We narrowed to 7,774 results for: ALP
-
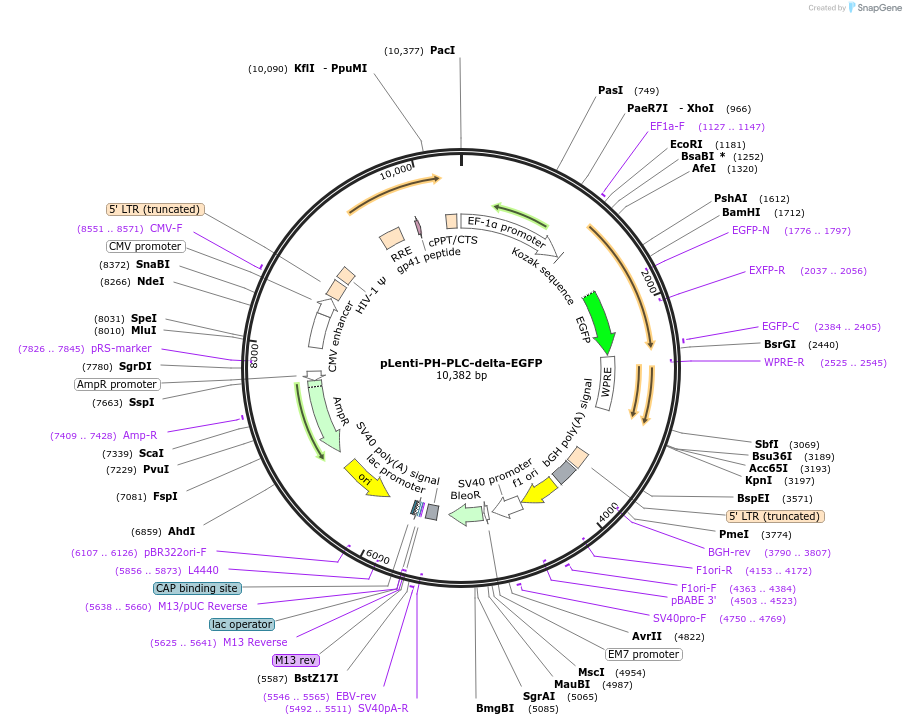
Plasmid#136998PurposeLentiviral EGFP lipid sensor for PI(4,5)P2DepositorInsertPH-PLC-delta-EGFP
UseLentiviralTagsEGFPPromoterEF1 alphaAvailable SinceApril 17, 2020AvailabilityAcademic Institutions and Nonprofits only -
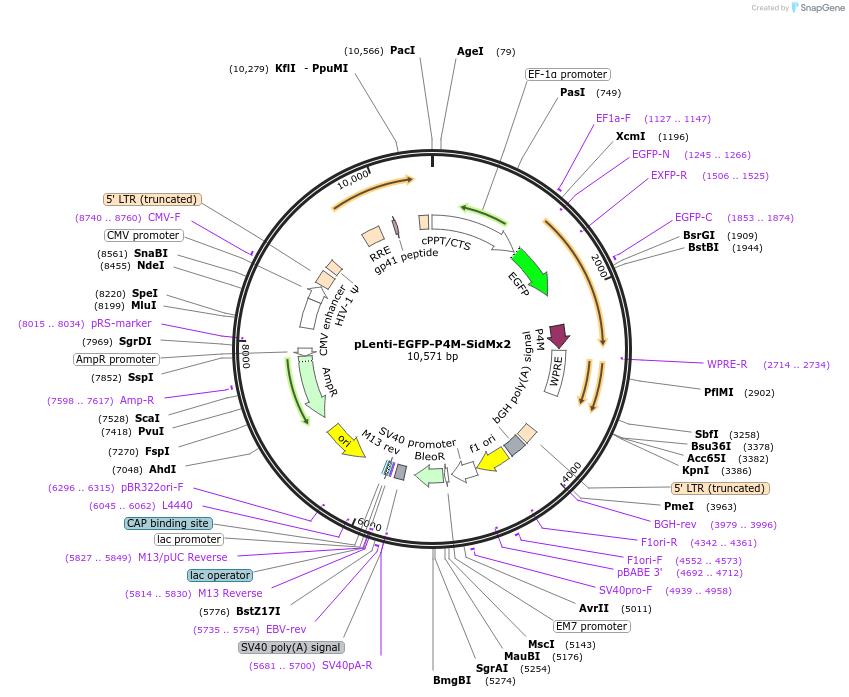
pLenti-EGFP-P4M-SidMx2
Plasmid#136997PurposeLentiviral EGFP lipid sensor for PI(4)PDepositorInsertEGFP-P4M-SidMx2
UseLentiviralTagsEGFPPromoterEF1 alphaAvailable SinceApril 17, 2020AvailabilityAcademic Institutions and Nonprofits only -
