We narrowed to 8,660 results for: GAL
-
Plasmid#154038PurposeDiatom promoter for P. tricornutum; uLoop specific syntax; AC overhangs; Lvl 0DepositorInsertPromoter Nub
UseSynthetic BiologyAvailable SinceDec. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
pL0_EF_tSVP
Plasmid#154041PurposeDiatom terminator for P. tricornutum; uLoop specific syntax; EF overhangs; Lvl 0DepositorInsertTerminator SVP
UseSynthetic BiologyAvailable SinceDec. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
pL0_AC_pPbt
Plasmid#154042PurposeDiatom promoter for P. tricornutum; uLoop specific syntax; AC overhangs; Lvl 0DepositorInsertPromoter Pbt
UseSynthetic BiologyAvailable SinceDec. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
AlACR2_EYFP_pcDNA3.1
Plasmid#161023PurposeExpresses AlACR2 in mammalian cellsDepositorInsertAlACR2
TagsEYFPExpressionMammalianAvailable SinceNov. 17, 2020AvailabilityAcademic Institutions and Nonprofits only -
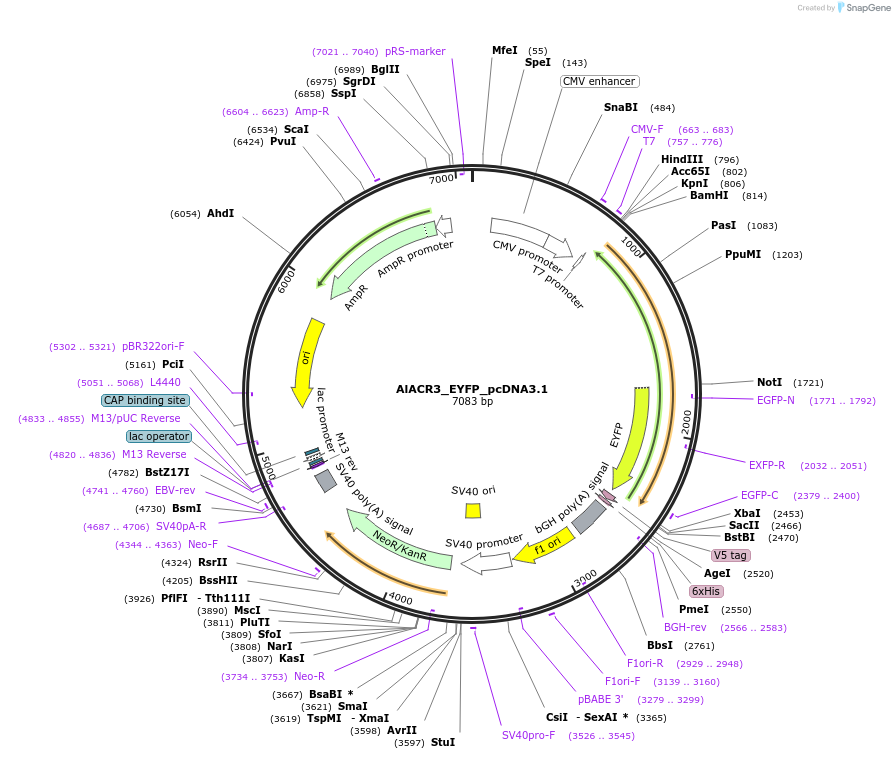
AlACR3_EYFP_pcDNA3.1
Plasmid#161024PurposeExpresses AlACR3 in mammalian cellsDepositorInsertAlACR3
TagsEYFPExpressionMammalianAvailable SinceNov. 17, 2020AvailabilityAcademic Institutions and Nonprofits only -
