We narrowed to 9,444 results for: Pol;
-
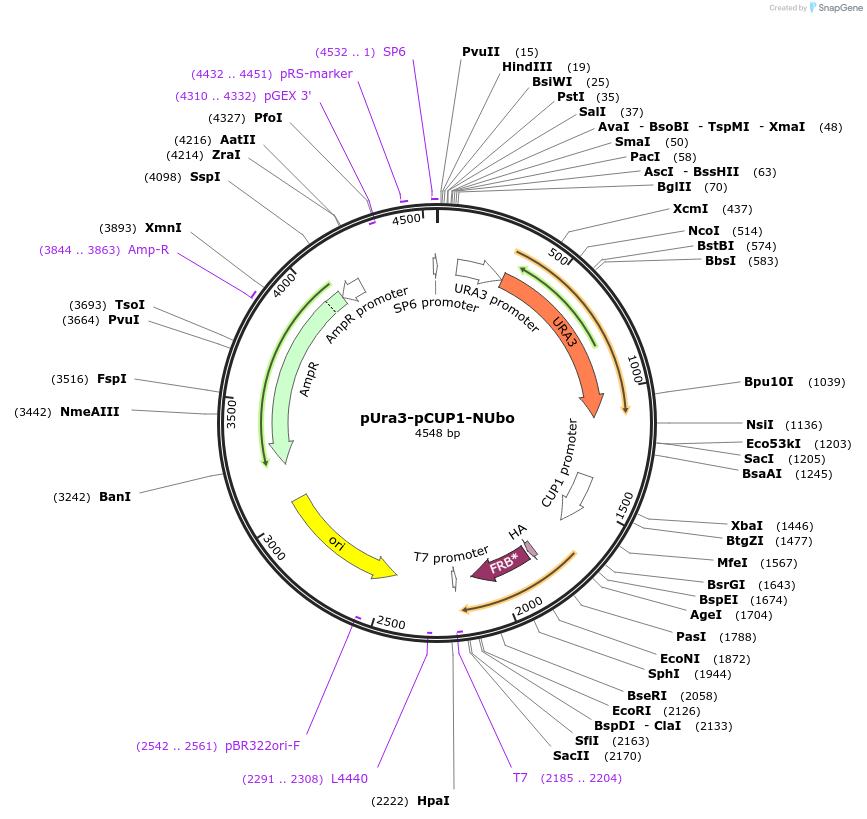
Plasmid#212760PurposeN-terminal PCR-based tagging with pCUP1-NUbo in budding yeast with URA3 selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 25, 2024AvailabilityAcademic Institutions and Nonprofits only
-
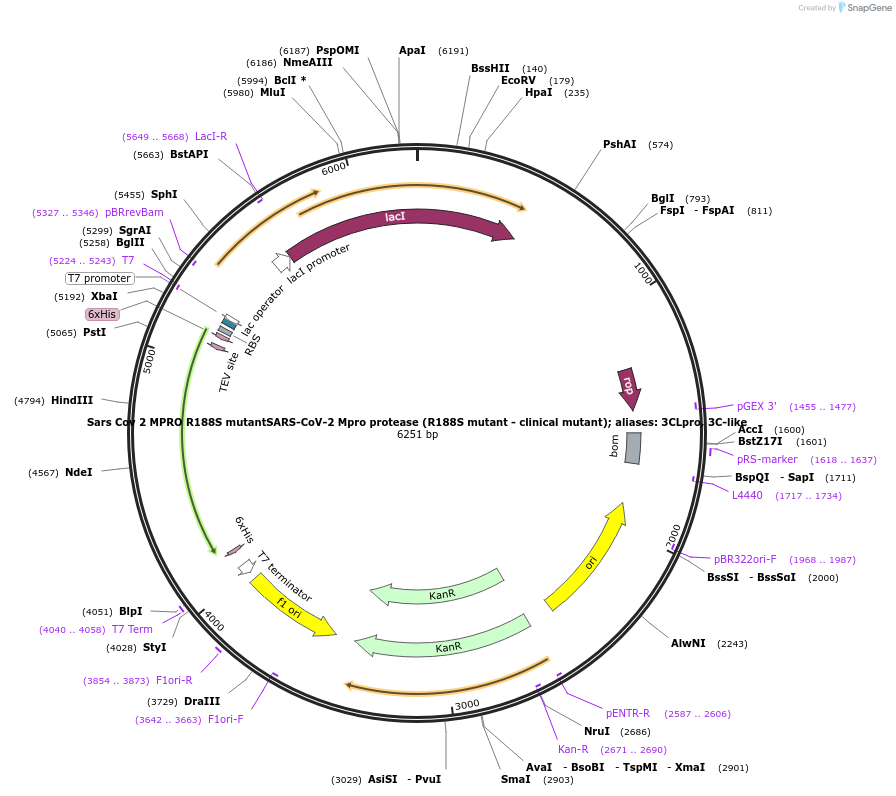
Sars Cov 2 MPRO R188S mutantSARS-CoV-2 Mpro protease (R188S mutant - clinical mutant); aliases: 3CLpro, 3C-like
Plasmid#204785PurposeBacterial expression of Sars Cov 2 MPRO R188S mutantDepositorAvailable SinceJan. 23, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
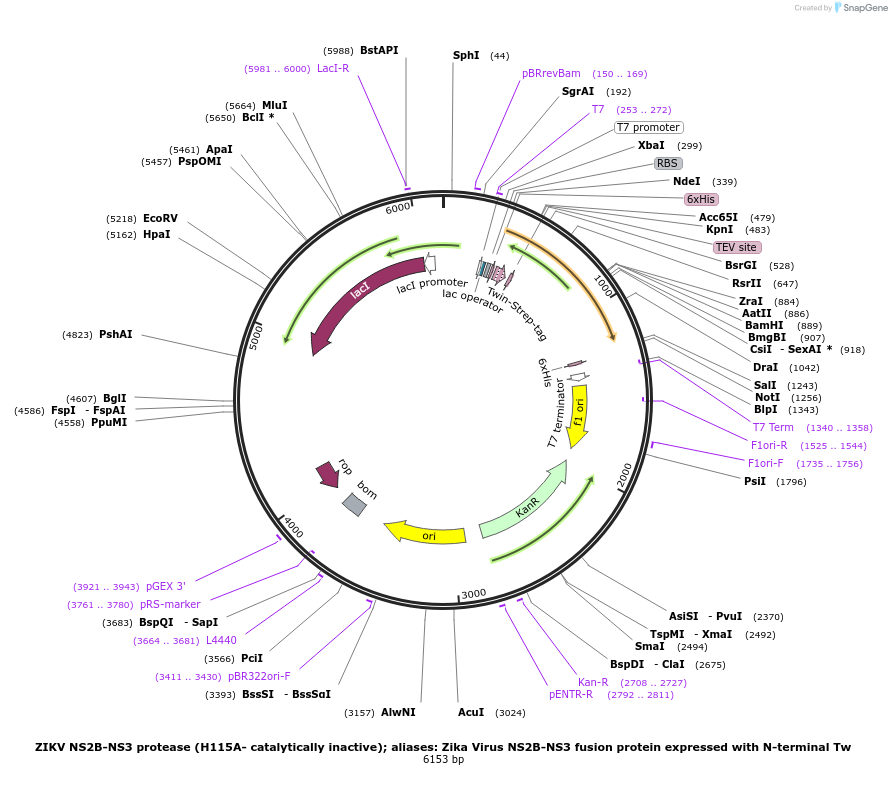
ZIKV NS2B-NS3 protease (H115A- catalytically inactive); aliases: Zika Virus NS2B-NS3 fusion protein expressed with N-terminal Tw