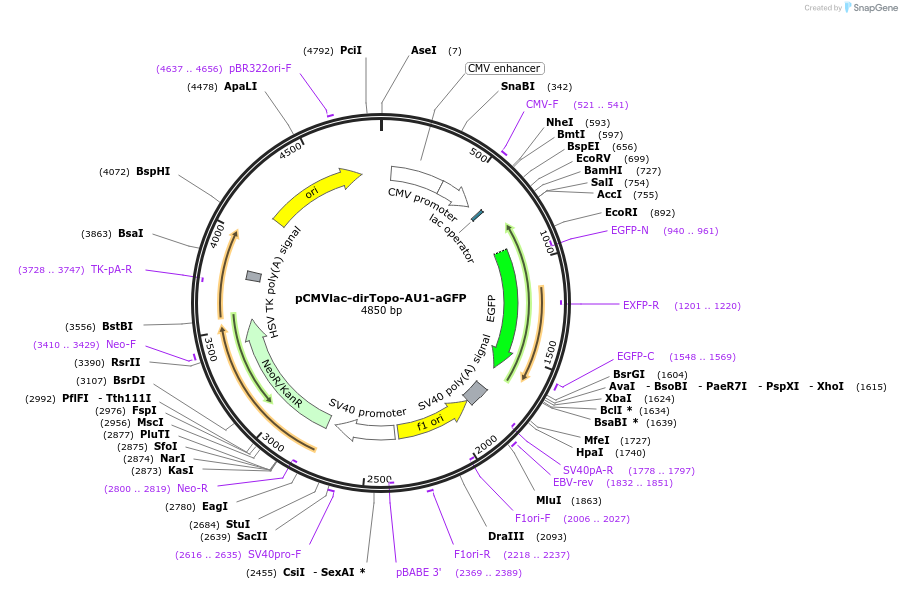
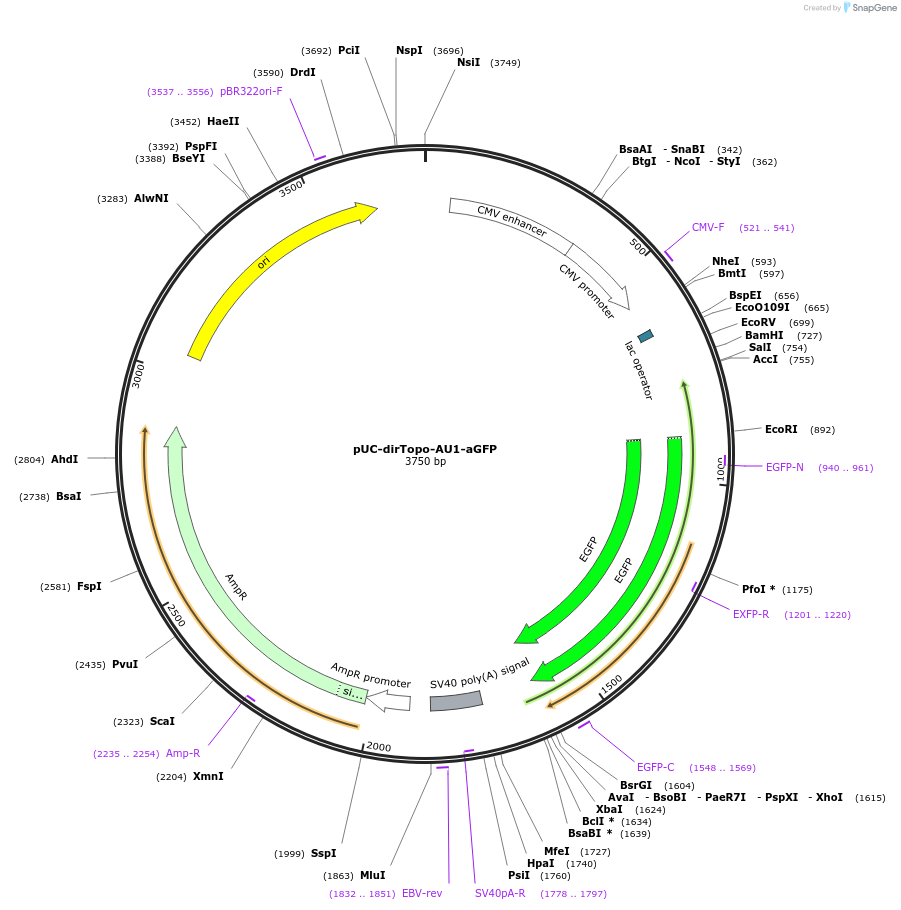
pCMVlac-dirTopo-AU1-aGFP
Plasmid#61971PurposeA directional cloning vector of PCR-amplified cDNAs for mammalian expressionDepositorInsertslacZ alpha
EGFP
ExpressionMammalianMutationIt contains silent mutationsAvailable SinceFeb. 4, 2015AvailabilityAcademic Institutions and Nonprofits only -
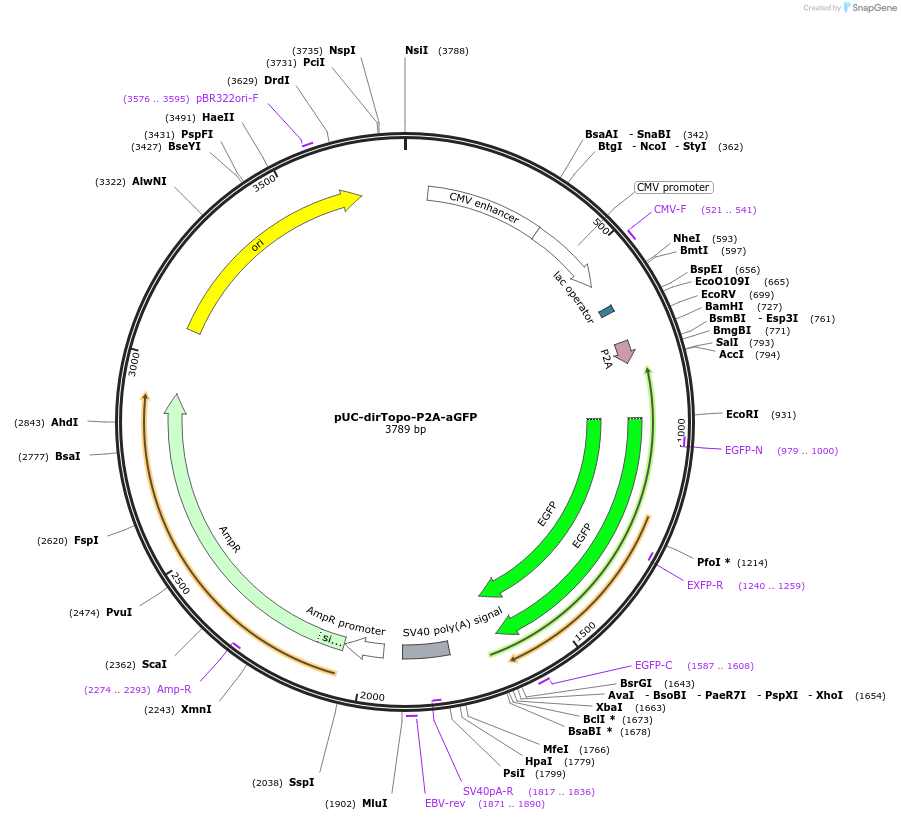
pUC-dirTopo-P2A-aGFP
