We narrowed to 11,399 results for: nar
-
Plasmid#82735PurposeGateway Donor vector containing SERPINB5, part of the Target Accelerator Plasmid Collection.DepositorInsertSERPINB5 (SERPINB5 Human)
UseGateway entry vectorMutationp. D141A; p. 176S>P; p.190_192delTDTins25amino…PromoterNoneAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
pDONR223_SERPINB5_p.A7T
Plasmid#82736PurposeGateway Donor vector containing SERPINB5, part of the Target Accelerator Plasmid Collection.DepositorInsertSERPINB5 (SERPINB5 Human)
UseGateway entry vectorMutationp. A7T; p.176S>P; p. 190_192delTDTins25; p.194…PromoterNoneAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
pDONR223_SERPINB5_p.A42F
Plasmid#82738PurposeGateway Donor vector containing SERPINB5, part of the Target Accelerator Plasmid Collection.DepositorInsertSERPINB5 (SERPINB5 Human)
UseGateway entry vectorMutationp.A42F; p.176S>P; p.190_192delTDTins25; p.194_…PromoterNoneAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
pDONR223_SERPINB5_p.A165T
Plasmid#82741PurposeGateway Donor vector containing SERPINB5, part of the Target Accelerator Plasmid Collection.DepositorInsertSERPINB5 (SERPINB5 Human)
UseGateway entry vectorMutationp. A165T; p.176S>P; p.190_192delTDTins25; p.19…PromoterNoneAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
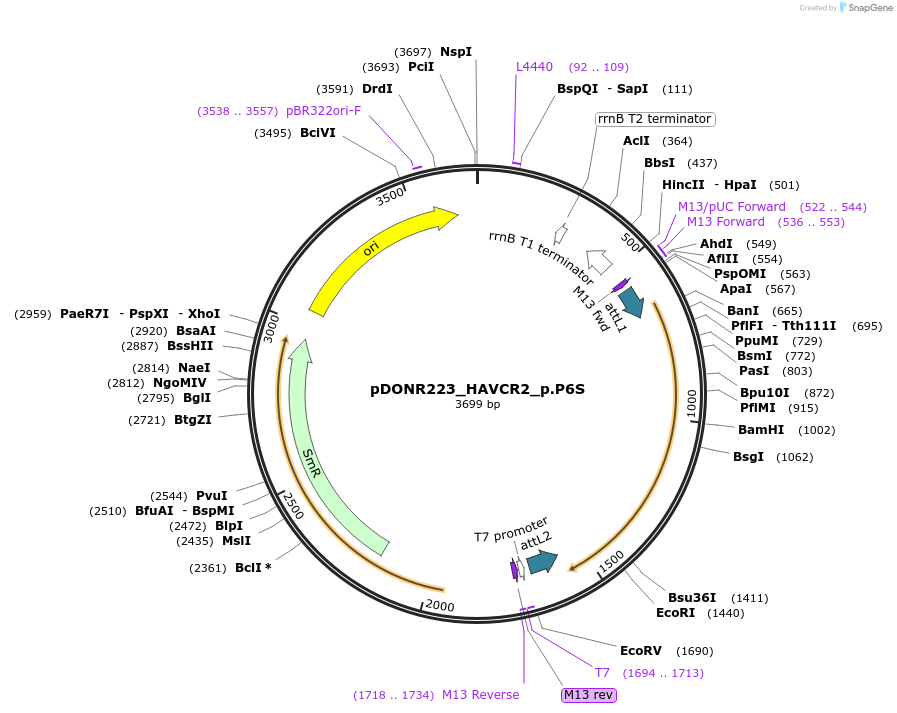
pDONR223_HAVCR2_p.P6S
Plasmid#82742PurposeGateway Donor vector containing HAVCR2, part of the Target Accelerator Plasmid Collection.DepositorAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
