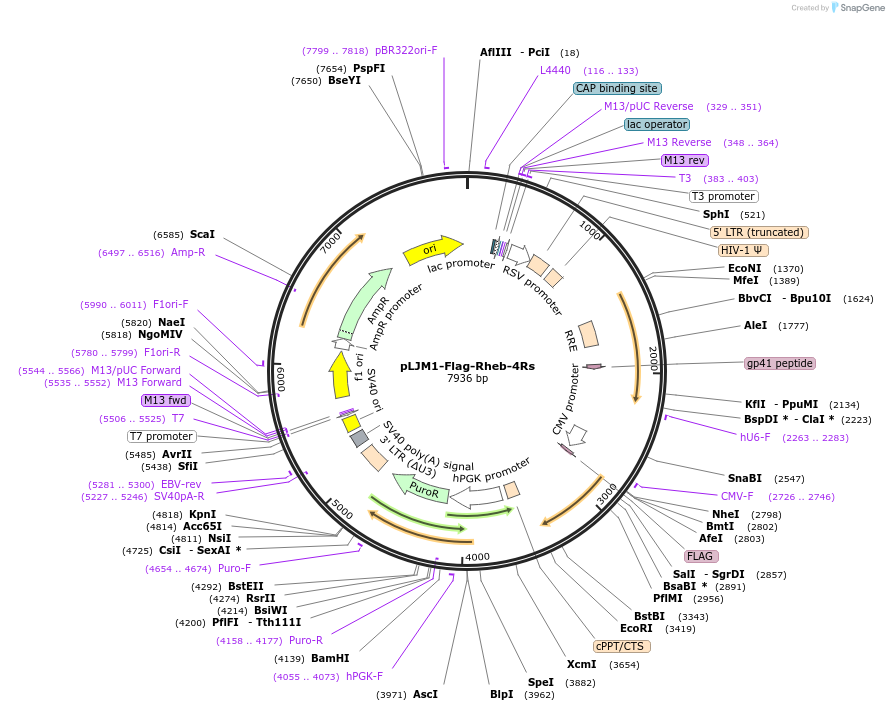
pLJM1-Flag-Rheb-4Rs
Plasmid#165022Purposeexpression of Flag-Rheb-4Rs mutant protein in mammalian cells (the lysine residues on the position 109, 135, 151 and 178 on Rheb protein are mutated to arginines)DepositorInsertRheb-4Rs (RHEB Human)
UseLentiviralTagsflagExpressionMammalianMutationchange lysine residues on position 109, 135, 151,…Available SinceMarch 10, 2021AvailabilityAcademic Institutions and Nonprofits only -
pMXs-puro HA-Atg14*dCC
Plasmid#38266DepositorInsertautophagy related genes 14 (ATG14 Human)
UseRetroviralTags3xHAExpressionMammalianMutationchanged A618T, A621T, C624A and T627C for siRNA r…PromoterLTRAvailable SinceOct. 30, 2012AvailabilityAcademic Institutions and Nonprofits only -
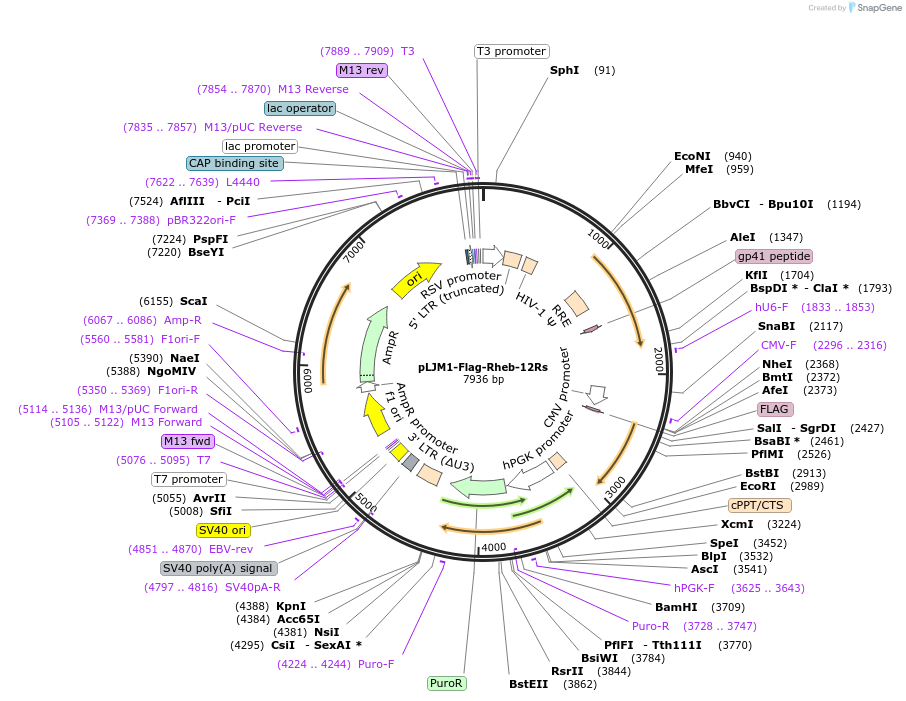
pLJM1-Flag-Rheb-12Rs
Plasmid#165023Purposeexpression of Flag-Rheb-12Rs mutant protein in mammalian cells (all the lysine residues on Rheb except the K19 and K120 are mutated to arginines).DepositorInsertRheb-12Rs (RHEB Human)