Plasmid#204791PurposeBacterial expression of a fusion of Zika NS2B-NS3 proteinsDepositorInsertZika NS2B-NS3 fusion
Tags6 HisExpressionBacterialAvailable SinceJan. 16, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
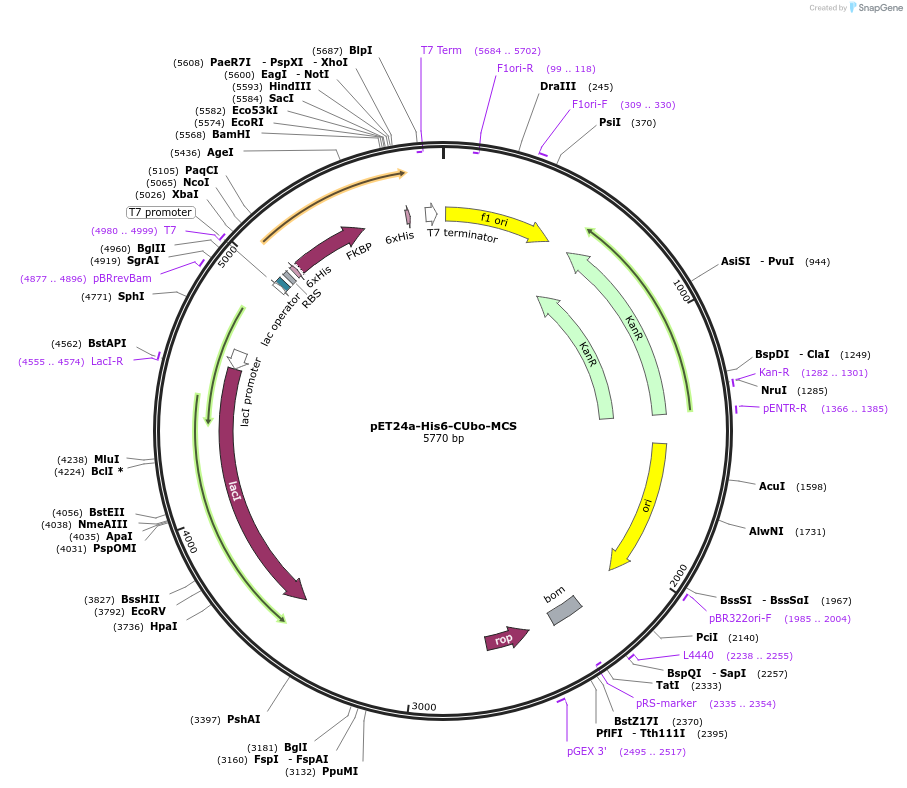
pET24a-His6-CUbo-MCS
Plasmid#212725PurposeBacterial expression of a protein of interest with N-terminal His6-CUbo-fusion, contains multiple cloning siteDepositorTypeEmpty backboneExpressionBacterialAvailable SinceJan. 9, 2024AvailabilityAcademic Institutions and Nonprofits only -
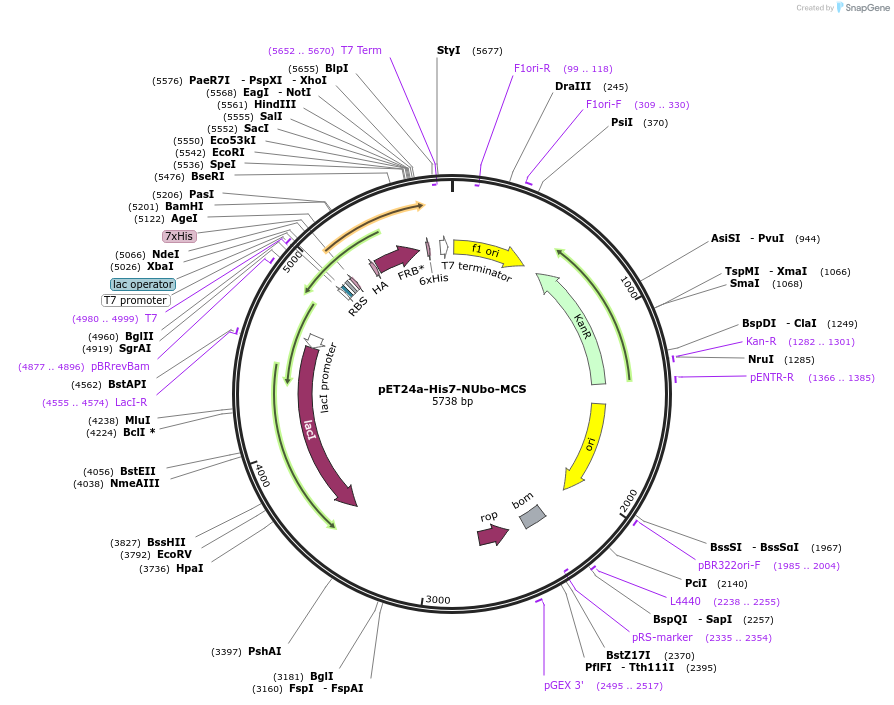
pET24a-His7-NUbo-MCS
Plasmid#212724PurposeBacterial expression of a protein of interest with N-terminal His7-NUbo-fusion, contains multiple cloning siteDepositorTypeEmpty backboneExpressionBacterialAvailable SinceJan. 