Plasmid#61974PurposeA directional cloning vector of PCR-amplified cDNAs for mammalian expressionDepositorInsertslacZ alpha
EGFP
ExpressionMammalianMutationIt contains silent mutationsAvailable SinceFeb. 4, 2015AvailabilityAcademic Institutions and Nonprofits only -
pUC-dirTopo-AU1-aGFP
Plasmid#61973PurposeA directional cloning vector of PCR-amplified cDNAs for mammalian expressionDepositorInsertslacZ alpha
EGFP
ExpressionMammalianMutationIt contains silent mutationsAvailable SinceFeb. 4, 2015AvailabilityAcademic Institutions and Nonprofits only -
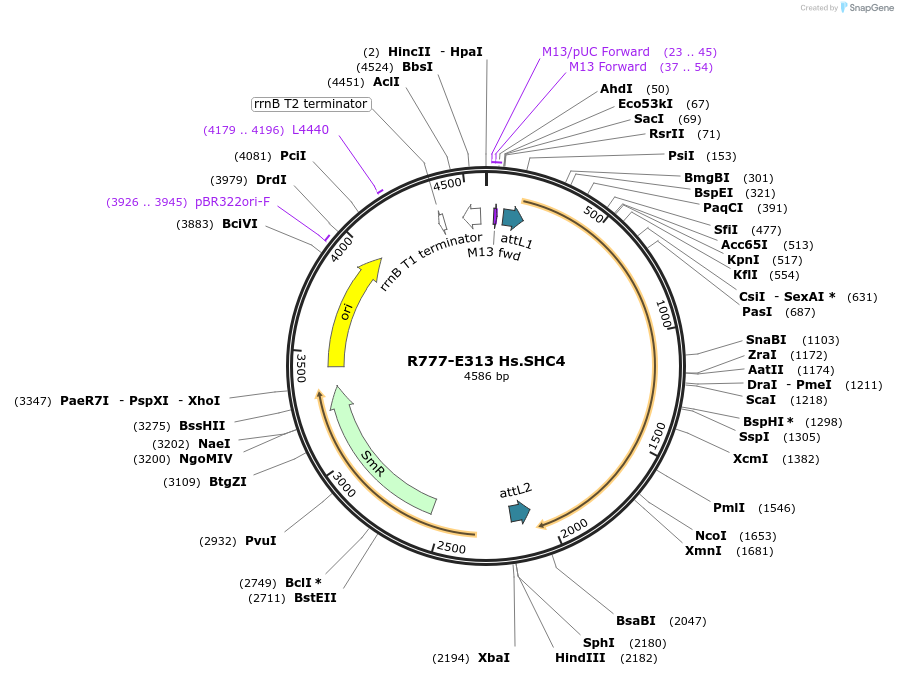
R777-E313 Hs.SHC4
Plasmid#70597PurposeGateway ORF clone of human SHC4 [NM_203349.3] with stop codon (for native or N-terminal fusions)DepositorInsertSHC4 (SHC4 Human)
UseGateway entry cloneAvailable SinceMarch 1, 2016AvailabilityAcademic Institutions and Nonprofits only -
EGFP-Cx43-7
Plasmid#56498PurposeLocalization: Gap Junctions, Excitation: 493, Emission: 509DepositorInsertCx43
TagsEGFPExpressionMammalianPromoterCMVAvailable SinceJan. 6, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