UseLentiviralTagsflagExpressionMammalianMutationAll the lysine residues except K19 and K120 are c…Available SinceMarch 10, 2021AvailabilityAcademic Institutions and Nonprofits only -
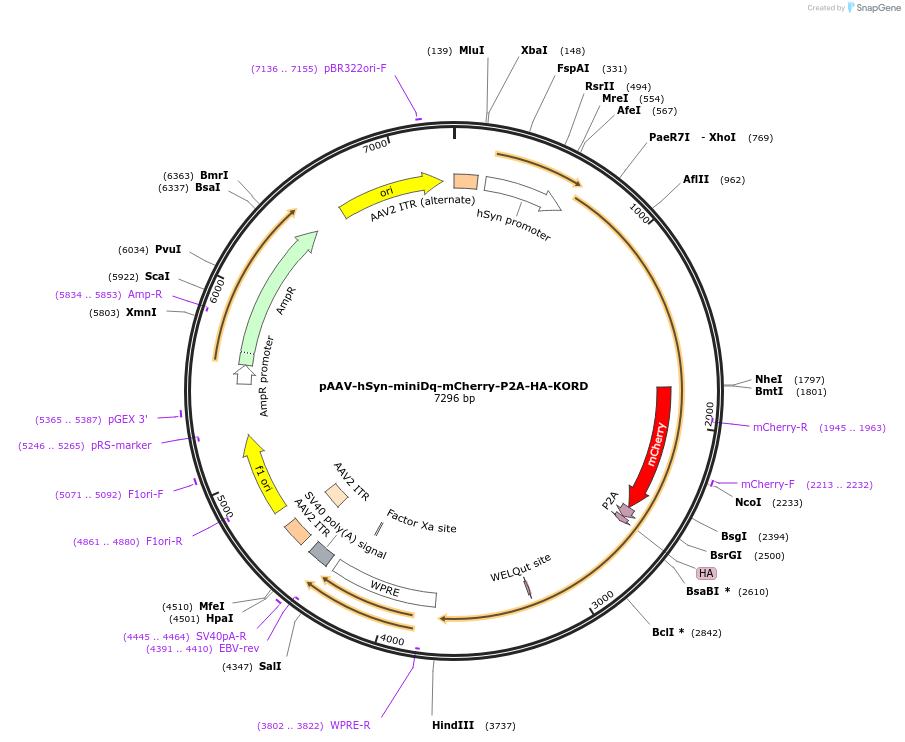
pAAV-hSyn-miniDq-mCherry-P2A-HA-KORD
Plasmid#204359PurposeAAV vector for coexpression of miniDq and KORD under the control of human synapsin promoterDepositorInsertminiDq-mCherry-P2A-HA-KORD
UseAAVTagsHA (for KORD) and mCherry (for miniDq)MutationFor miniDq, the third intracellular loop (ICL3) o…PromoterhSynAvailable SinceOct. 22, 2024AvailabilityAcademic Institutions and Nonprofits only -
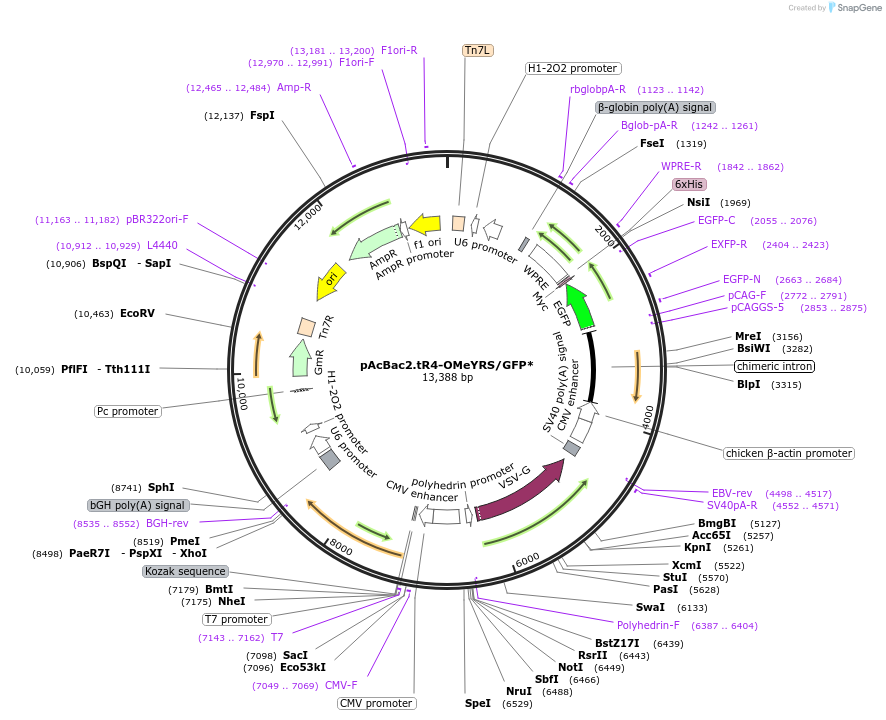
pAcBac2.tR4-OMeYRS/GFP*
Plasmid#50831Purposebaculovirus-based delivery system that enables the efficient incorporation of unnatural amino acids into proteins in mammalian cellsDepositorInsertstwo-copy Tyr tRNA cassette
EGFP*
OMeYRS
two copy Tyr tRNA cassette