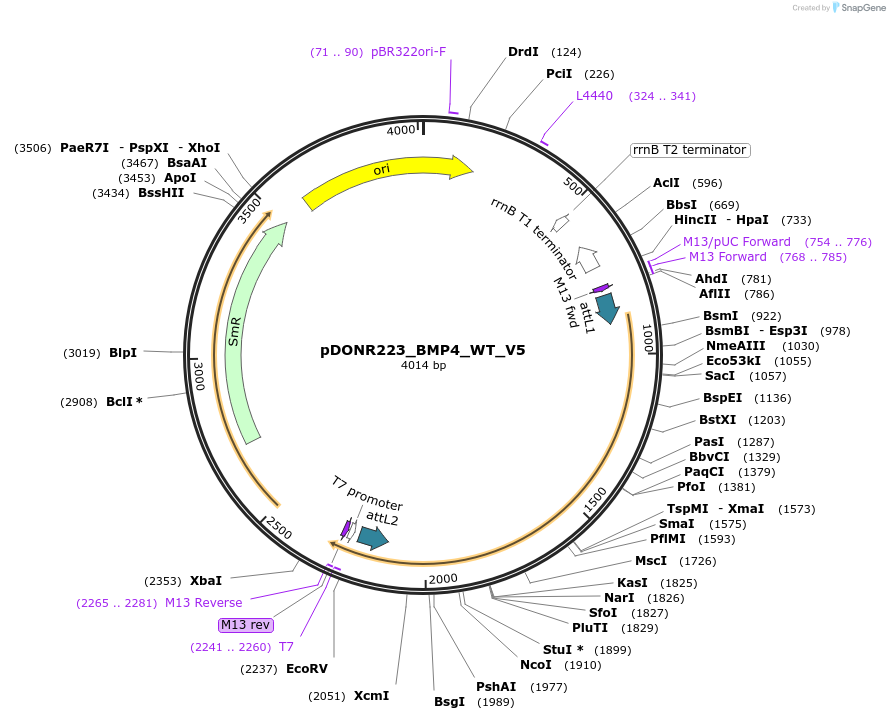
pDONR223_BMP4_WT_V5
Plasmid#82937PurposeGateway Donor vector containing BMP4 , used as an over expression control for gene expression analysis. Part of the Target Accelerator Plasmid Collection.DepositorAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
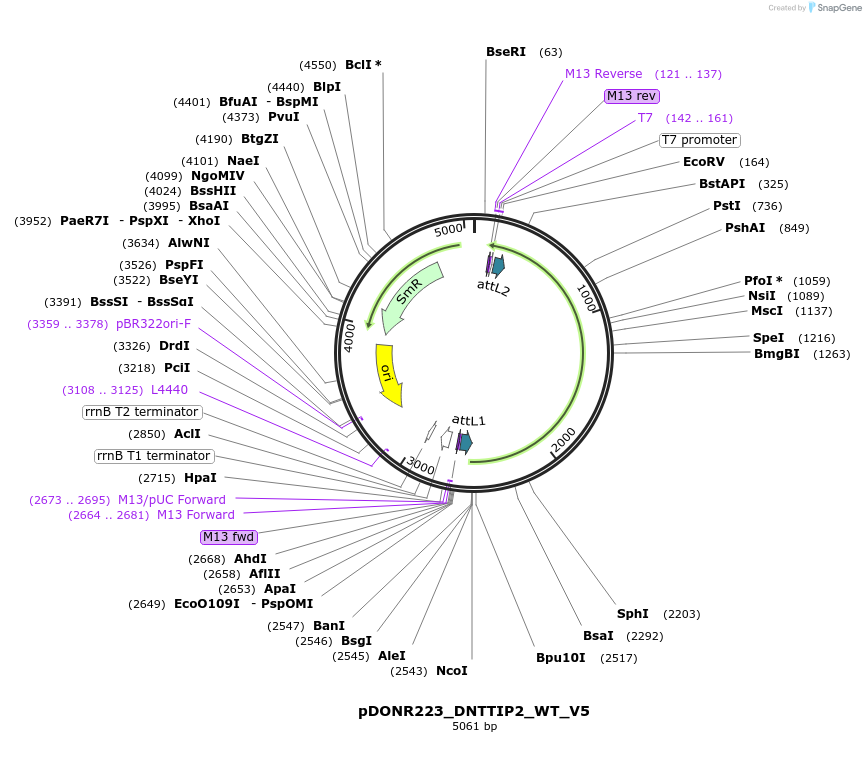
pDONR223_DNTTIP2_WT_V5
Plasmid#82939PurposeGateway Donor vector containing DNTTIP2, used as an over expression control for gene expression analysis. Part of the Target Accelerator Plasmid Collection.DepositorAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
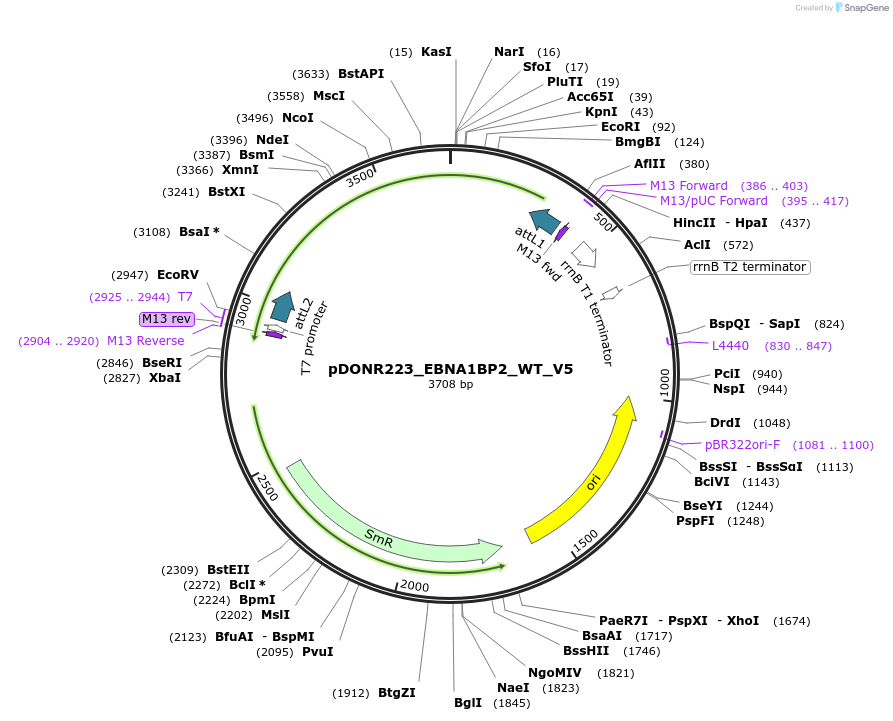
pDONR223_EBNA1BP2_WT_V5