9, 2024AvailabilityAcademic Institutions and Nonprofits only -
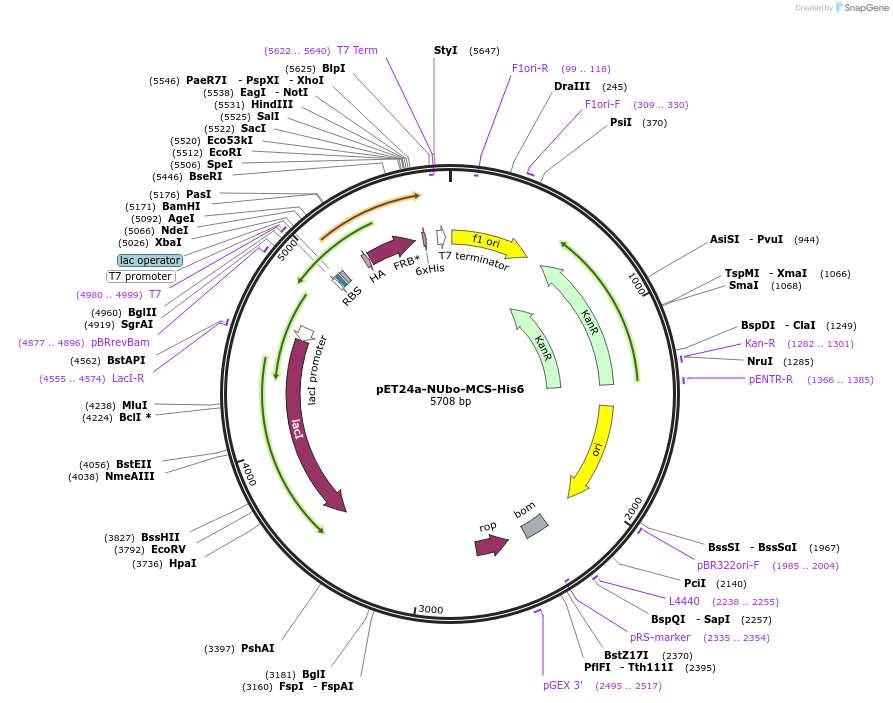
pET24a-NUbo-MCS-His6
Plasmid#212723PurposeBacterial expression of a protein of interest with N-terminal NUbo- and C-terminal His6-fusions, contains multiple cloning siteDepositorTypeEmpty backboneExpressionBacterialAvailable SinceJan. 9, 2024AvailabilityAcademic Institutions and Nonprofits only -
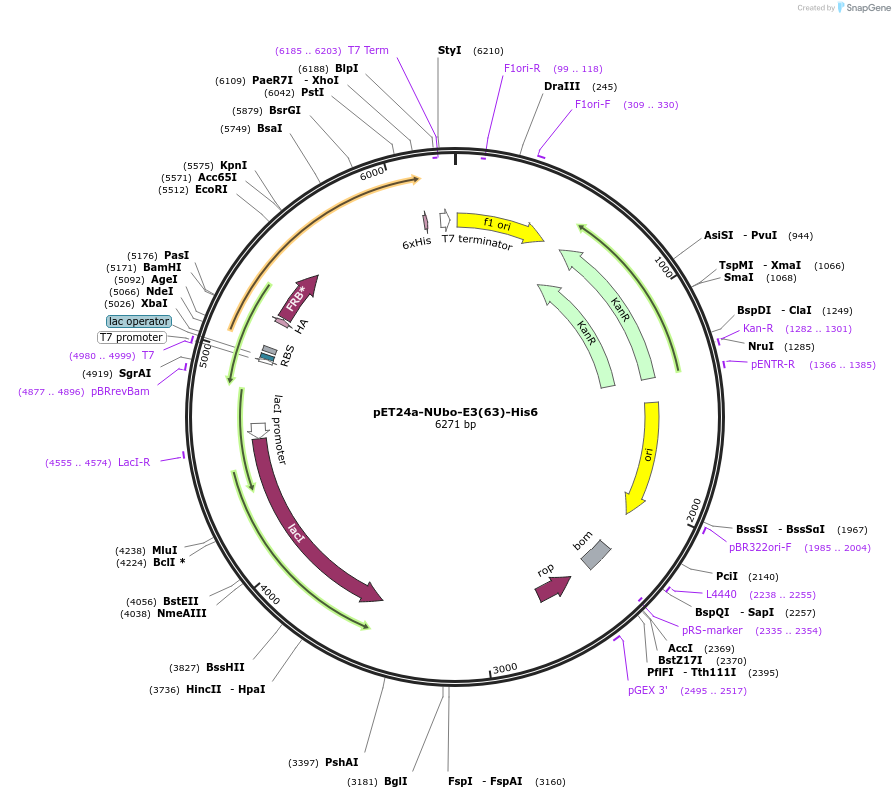
pET24a-NUbo-E3(63)-His6
Plasmid#212717PurposeBacterial expression of NUbo-E3(63)-His6DepositorInsertE3(63)
Tags6xHis and NUboExpressionBacterialAvailable SinceJan. 8, 2024AvailabilityAcademic Institutions and Nonprofits only -