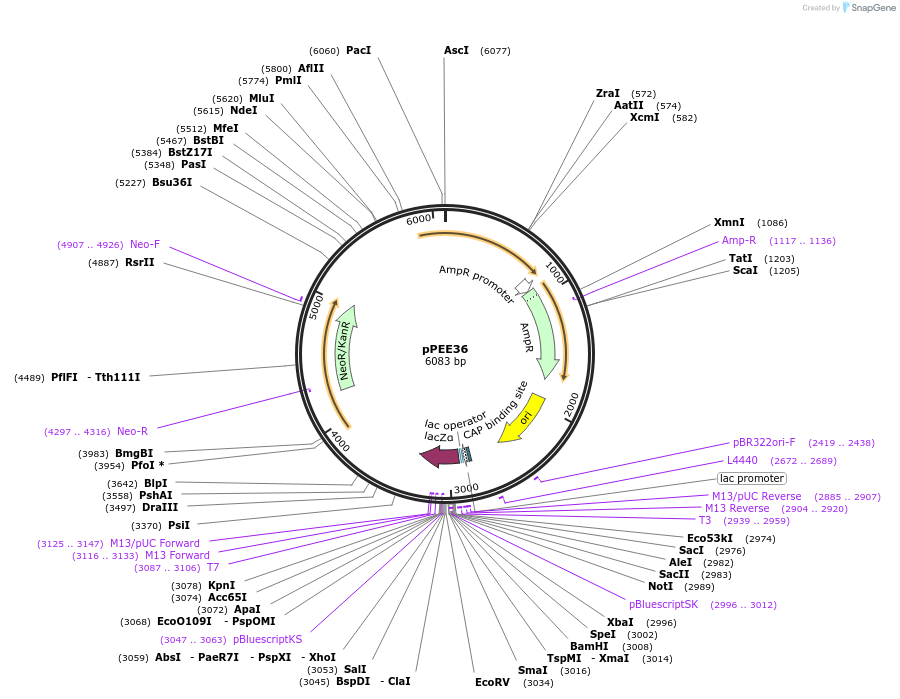
pPEE36
Plasmid#128376PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 7, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceApril 28, 2020AvailabilityAcademic Institutions and Nonprofits only -
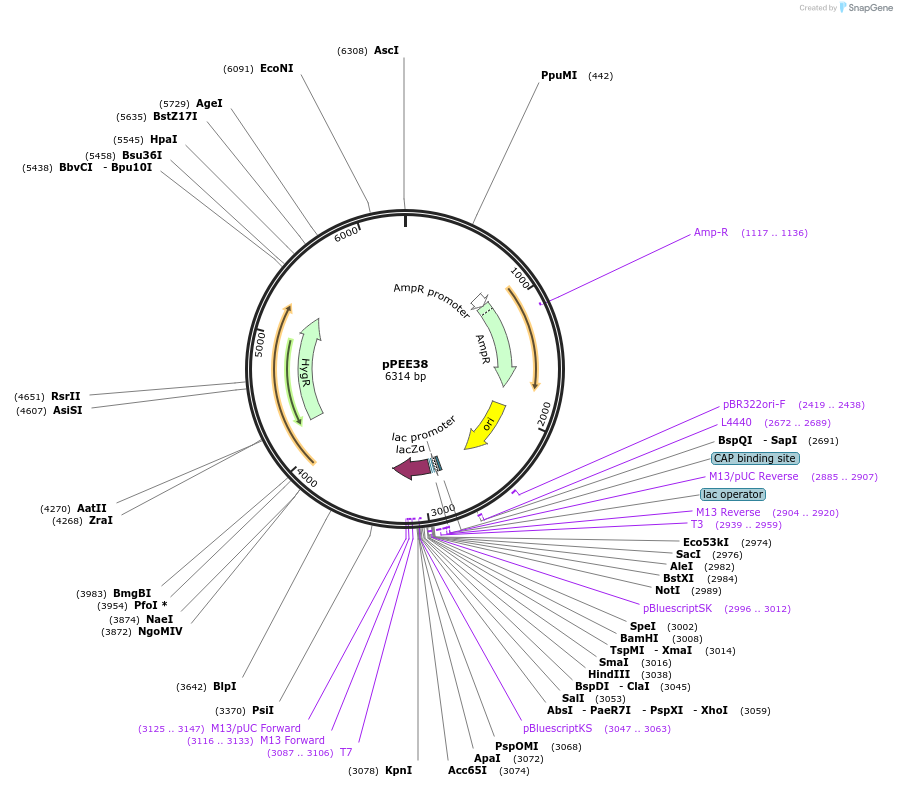
pPEE38
Plasmid#128378PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 4, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceJan. 8, 2020AvailabilityAcademic Institutions and Nonprofits only -
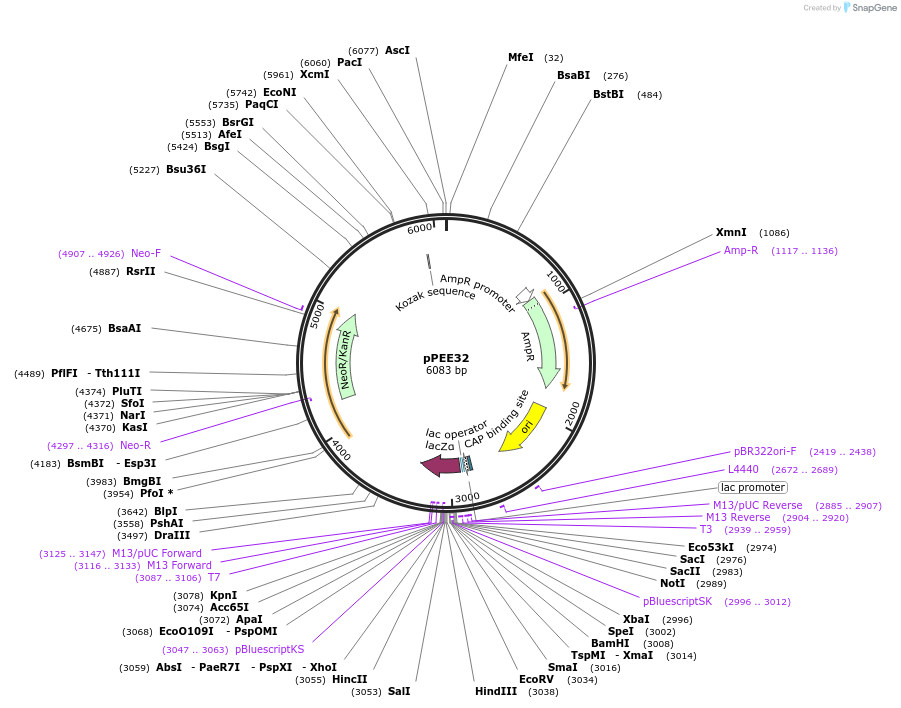
pPEE32