Plasmid#82932PurposeGateway Donor vector containing EBNA1BP2, used as an over expression control for gene expression analysis. Part of the Target Accelerator Plasmid Collection.DepositorAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
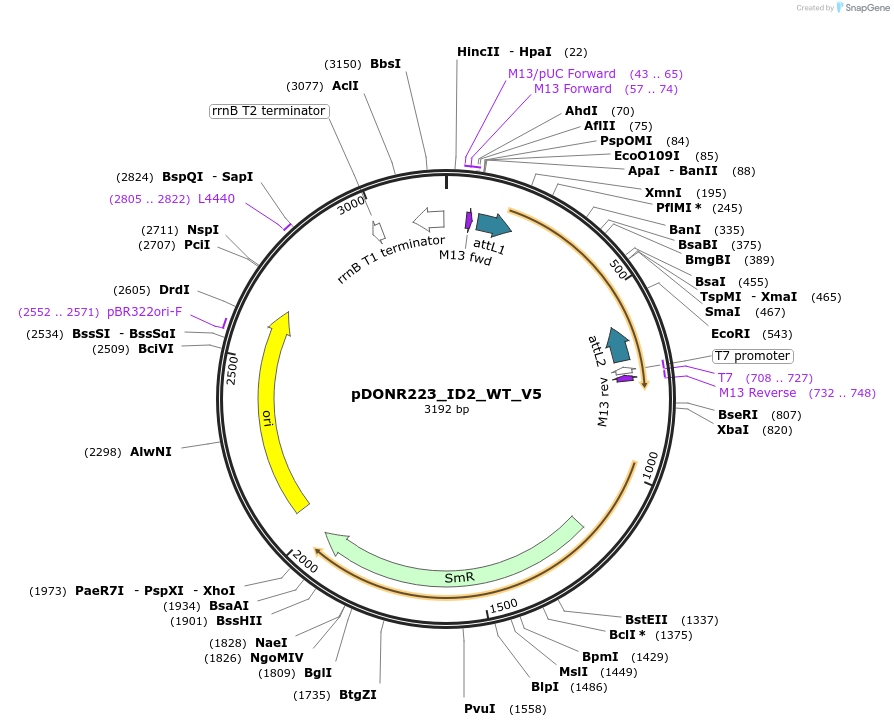
pDONR223_ID2_WT_V5
Plasmid#82960PurposeGateway Donor vector containing ID2, used as an over expression control for gene expression analysis. Part of the Target Accelerator Plasmid Collection.DepositorAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
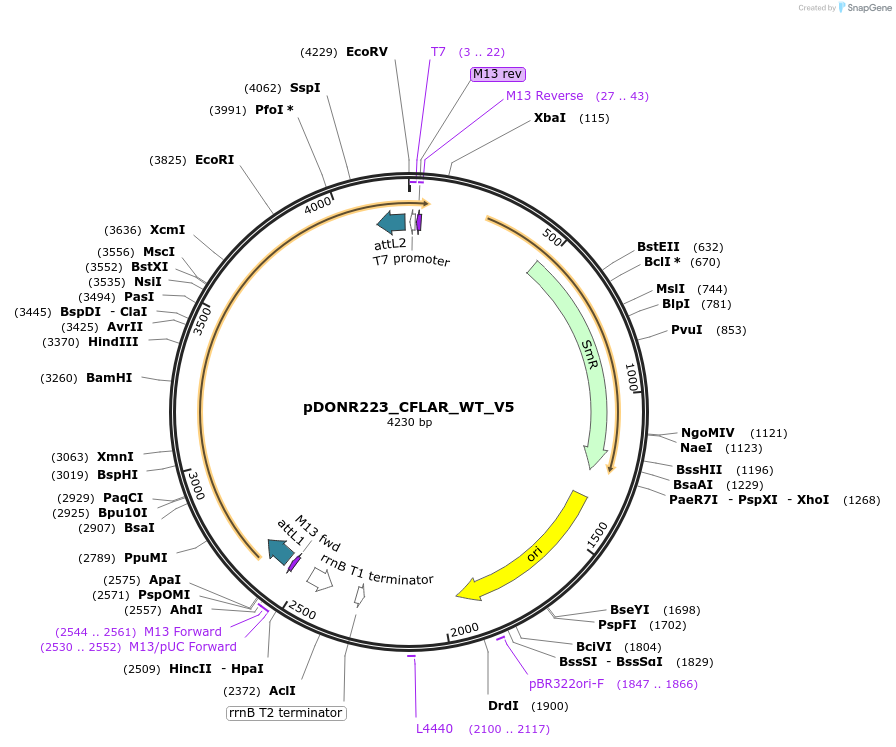
pDONR223_CFLAR_WT_V5
Plasmid#82936PurposeGateway Donor vector containing CFLAR, used as an over expression control for gene expression analysis. Part