GST-TRIM24ΔRING
Plasmid#28148DepositorInsertTRIM24 (TRIM24 Human)
TagsGSTExpressionMammalianMutationDeletion of the RING domain: aa 1-139Available SinceFeb. 10, 2012AvailabilityAcademic Institutions and Nonprofits only -
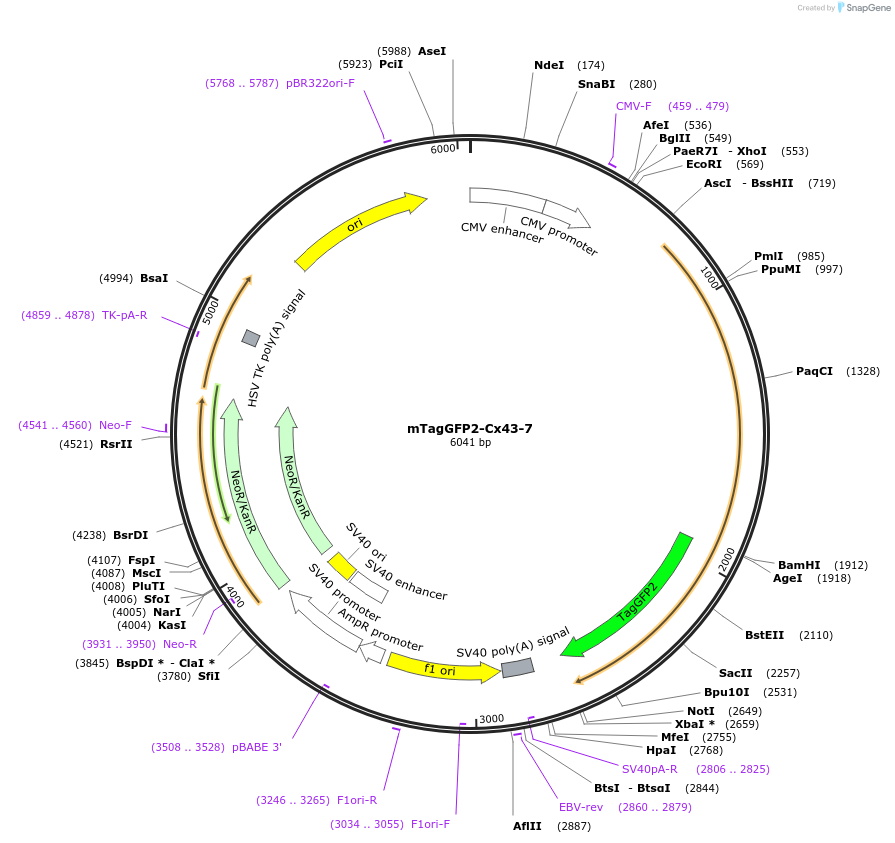
mTagGFP2-Cx43-7
Plasmid#56414PurposeLocalization: Gap Junctions, Excitation: 483, Emission: 506DepositorInsertCx43
TagsmTagGFP2ExpressionMammalianPromoterCMVAvailable SinceApril 1, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