Plasmid#128372PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 3, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceJan. 8, 2020AvailabilityAcademic Institutions and Nonprofits only -
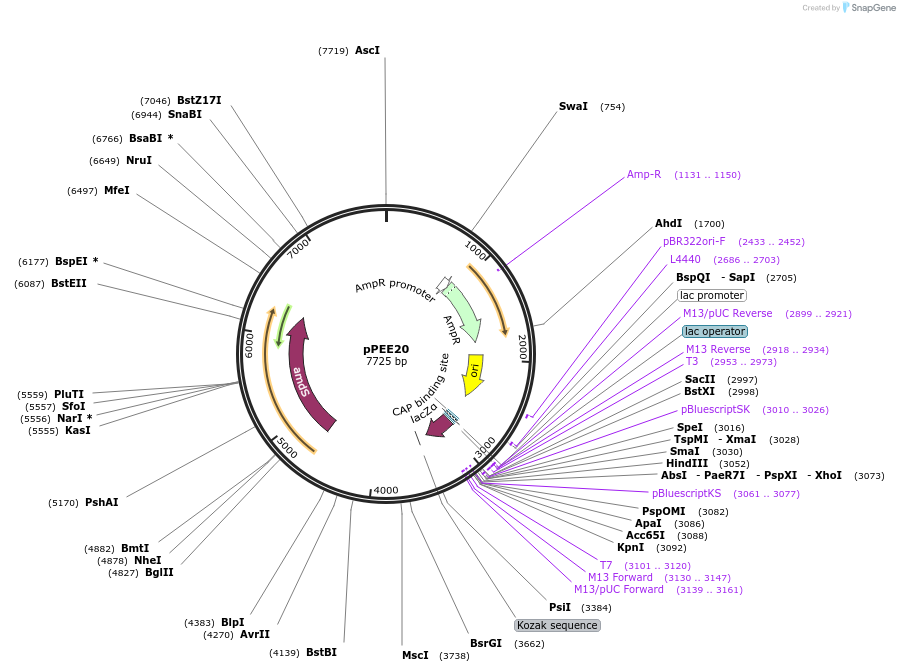
pPEE20
Plasmid#128363PurposeDominant marker in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 4, a small gene-free region. Contains the amdS2 counterselectable marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterTEF1Available SinceOct. 11, 2019AvailabilityAcademic Institutions and Nonprofits only -
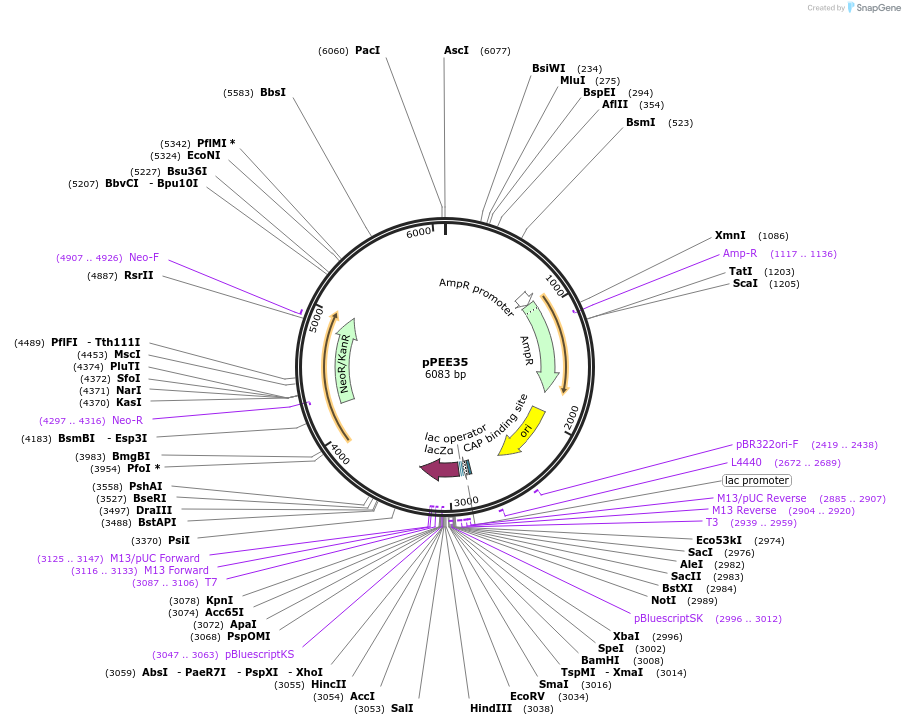
pPEE35