of the Target Accelerator Plasmid Collection.DepositorAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
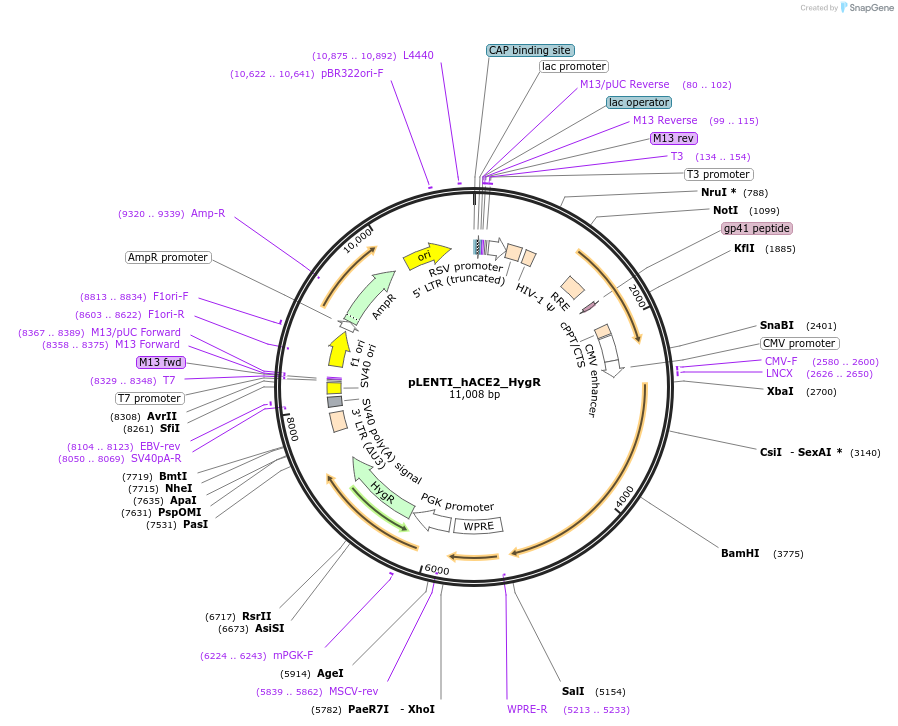
pLENTI_hACE2_HygR
Plasmid#155296PurposeLentiviral vector to generate hACE2 stable expressing cell line, Hygromycin resistanceDepositorAvailable SinceAug. 5, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
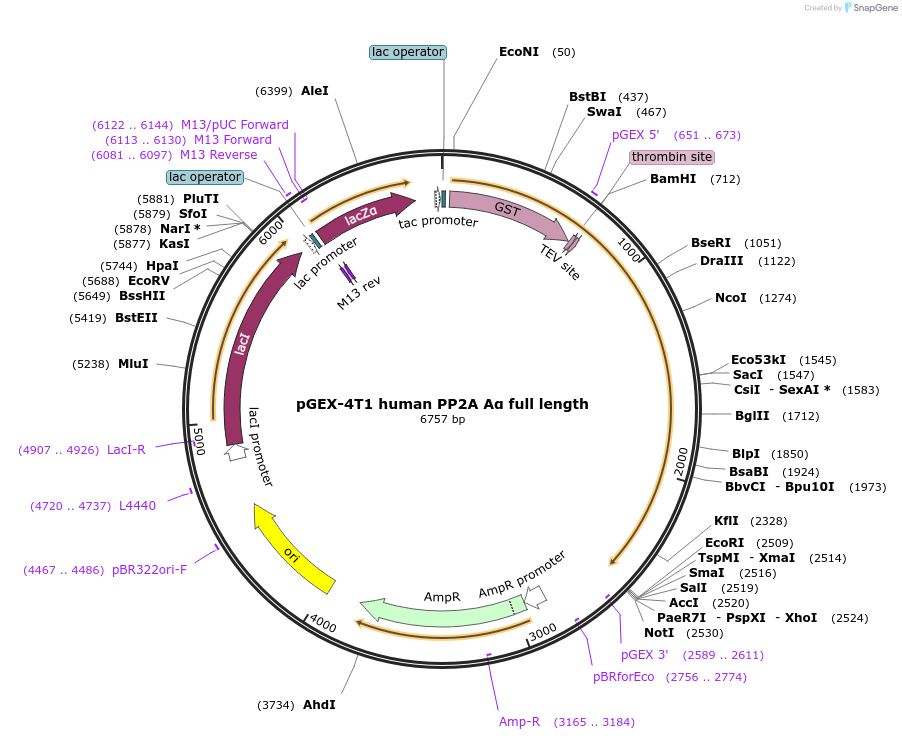
pGEX-4T1 human PP2A Aα full length
Plasmid#132634Purposeexpresses human PP2A Aα full length in E. coliDepositorInsertPP2A Aα (PPP2R1A Human)
ExpressionBacterialAvailable SinceOct. 17, 2019AvailabilityAcademic Institutions and Nonprofits only -
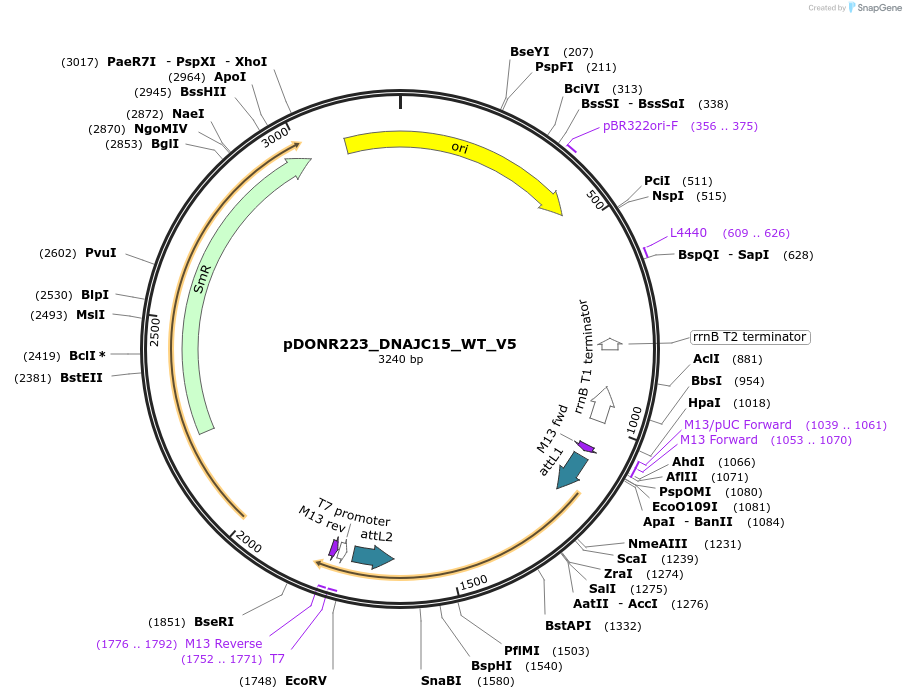
pDONR223_DNAJC15_WT_V5