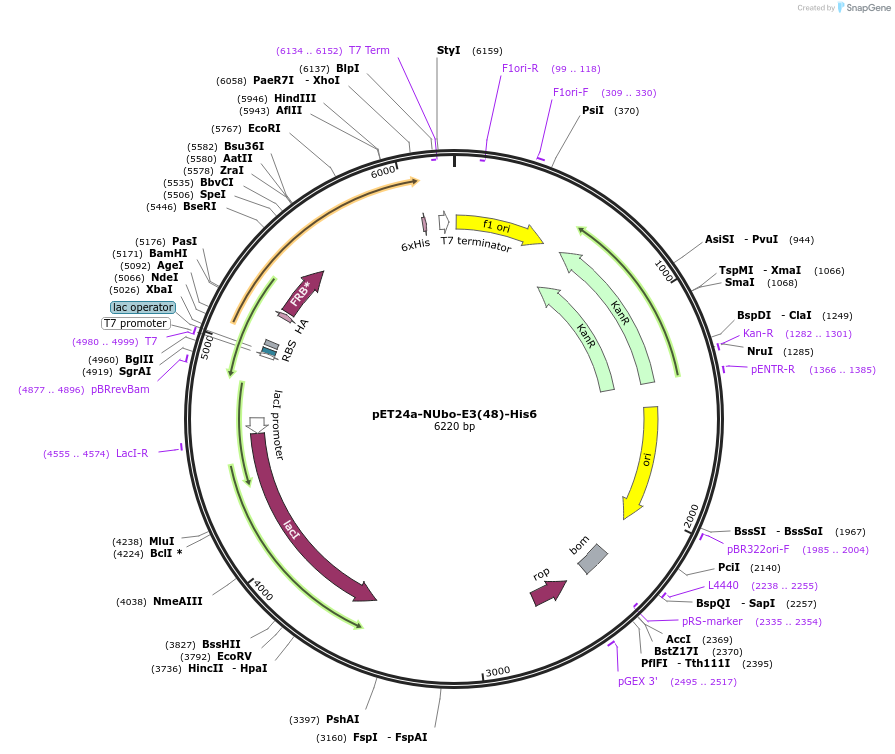
pET24a-NUbo-E3(48)-His6
Plasmid#212714PurposeBacterial expression of NUbo-E3(48)-His6DepositorInsertE3(48)
Tags6xHis and NUboExpressionBacterialAvailable SinceJan. 8, 2024AvailabilityAcademic Institutions and Nonprofits only -
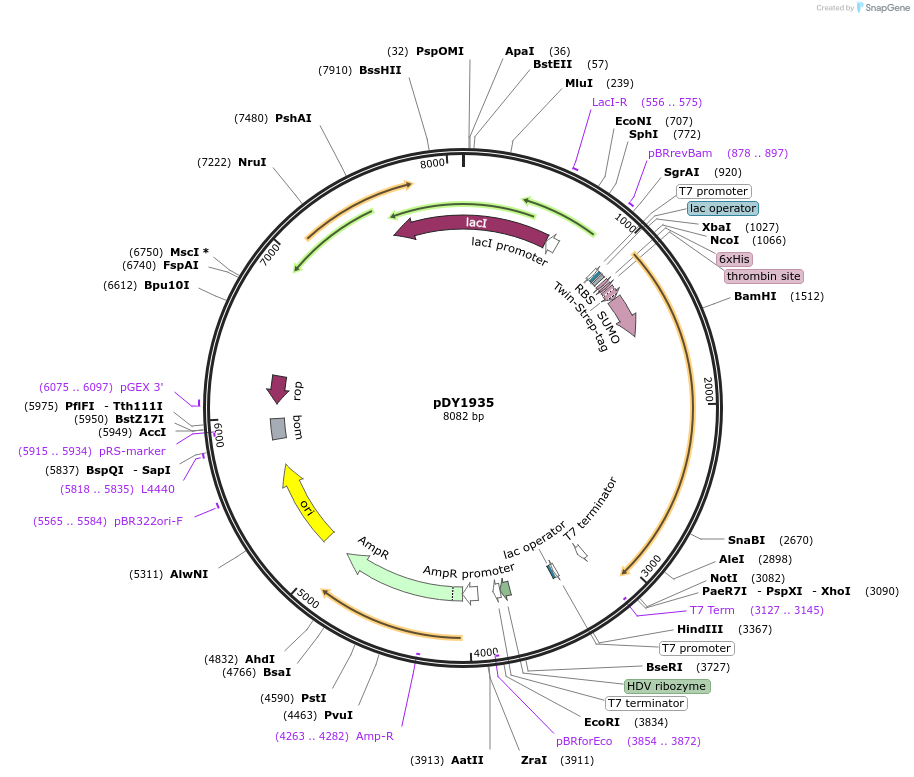
pDY1935
Plasmid#212315PurposeExpress ApmFnuc protein with sumotagDepositorInsertIsvmimi
Tags6xHis-thrombin site-TwinStrepTag-SUMOExpressionBacterialAvailable SinceJan. 3, 2024AvailabilityAcademic Institutions and Nonprofits only -
WNV NS2B-NS3 protease (based on PDB 2IJO, catalytically active); aliases: West Nile Virus NS2B-NS3 fusion protein
Plasmid#204799PurposeBacterial expression of a fusion of West Nile NS2B-NS3 proteinsDepositorInsertWest Nile NS2B-NS3 fusion