Plasmid#128375PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 6, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
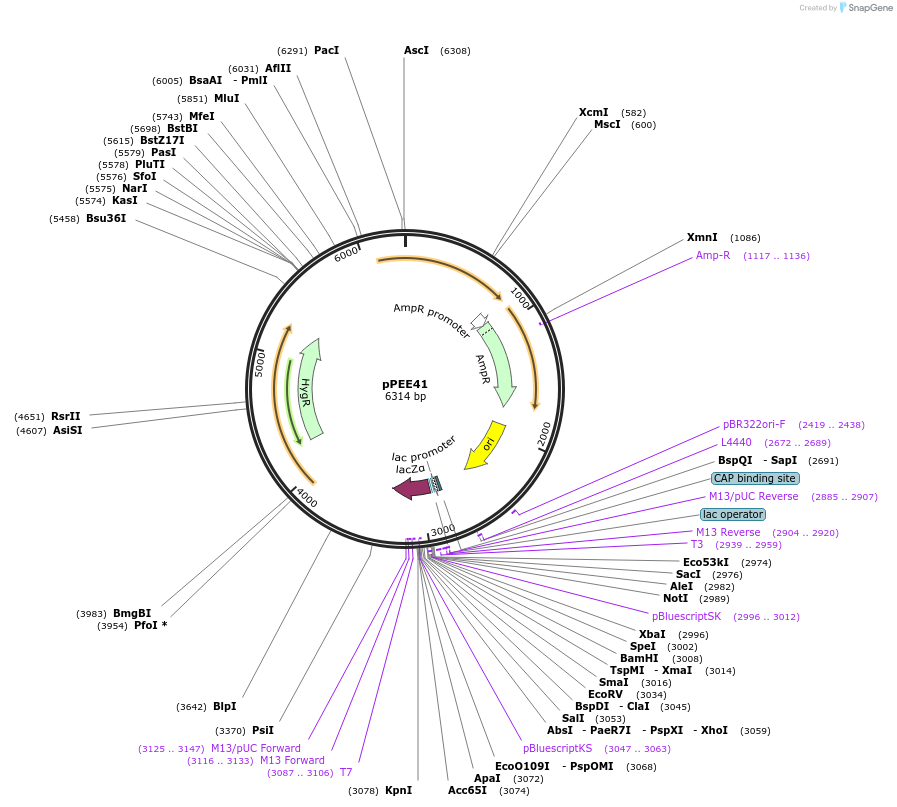
pPEE41
Plasmid#128381PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 7, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
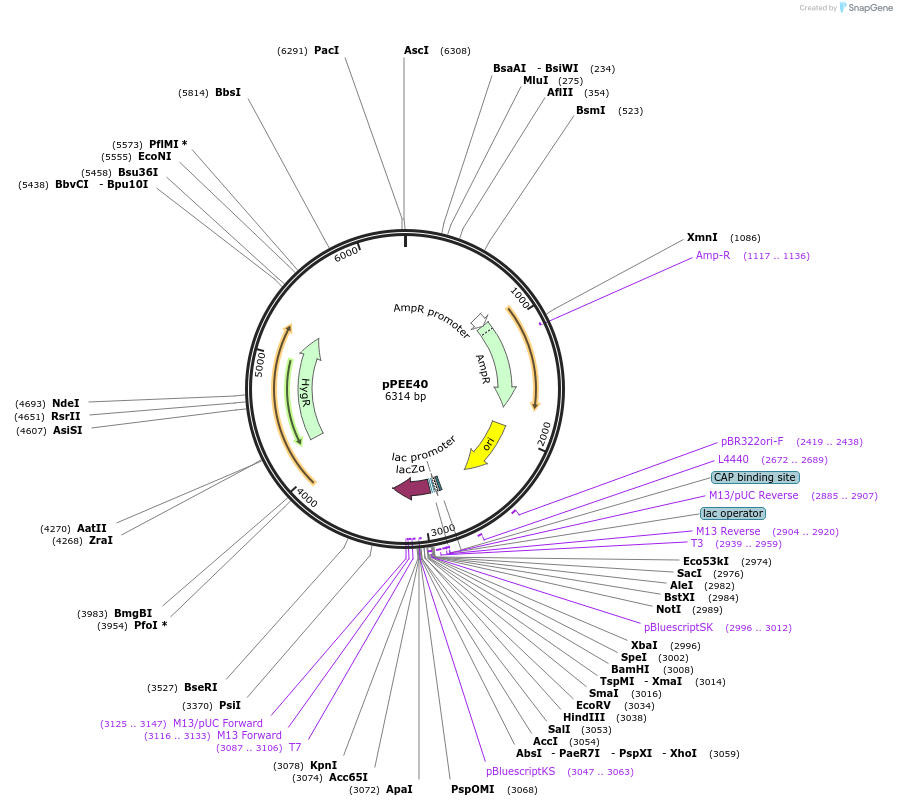
pPEE40
Plasmid#128380PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 6, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
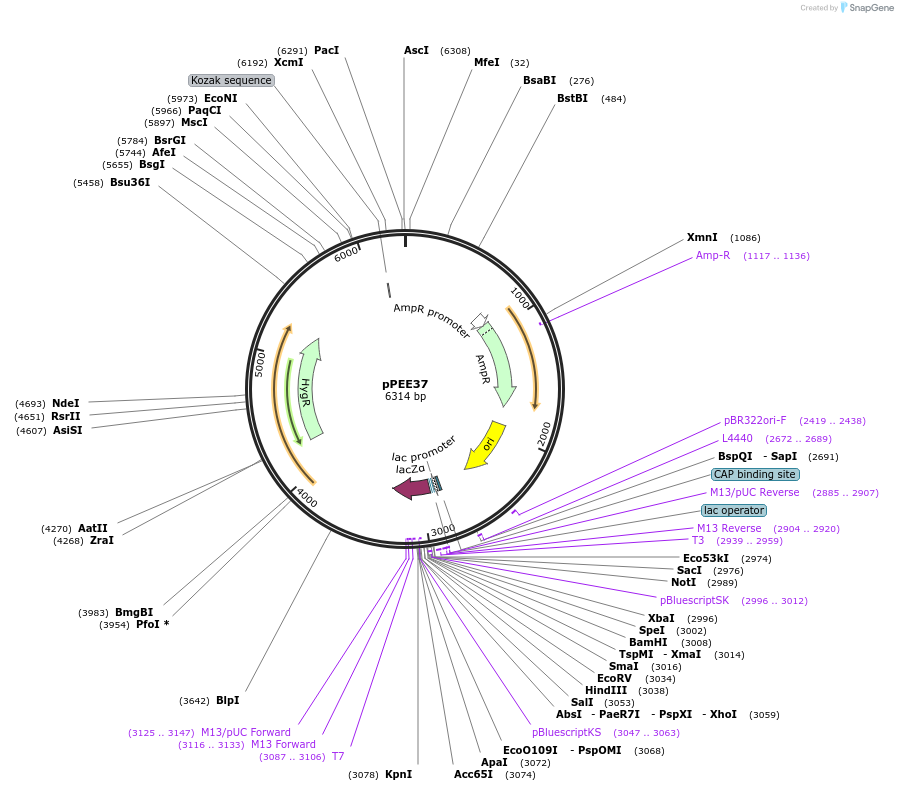
pPEE37
Plasmid#128377PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 3, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