UseBaculoviralTagsHis, Myc-6xHis, and WPREExpressionInsectMutationY39TAG and able to charge various unnatural amino…PromoterCAG, CMV, and U6 and H1Available SinceJune 3, 2015AvailabilityAcademic Institutions and Nonprofits only -
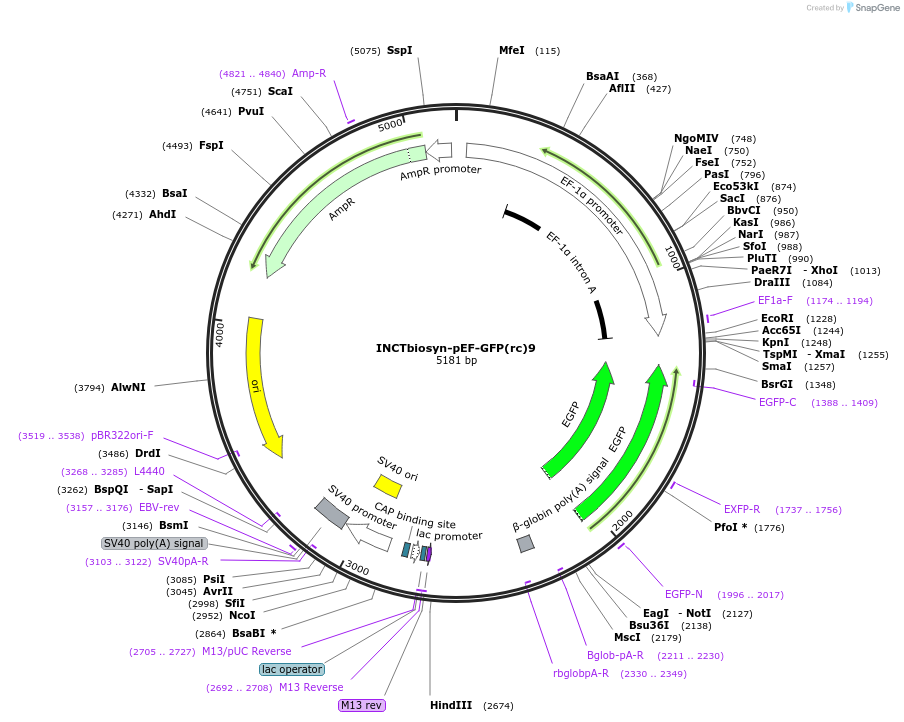
INCTbiosyn-pEF-GFP(rc)9
Plasmid#127508PurposePlasmid has an EF1 alpha promoter in a forward sequence orientation and an inverted EGFP coding sequence (reverse complement) flanked by attB and attP integrase 9 attachment sites.DepositorInsertEGFP reverse complement coding sequence flanked by attB/attP Integrase 9 attachment sites.
ExpressionMammalianAvailable SinceMay 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
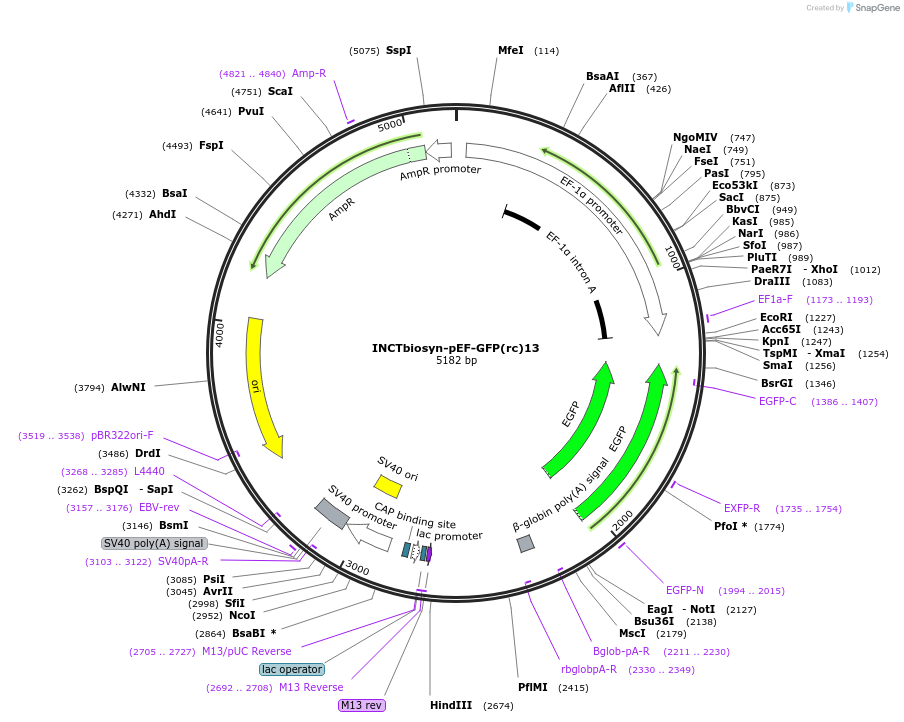
INCTbiosyn-pEF-GFP(rc)13
Plasmid#127509PurposePlasmid has an EF1 alpha promoter in a forward sequence orientation and an inverted EGFP coding sequence (reverse complement) flanked by attB and attP integrase 13 attachment sites.DepositorInsertEGFP reverse complement coding sequence flanked by attB/attP Integrase 13 attachment sites.