ExpressionBacterialAvailable SinceDec. 11, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
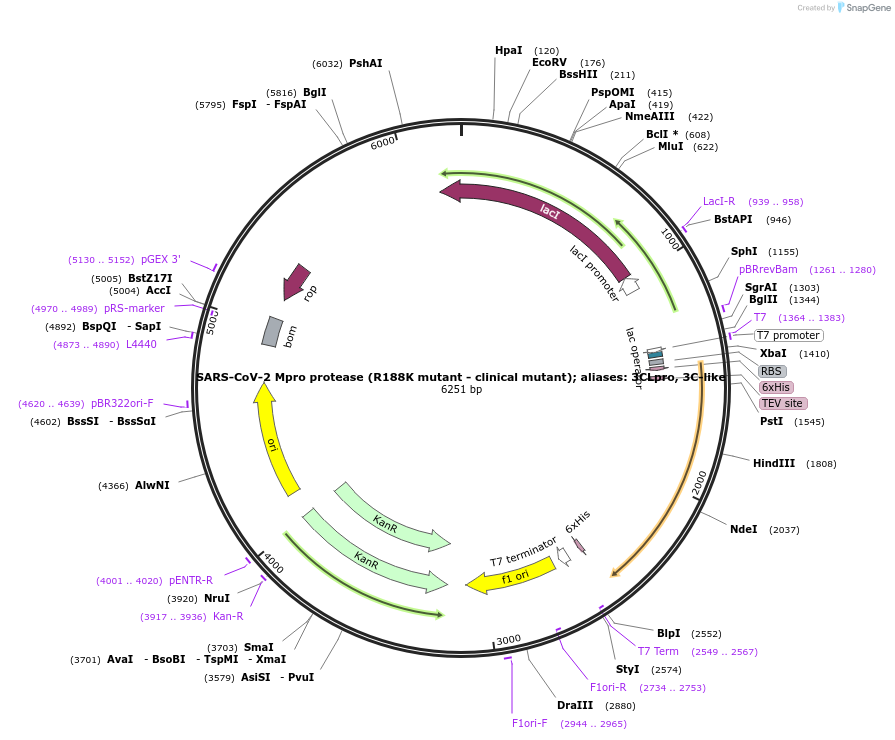
SARS-CoV-2 Mpro protease (R188K mutant - clinical mutant); aliases: 3CLpro, 3C-like
Plasmid#204770PurposeBacterial expression of Sars Cov 2 MPRO R188K mutantDepositorAvailable SinceNov. 30, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
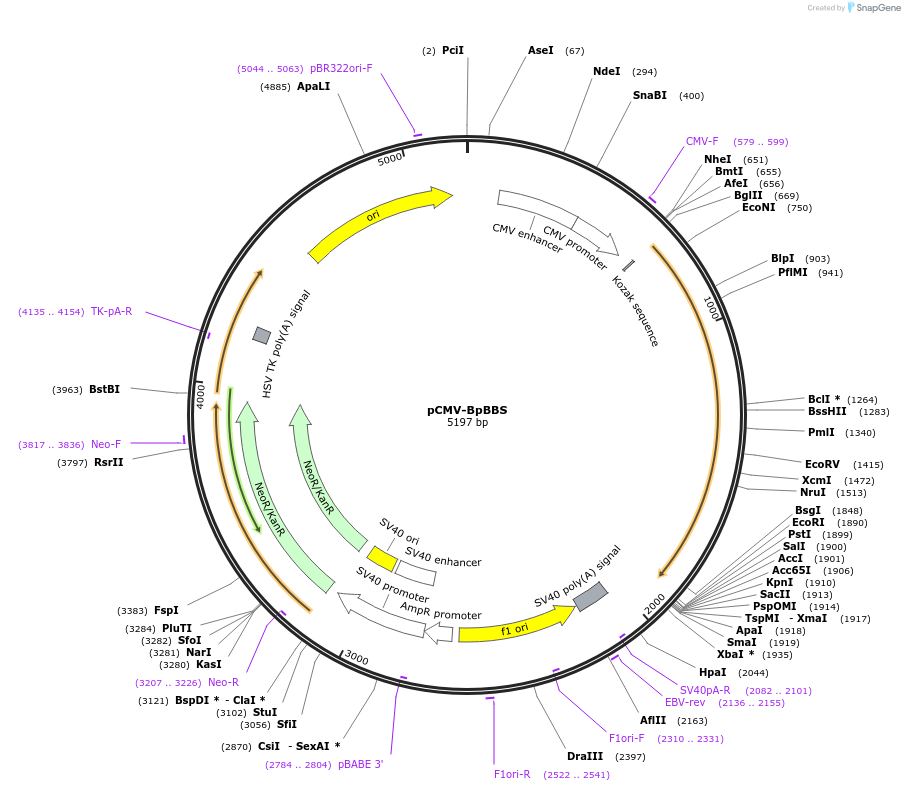
pCMV-BpBBS
Plasmid#199468PurposeExpression of the biliverdin-binding serpin of Boana punctata treefrog (BpBBS) in mammalian cells.DepositorInsertBpBBS
ExpressionMammalianAvailable SinceMay 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
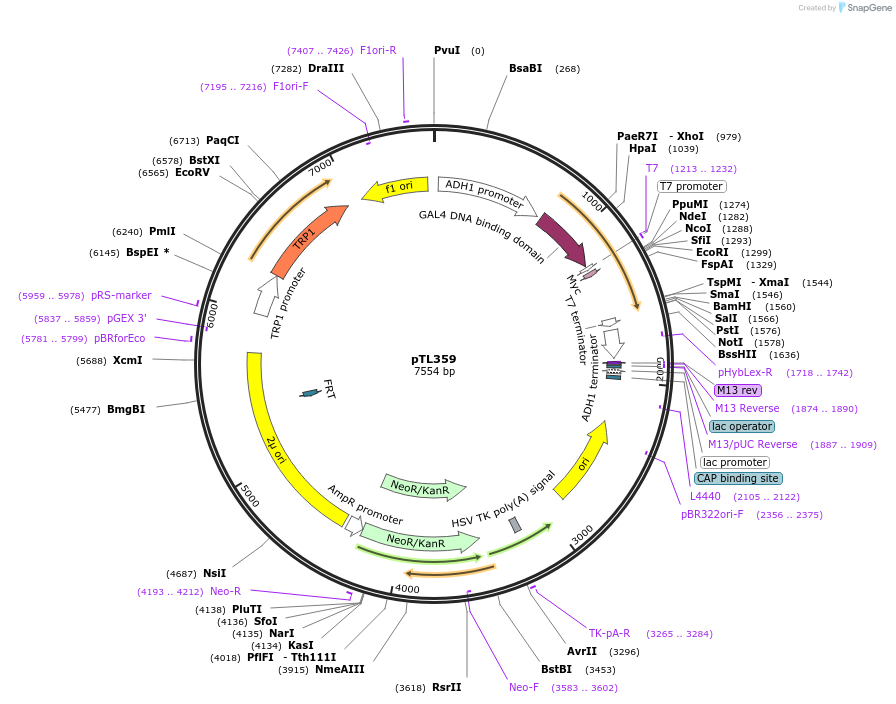
pTL359