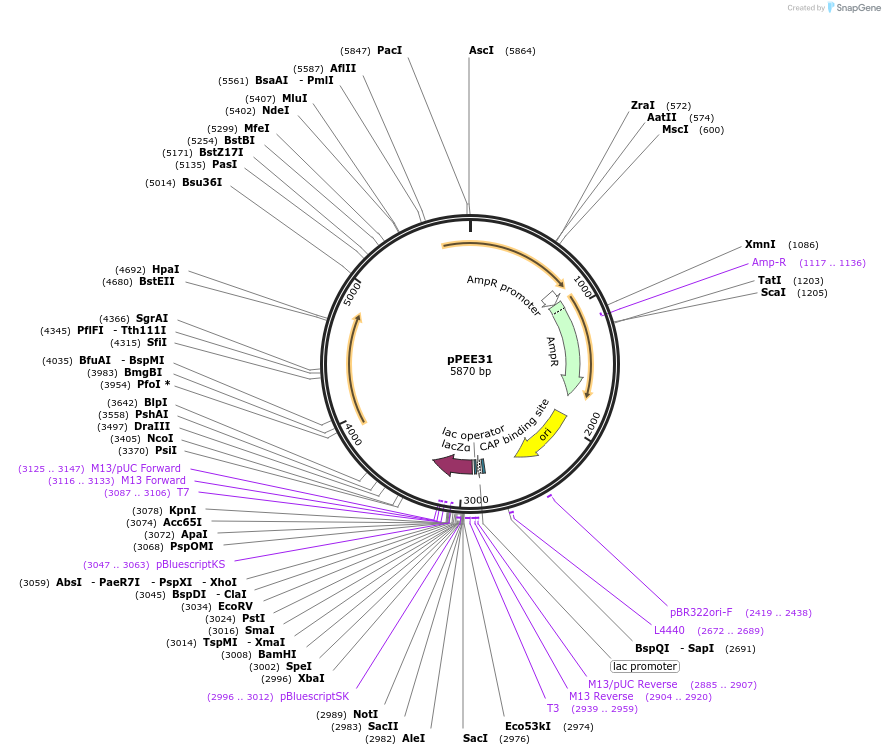
pPEE31
Plasmid#128371PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 7, a small gene-free region. Contains the nourseothricin resistance marker (NAT)DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
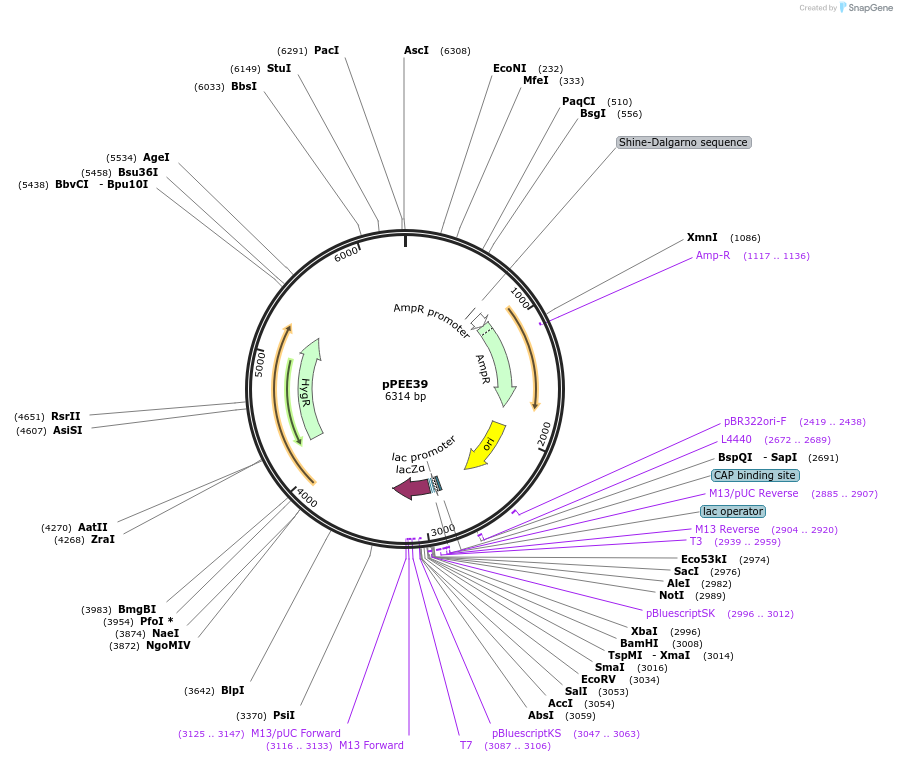
pPEE39
Plasmid#128379PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 5, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
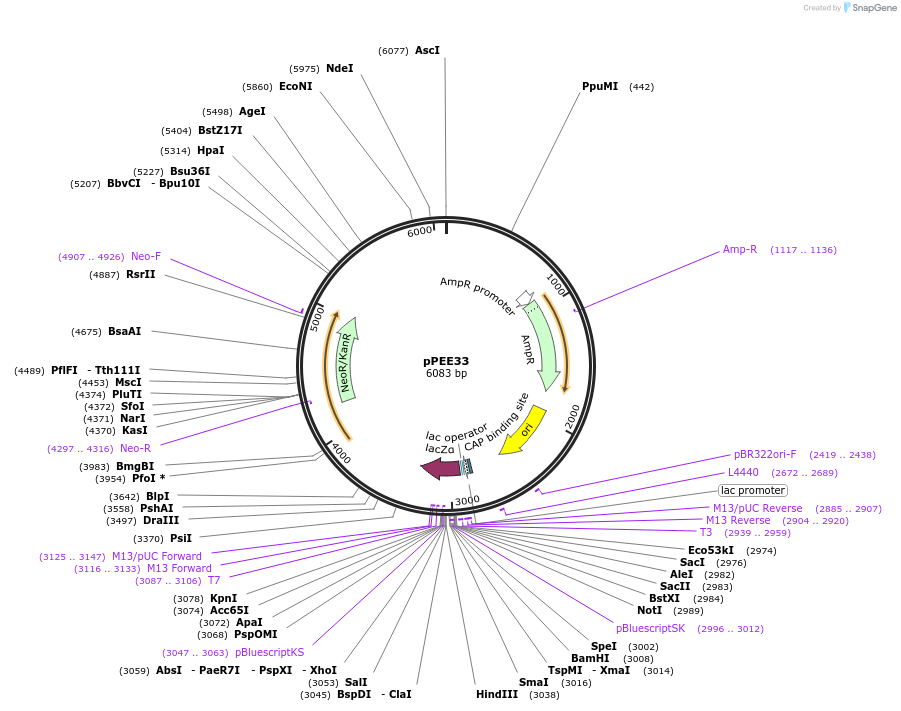
pPEE33
Plasmid#128373PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 4, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