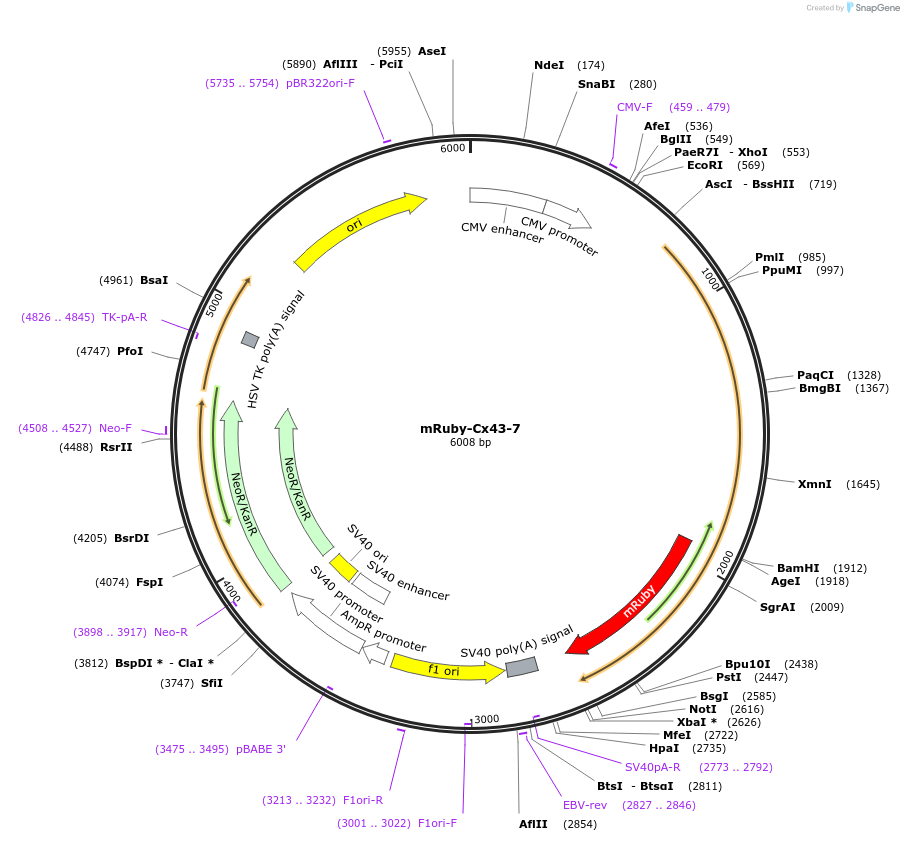
mRuby-Cx43-7
Plasmid#55856PurposeLocalization: Gap Junctions, Excitation: 558, Emission: 605DepositorInsertCx43
TagsmRubyExpressionMammalianPromoterCMVAvailable SinceMarch 12, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
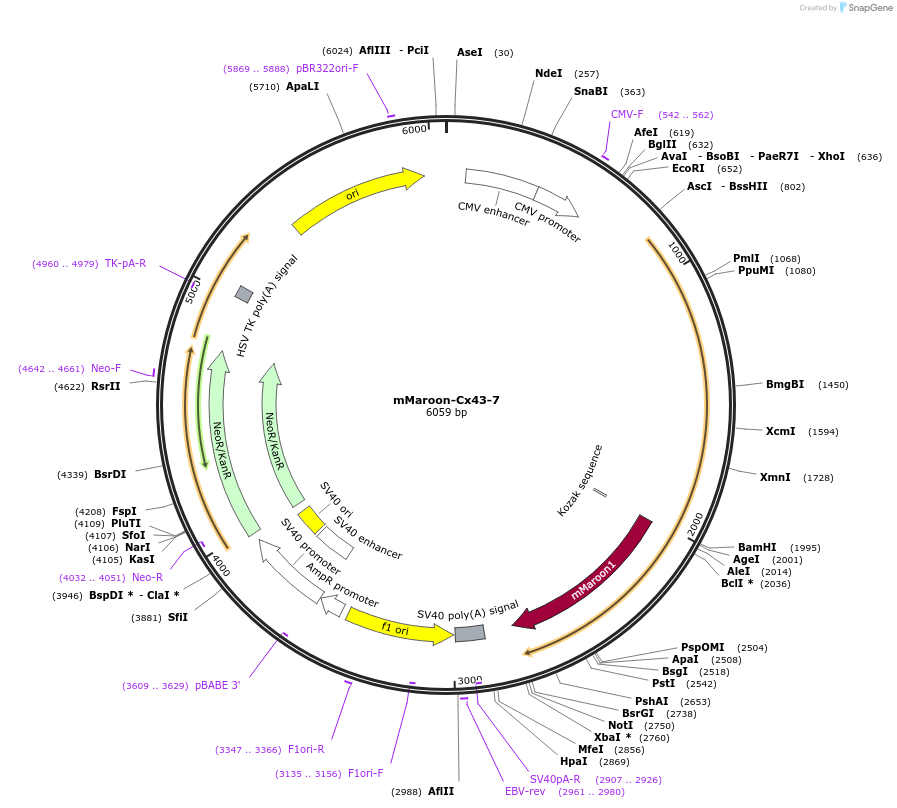
mMaroon-Cx43-7
Plasmid#56198PurposeLocalization: Gap Junctions, Excitation: 610, Emission: 652DepositorInsertCx43
TagsmMaroonExpressionMammalianPromoterCMVAvailable SinceMarch 12, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