Plasmid#168420PurposepGBKT7 yeast two-hybrid plasmidDepositorInsertOsaHIP41
ExpressionYeastAvailable SinceNov. 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
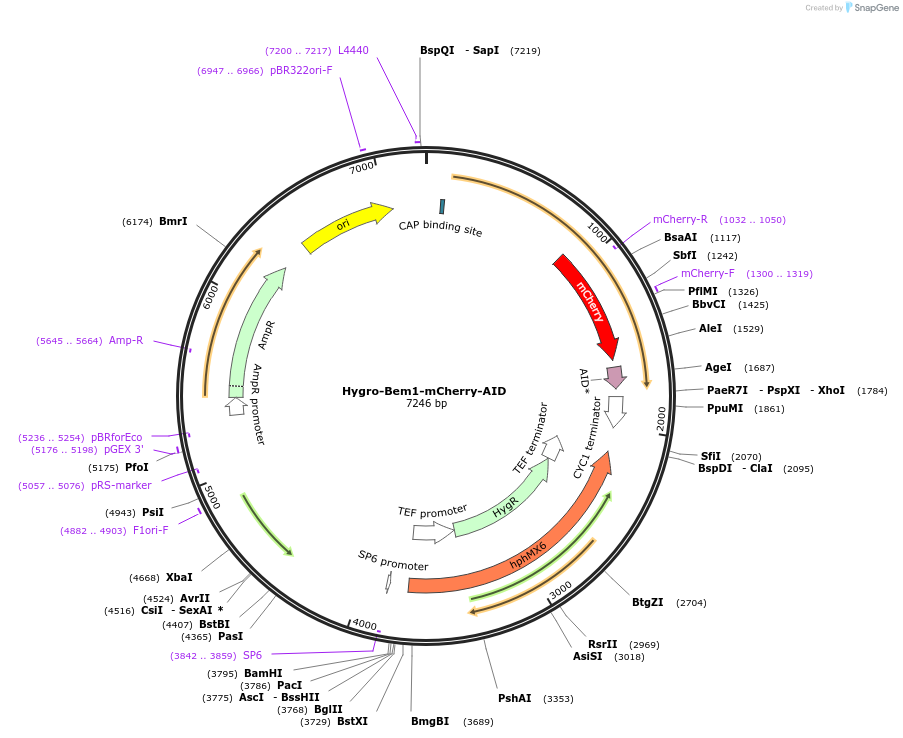
Hygro-Bem1-mCherry-AID
Plasmid#173925PurposePlasmid designed for the genomic tagging of saccharomyces cerevisiae Bem1 with mCherry-AIDDepositorInsertsPartial Bem1 C-terminus
hygromycin marker
Bem1 downstream sequence
TagsmCherry- AIDAvailable SinceNov. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
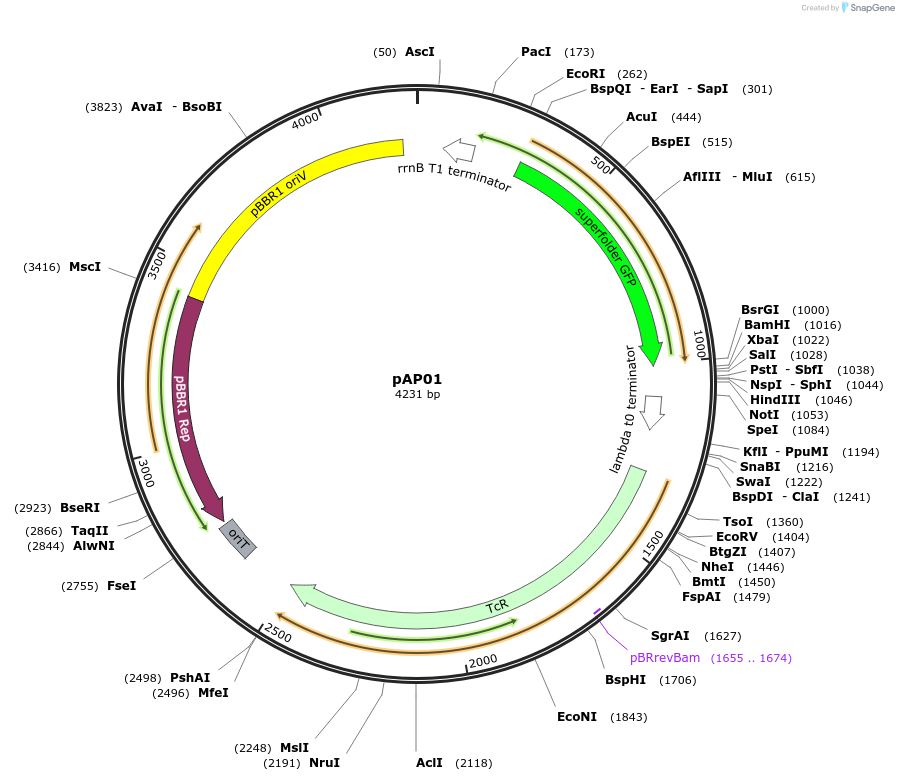
pAP01
Plasmid#186260PurposesfGFP under light light responsive PEL222 promoterDepositorInsertsfGFP
ExpressionBacterialPromoterPEL222 (GGTAGCCTTTAGTCCATGTTAGCGAAGAAAATGGTTTGTTA…Available SinceAug. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