ExpressionMammalianAvailable SinceMay 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
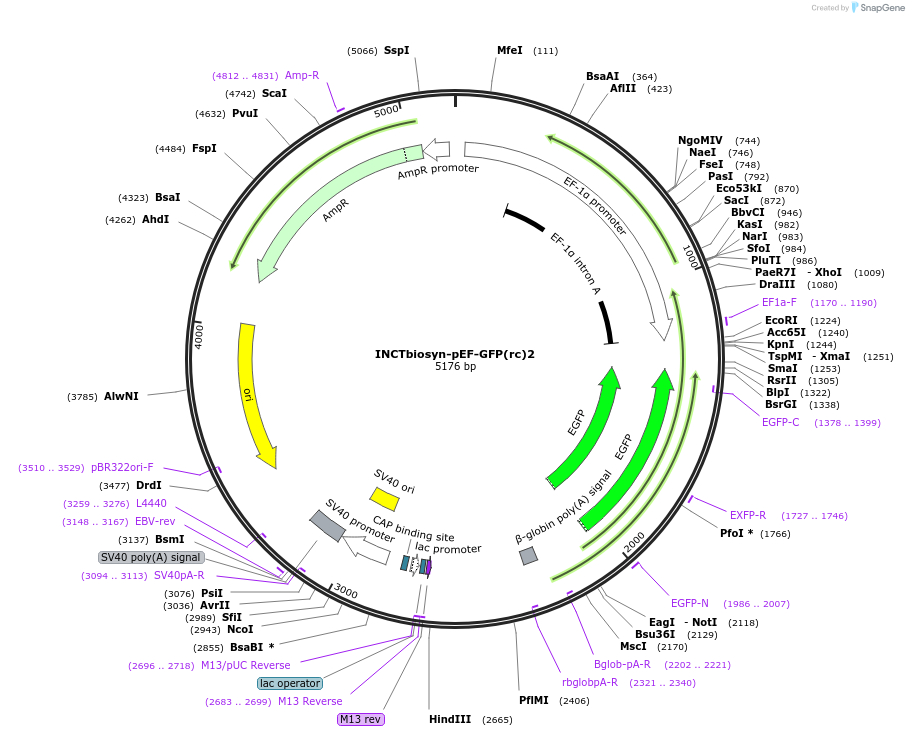
INCTbiosyn-pEF-GFP(rc)2
Plasmid#127504PurposePlasmid has an EF1 alpha promoter in a forward sequence orientation and an inverted EGFP coding sequence (reverse complement) flanked by attB and attP integrase 2 attachment sites.DepositorInsertEGFP reverse complement coding sequence flanked by attB/attP Integrase 2 attachment sites.
ExpressionMammalianAvailable SinceMay 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
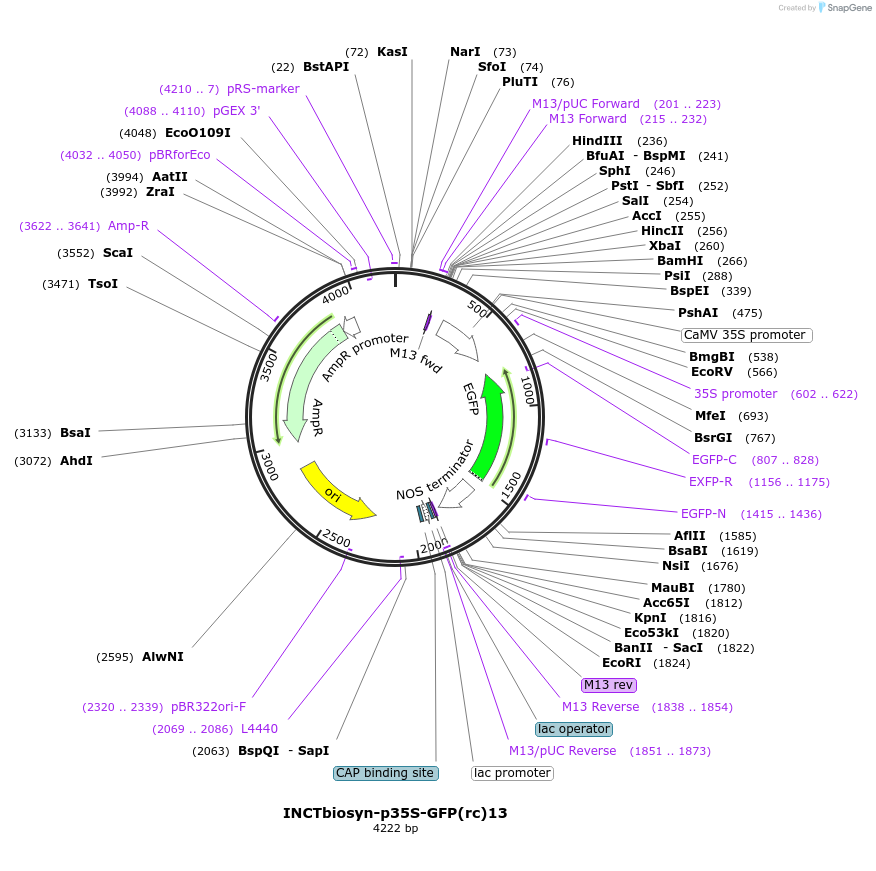
INCTbiosyn-p35S-GFP(rc)13
Plasmid#127526PurposePlasmid has a CaMV 35S promoter in a forward sequence orientation and an inverted EGFP coding sequence (reverse complement) flanked by attB and attP integrase 13 attachment sites.DepositorInsertEGFP reverse complement coding sequence flanked by attB/attP Integrase 13 attachment sites.