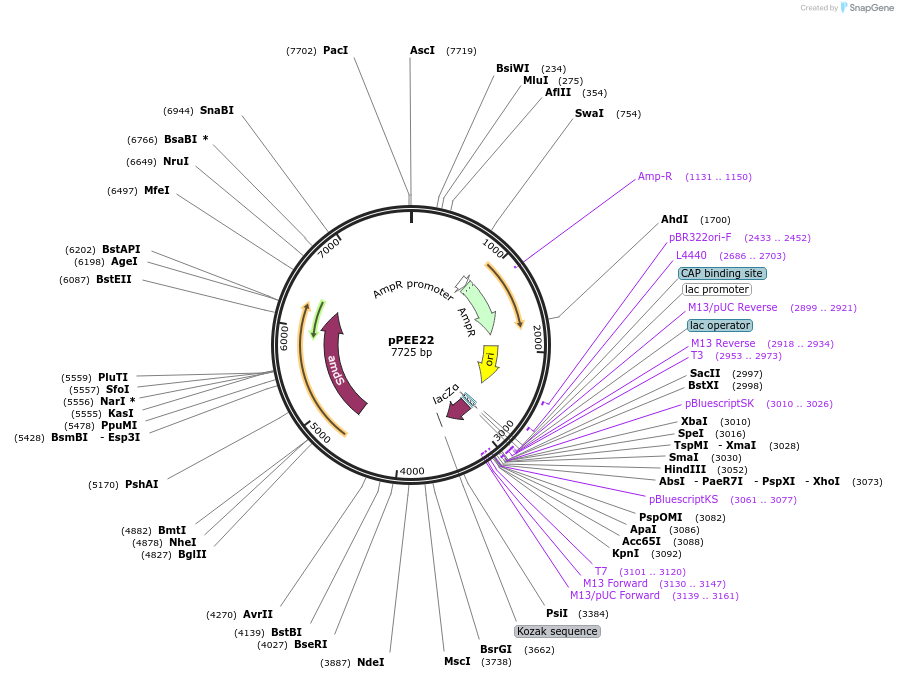
pPEE22
Plasmid#128365PurposeDominant marker in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 6, a small gene-free region. Contains the amdS2 counterselectable marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterTEF1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
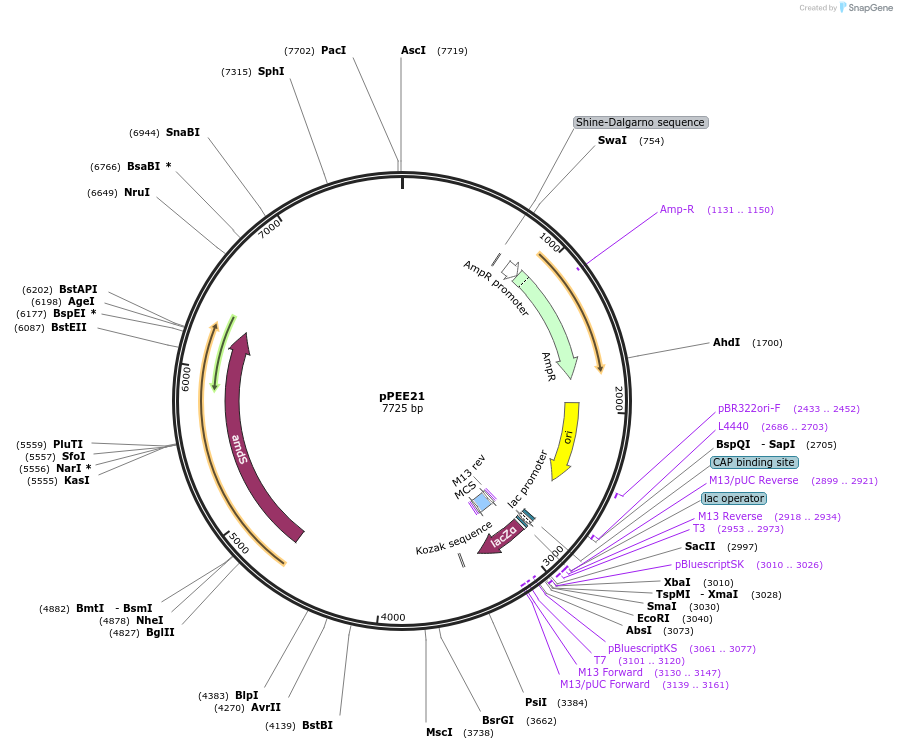
pPEE21
Plasmid#128364PurposeDominant marker in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 5, a small gene-free region. Contains the amdS2 counterselectable marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterTEF1Available SinceAug. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
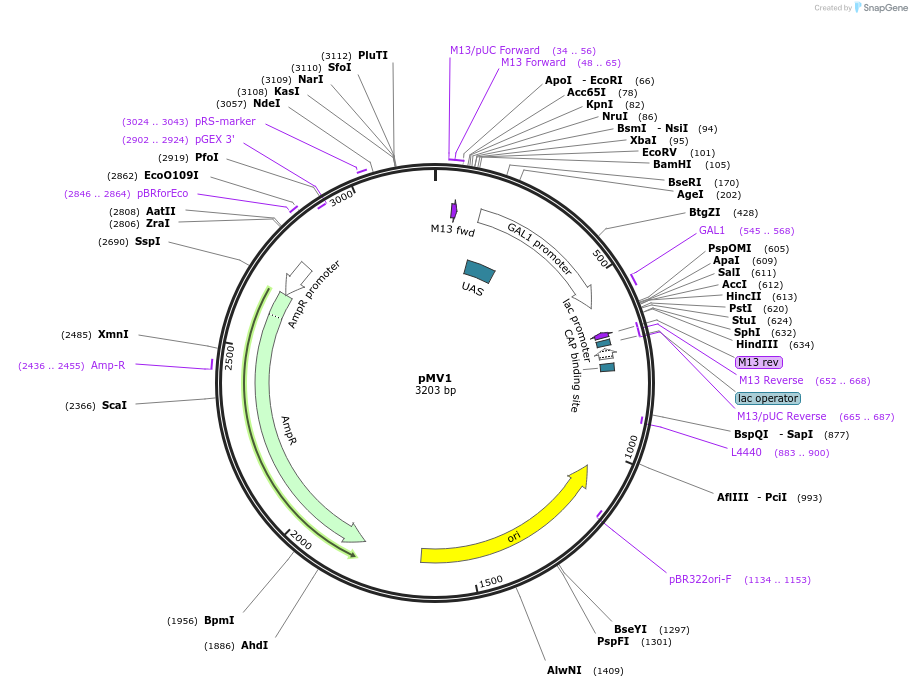
pMV1
Plasmid#128867PurposePosition 2 module- Gal1 promoterDepositorInsertGal1 promoter
UseSynthetic BiologyExpressionYeastAvailable SinceAug. 14, 2019AvailabilityAcademic Institutions and Nonprofits only -
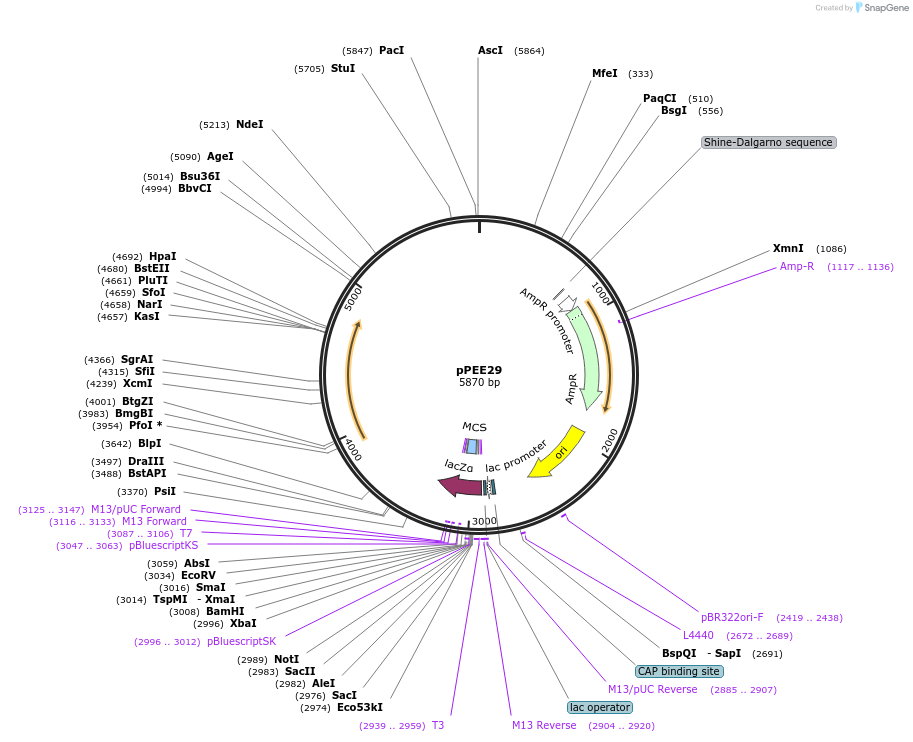
pPEE29
Plasmid#128369PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 5, a small gene-free region. Contains the nourseothricin resistance marker (NAT)DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only