Plasmid#82964PurposeGateway Donor vector containing DNAJC15, used as an over expression control for gene expression analysis. Part of the Target Accelerator Plasmid Collection.DepositorAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
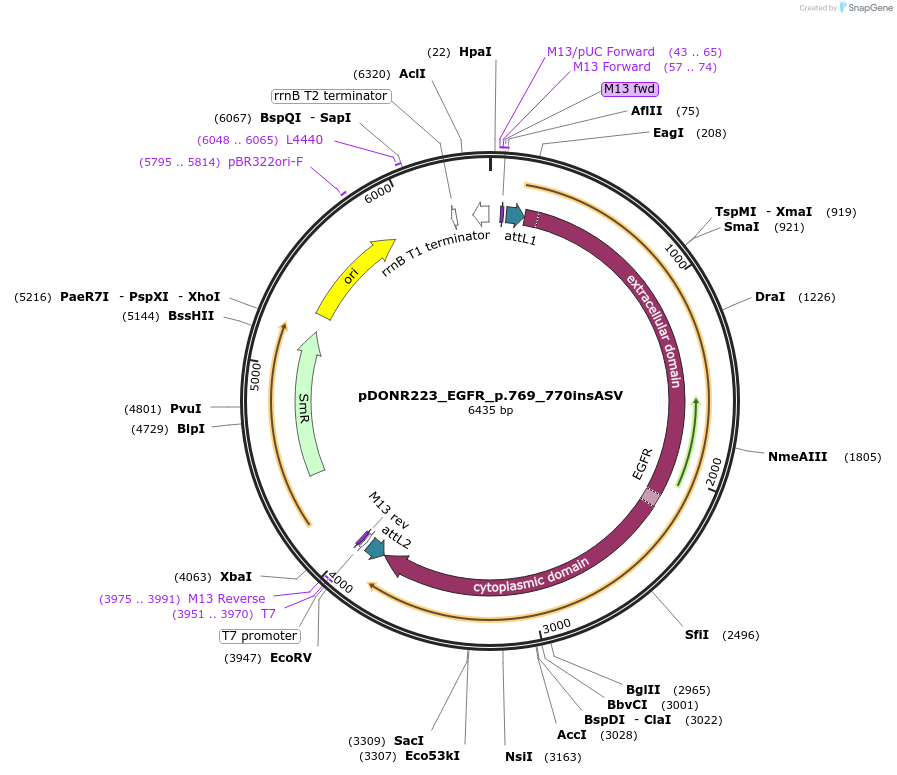
pDONR223_EGFR_p.769_770insASV
Plasmid#82880PurposeGateway Donor vector containing EGFR, part of the Target Accelerator Plasmid Collection.DepositorInsertEGFR (EGFR Human)
UseGateway entry vectorMutationASV insertion between AA 769 and 770PromoterNoneAvailable SinceOct. 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
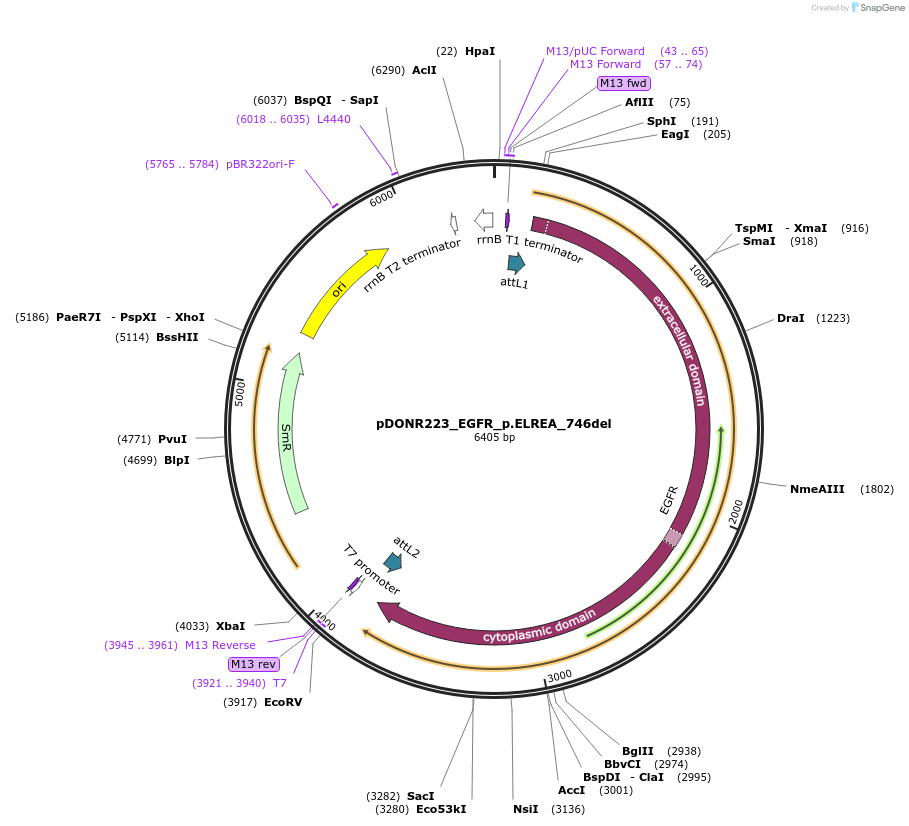
pDONR223_EGFR_p.ELREA_746del
Plasmid#82911PurposeGateway Donor vector containing EGFR, part of the Target Accelerator Plasmid Collection.DepositorInsertEGFR (EGFR Human)