ExpressionPlantAvailable SinceMay 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
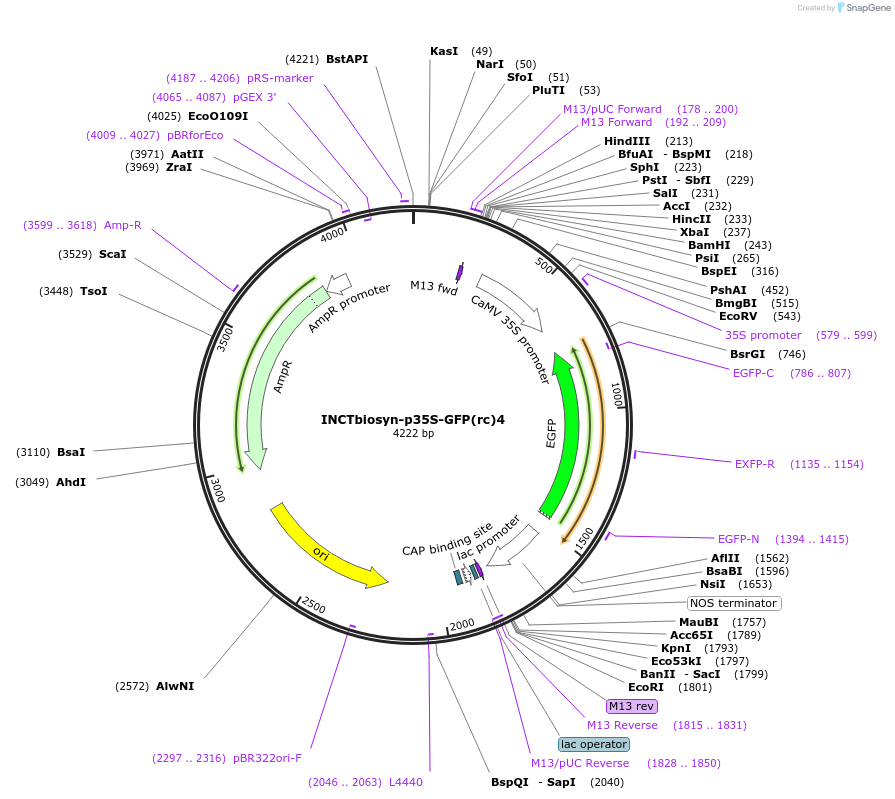
INCTbiosyn-p35S-GFP(rc)4
Plasmid#127522PurposePlasmid has a CaMV 35S promoter in a forward sequence orientation and an inverted EGFP coding sequence (reverse complement) flanked by attB and attP integrase 4 attachment sites.DepositorInsertEGFP reverse complement coding sequence flanked by attB/attP Integrase 4 attachment sites.
ExpressionPlantAvailable SinceMay 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
INCTbiosyn-p35S-GFP(rc)5
Plasmid#127523PurposePlasmid has a CaMV 35S promoter in a forward sequence orientation and an inverted EGFP coding sequence (reverse complement) flanked by attB and attP integrase 5 attachment sites.DepositorInsertEGFP reverse complement coding sequence flanked by attB/attP Integrase 5 attachment sites.