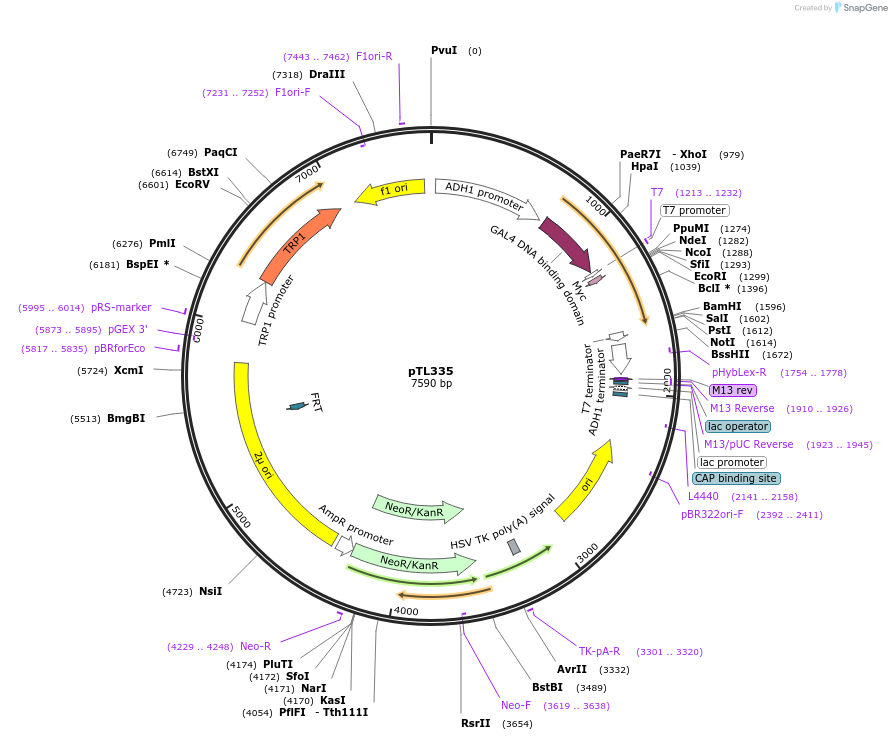
pTL335
Plasmid#168397PurposepGBKT7 yeast two-hybrid plasmidDepositorInsertOsaHIP24
ExpressionYeastAvailable SinceAug. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
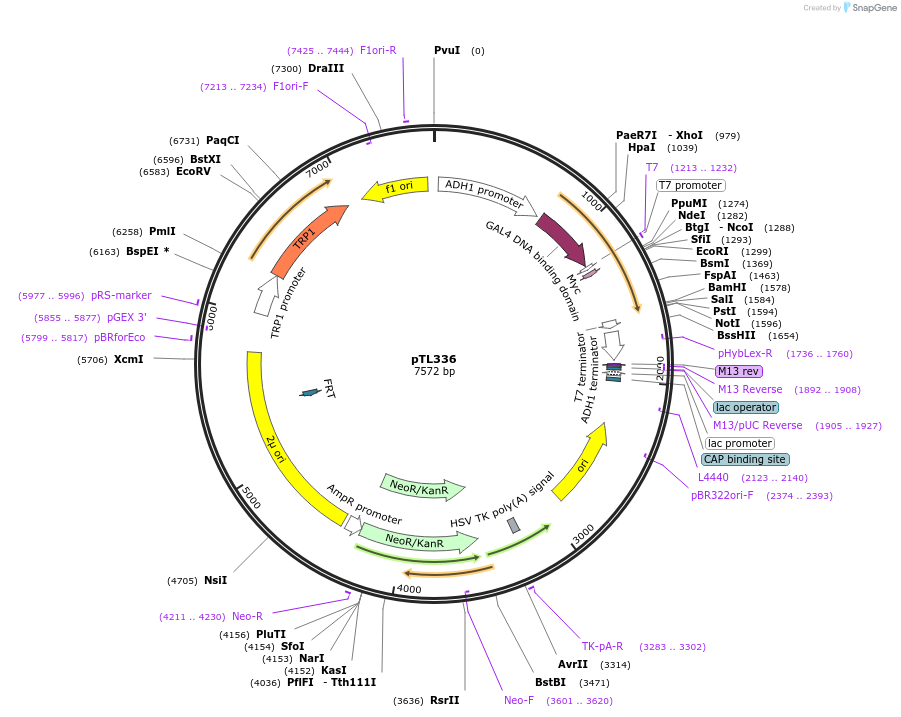
pTL336
Plasmid#168396PurposepGBKT7 yeast two-hybrid plasmidDepositorInsertOsaHIP34
ExpressionYeastAvailable SinceAug. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
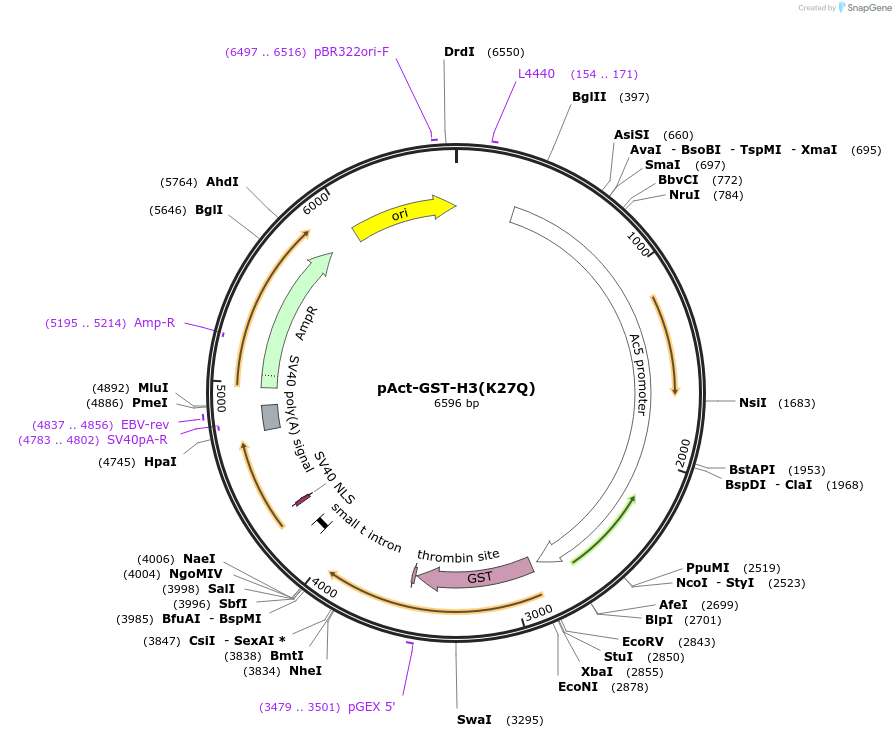
pAct-GST-H3(K27Q)
Plasmid#182843Purposea K27Q point mutation (AAG/CAG) of Drosophila histone H3 was generated from pAct-GST-H3. Expression in S2 cells.DepositorInsertGST-histone H3 fusion