UseGateway entry vectorMutationDeletion of residues 746-750 (ELREA)PromoterNoneAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
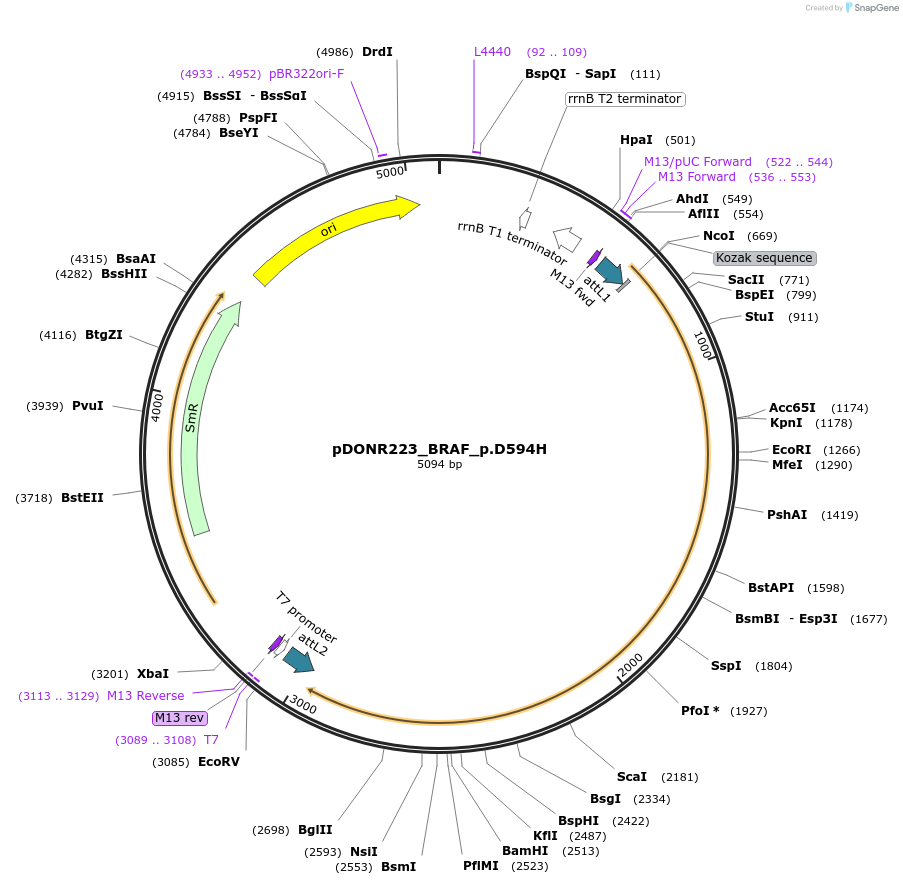
pDONR223_BRAF_p.D594H
Plasmid#82816PurposeGateway Donor vector containing BRAF, part of the Target Accelerator Plasmid Collection.DepositorAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
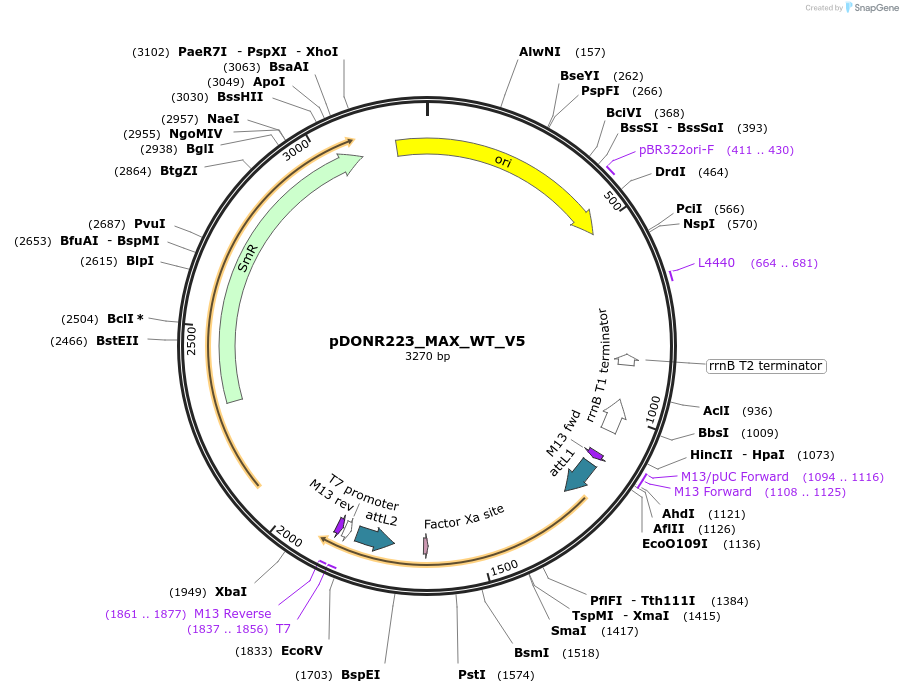
pDONR223_MAX_WT_V5
Plasmid#82944PurposeGateway Donor vector containing MAX, used as an over expression control for gene expression analysis. Part of the Target Accelerator Plasmid Collection.DepositorAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