ExpressionPlantAvailable SinceMay 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
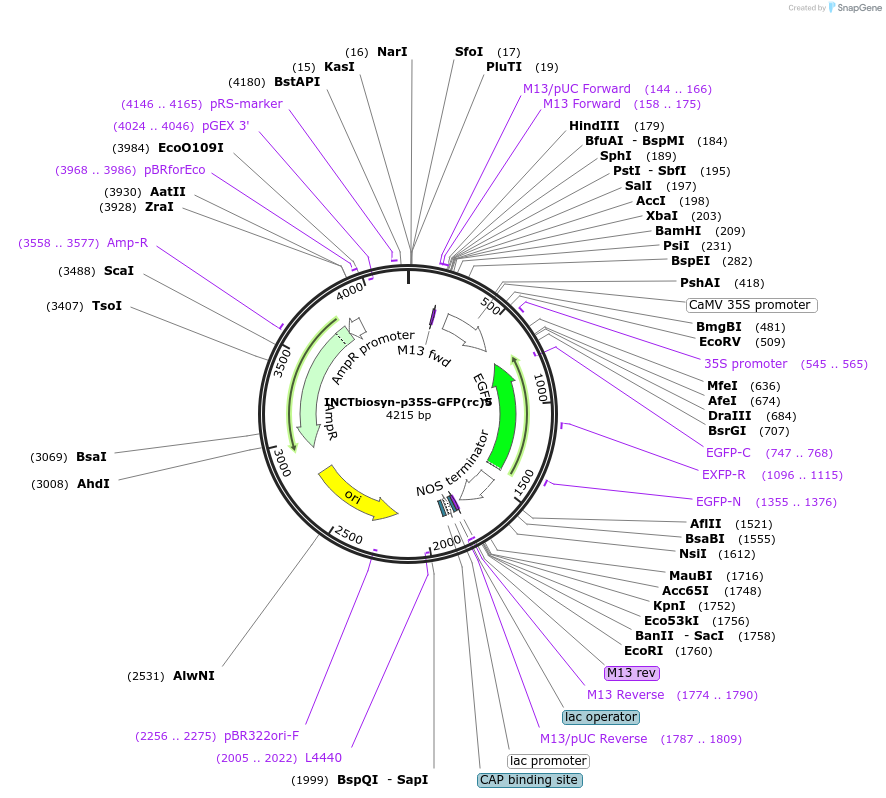
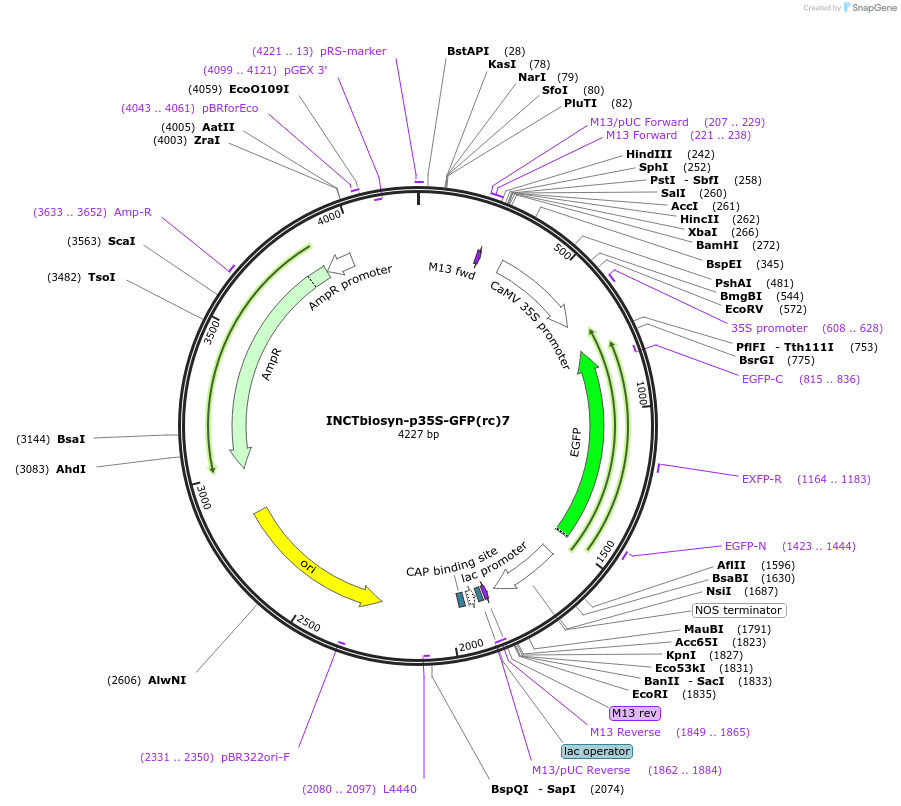
INCTbiosyn-p35S-GFP(rc)7
Plasmid#127524PurposePlasmid has a CaMV 35S promoter in a forward sequence orientation and an inverted EGFP coding sequence (reverse complement) flanked by attB and attP integrase 7 attachment sites.DepositorInsertEGFP reverse complement coding sequence flanked by attB/attP Integrase 7 attachment sites.
ExpressionPlantAvailable SinceMay 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
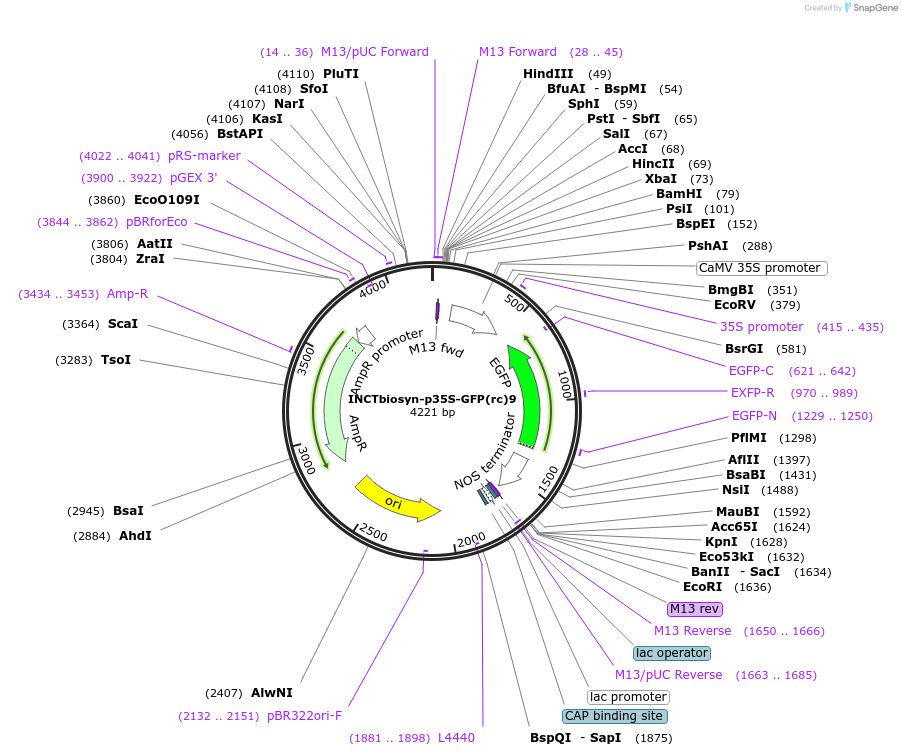
INCTbiosyn-p35S-GFP(rc)9
Plasmid#127525PurposePlasmid has a CaMV 35S promoter in a forward sequence orientation and an inverted EGFP coding sequence (reverse complement) flanked by attB and attP integrase 9 attachment sites.DepositorInsertEGFP reverse complement coding sequence flanked by attB/attP Integrase 9 attachment sites.