TagsGSTExpressionInsectPromoterActin5CAvailable SinceApril 21, 2022AvailabilityAcademic Institutions and Nonprofits only -
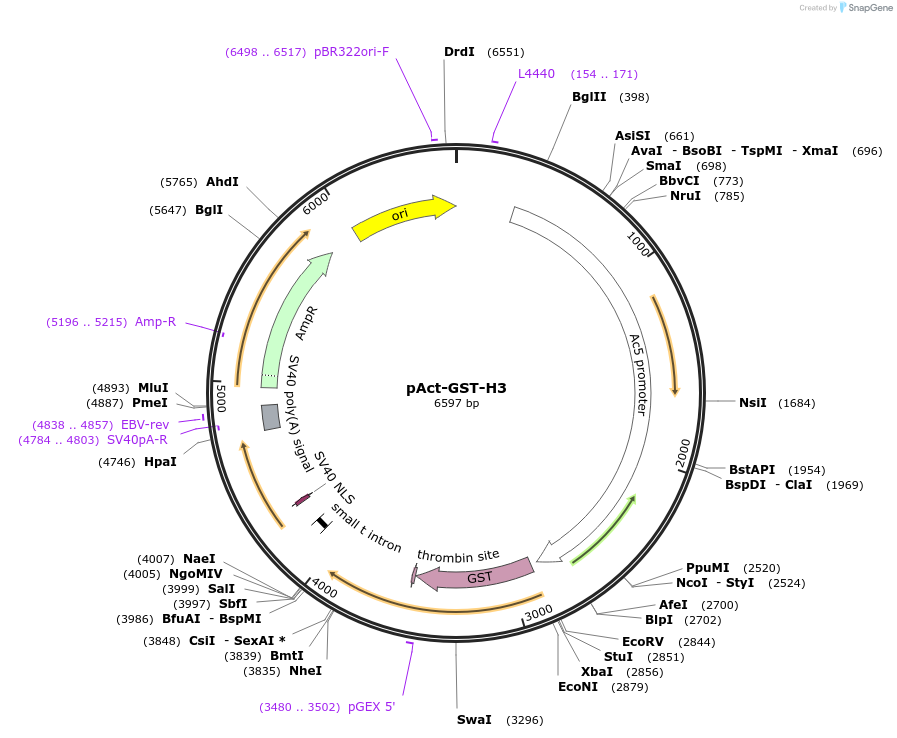
pAct-GST-H3
Plasmid#182842PurposeA PCR product encoding GST-H3 (Drosophila histone H3) was was inserted into a modified pAct-5c vector by Sac I and Sal I sites. Expression in S2 cells.DepositorInsertGST-histone H3 fusion
TagsGSTExpressionInsectPromoterActin5CAvailable SinceApril 21, 2022AvailabilityAcademic Institutions and Nonprofits only -
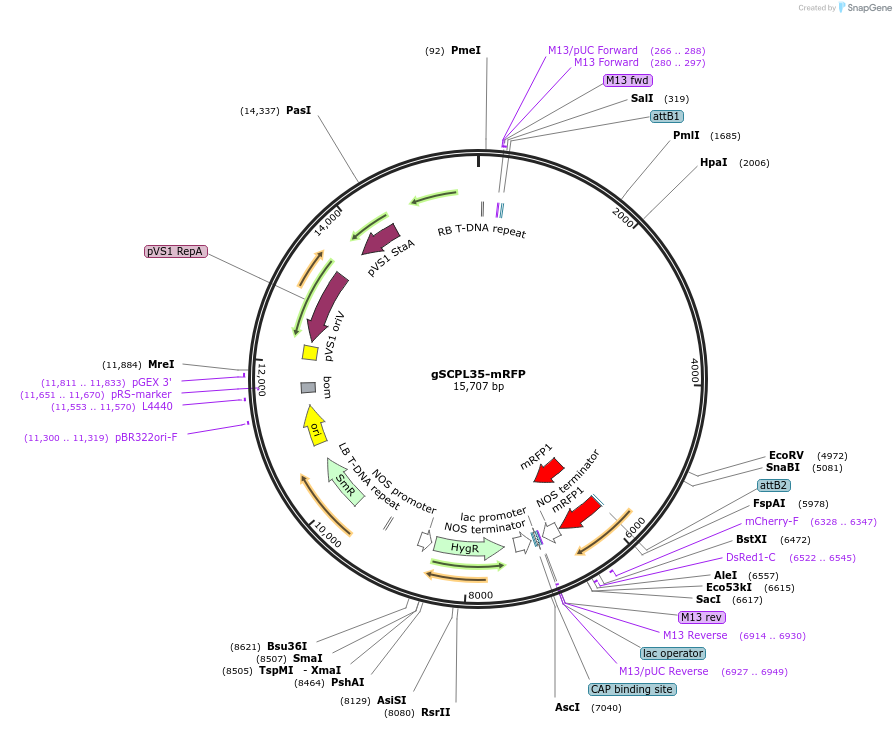
gSCPL35-mRFP
Plasmid#181960PurposeSCPL35 protein tagged with mRFP, under control of native promoterDepositorInsertSCPL35
TagsmRFPExpressionPlantPromoternative SCPL35Available SinceMarch 14, 2022AvailabilityAcademic Institutions and Nonprofits only