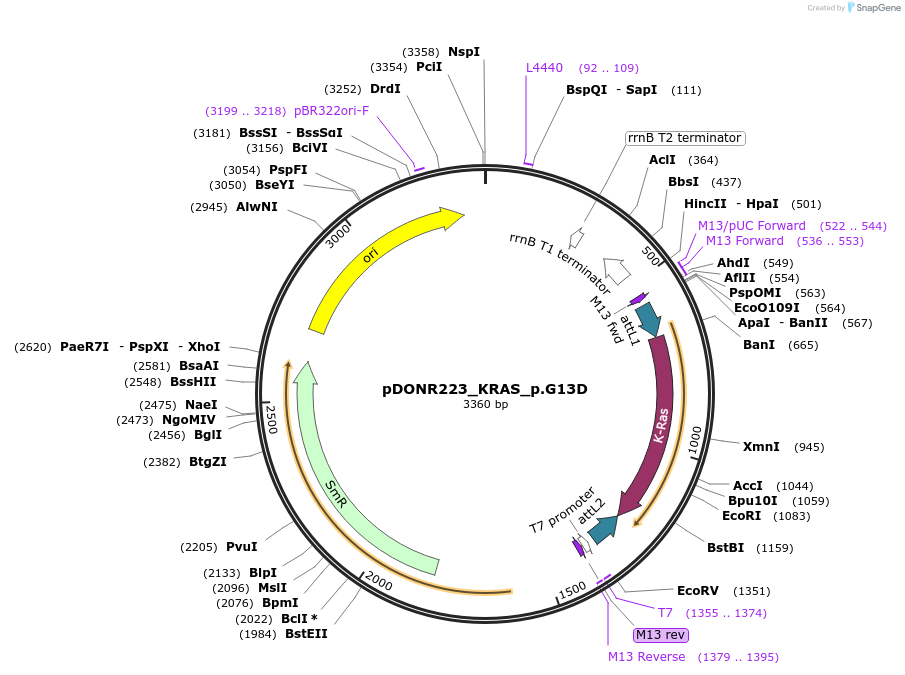
pDONR223_KRAS_p.G13D
Plasmid#82808PurposeGateway Donor vector containing KRAS, part of the Target Accelerator Plasmid Collection.DepositorAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
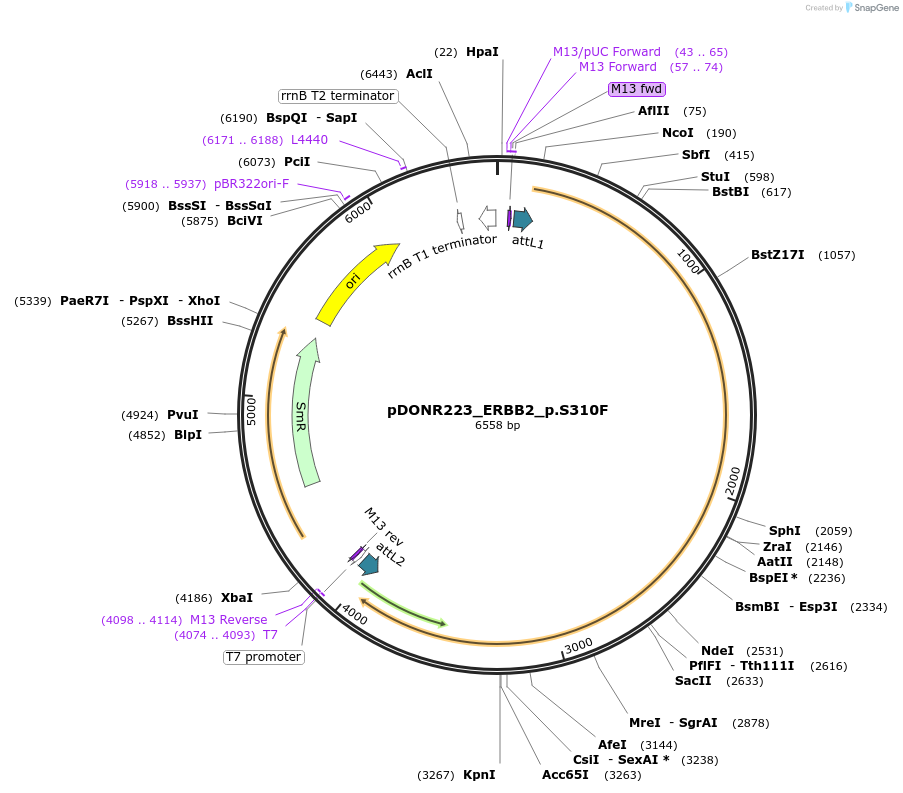
pDONR223_ERBB2_p.S310F
Plasmid#82854PurposeGateway Donor vector containing ERBB2, part of the Target Accelerator Plasmid Collection.DepositorAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
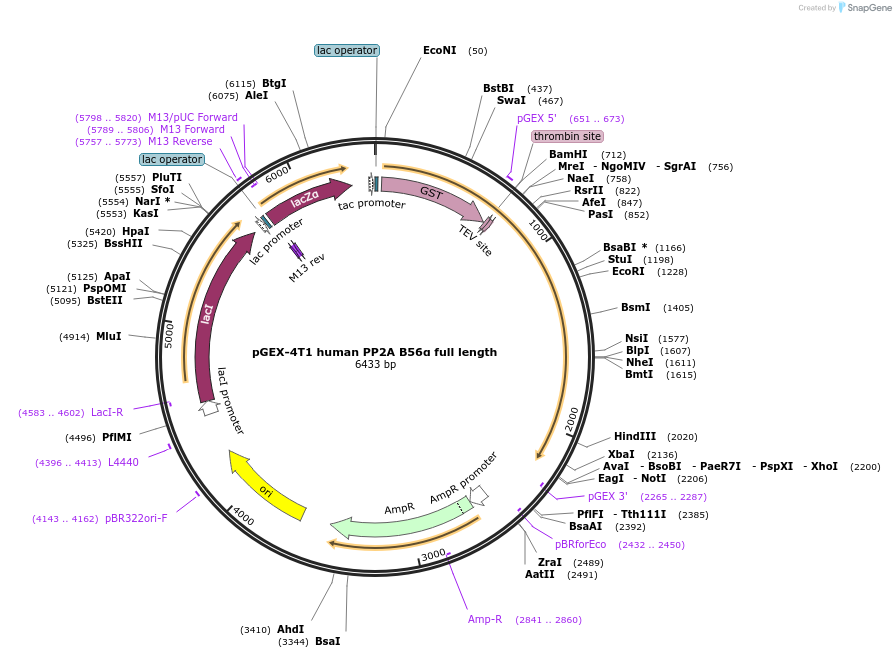
pGEX-4T1 human PP2A B56α full length
Plasmid#132635Purposeexpresses human PP2A B56α full length in E. coliDepositorInsertPP2A B56α
ExpressionBacterialAvailable SinceOct. 17, 2019AvailabilityAcademic Institutions and Nonprofits only