ExpressionPlantAvailable SinceMay 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
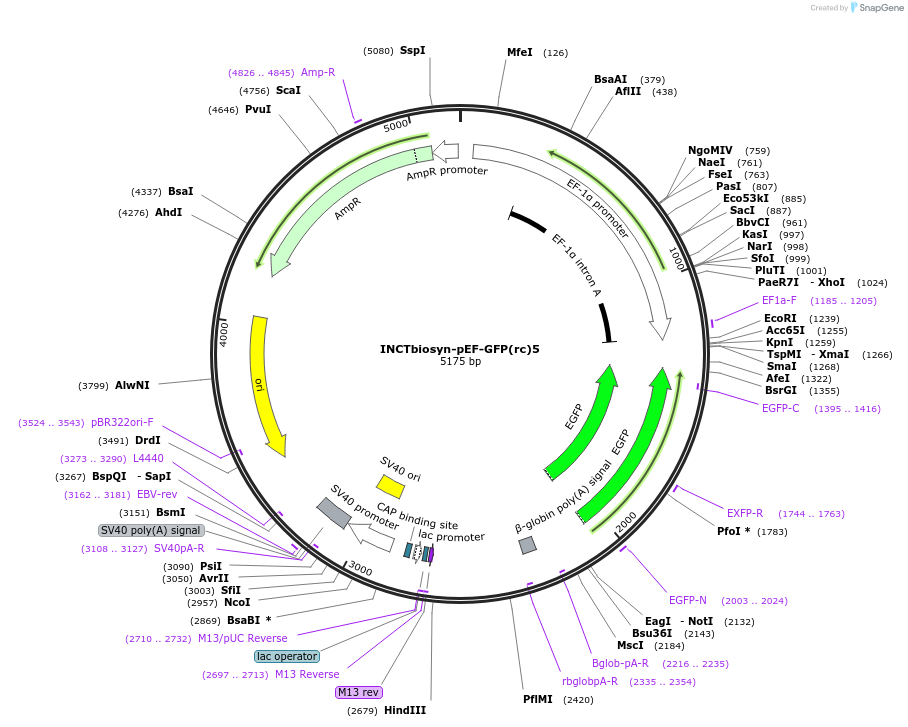
INCTbiosyn-pEF-GFP(rc)5
Plasmid#127506PurposePlasmid has an EF1 alpha promoter in a forward sequence orientation and an inverted EGFP coding sequence (reverse complement) flanked by attB and attP integrase 5 attachment sites.DepositorInsertEGFP reverse complement coding sequence flanked by attB/attP Integrase 5 attachment sites.
ExpressionMammalianAvailable SinceMay 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
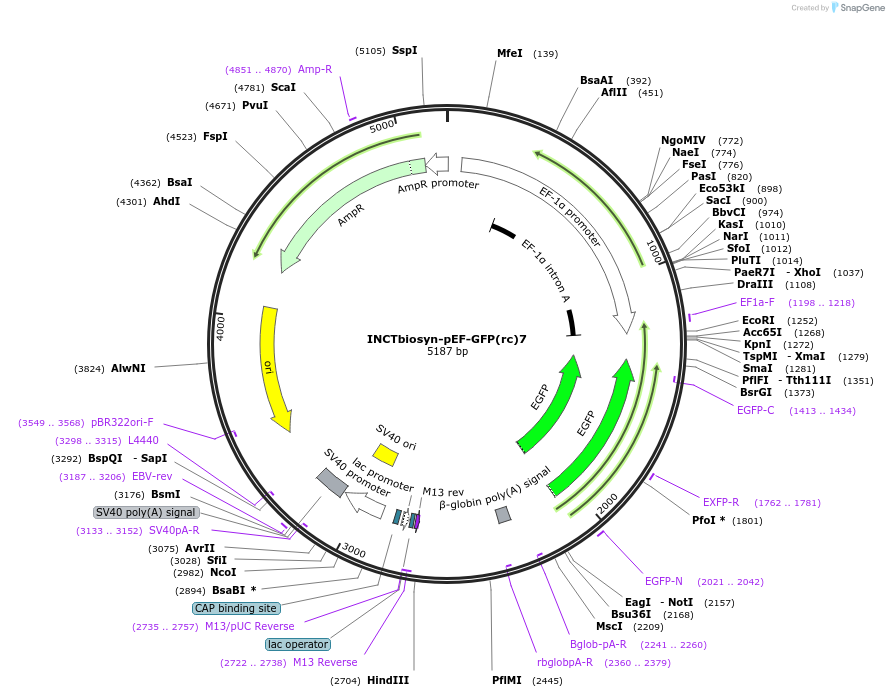
INCTbiosyn-pEF-GFP(rc)7
Plasmid#127507PurposePlasmid has an EF1 alpha promoter in a forward sequence orientation and an inverted EGFP coding sequence (reverse complement) flanked by attB and attP integrase 7 attachment sites.DepositorInsertEGFP reverse complement coding sequence flanked by attB/attP Integrase 7 attachment sites.
ExpressionMammalianAvailable SinceMay 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
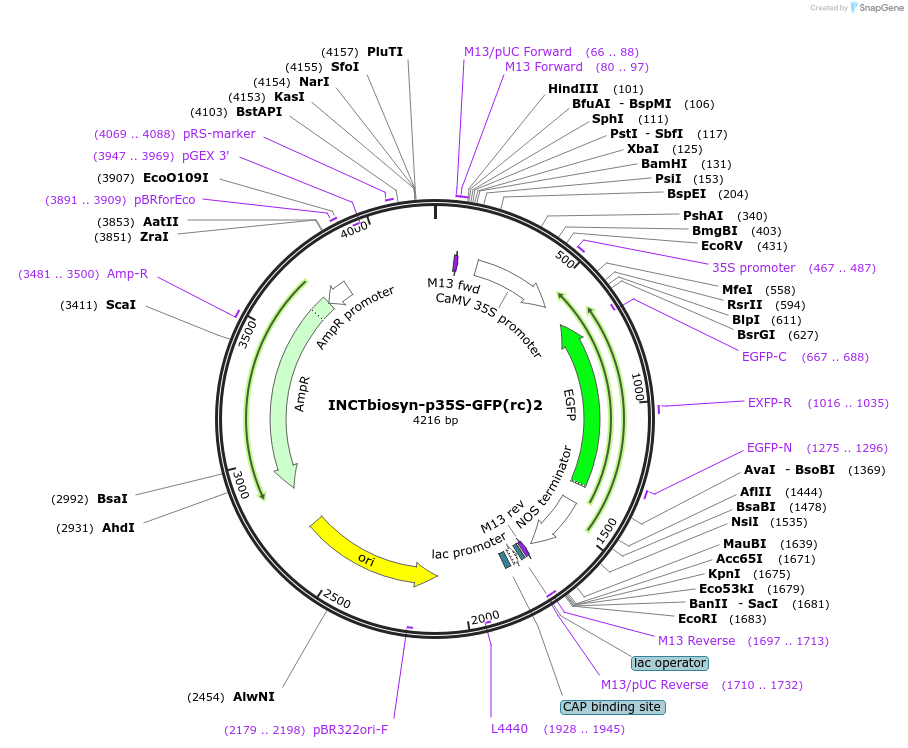
INCTbiosyn-p35S-GFP(rc)2
Plasmid#127521PurposePlasmid has a CaMV 35S promoter in a forward sequence orientation and an inverted EGFP coding sequence (reverse complement) flanked by attB and attP integrase 2 attachment sites.DepositorInsertEGFP reverse complement coding sequence flanked by attB/attP Integrase 2 attachment sites.
ExpressionPlantAvailable SinceMay 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
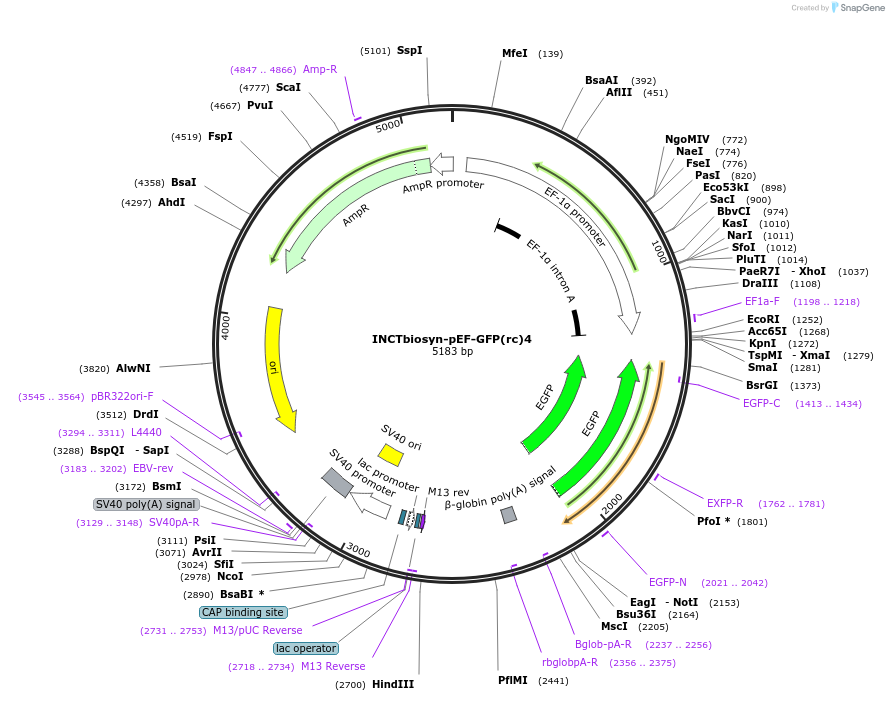
INCTbiosyn-pEF-GFP(rc)4
Plasmid#127505PurposePlasmid has an EF1 alpha promoter in a forward sequence orientation and an inverted EGFP coding sequence (reverse complement) flanked by attB and attP integrase 4 attachment sites.DepositorInsertEGFP reverse complement coding sequence flanked by attB/attP Integrase 4 attachment sites.
ExpressionMammalianAvailable SinceMay 6, 2020AvailabilityAcademic Institutions and Nonprofits only