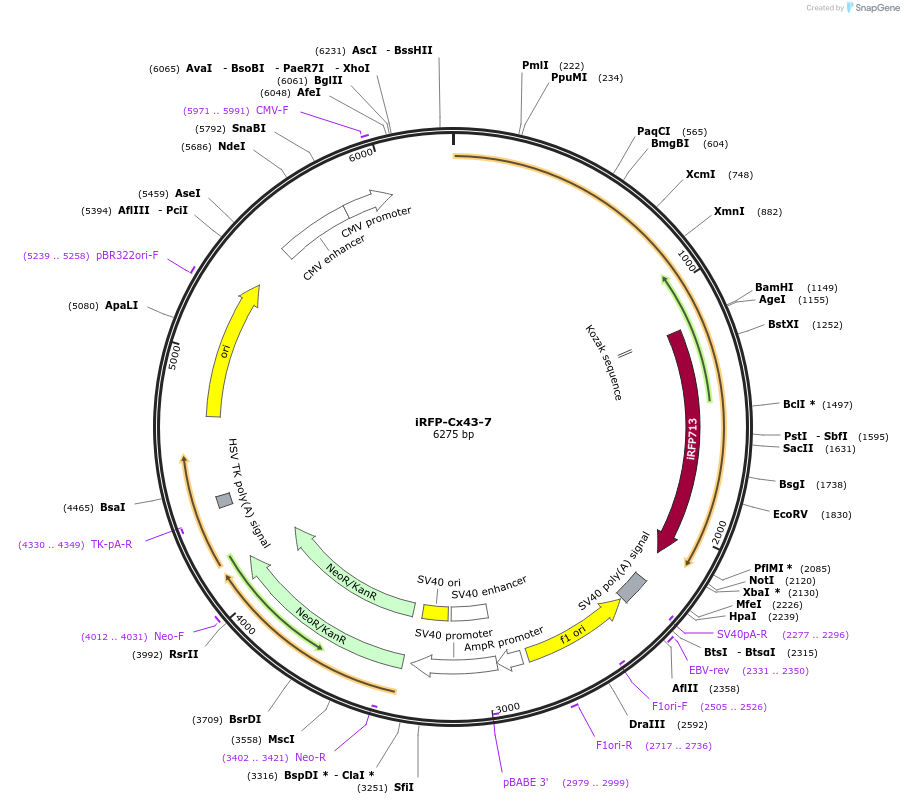
iRFP-Cx43-7
Plasmid#56276PurposeLocalization: Gap Junctions, Excitation: 690, Emission: 713DepositorInsertCx43
TagsiRFPExpressionMammalianPromoterCMVAvailable SinceApril 28, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
Sc Pol12-pRS406/GAL
Plasmid#241976PurposeOver-express Sc Pol12 (Sc pol alpha subunit) in yeast (integrated)DepositorInsertPol12 (POL12 Budding Yeast)
ExpressionYeastAvailable SinceAug. 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
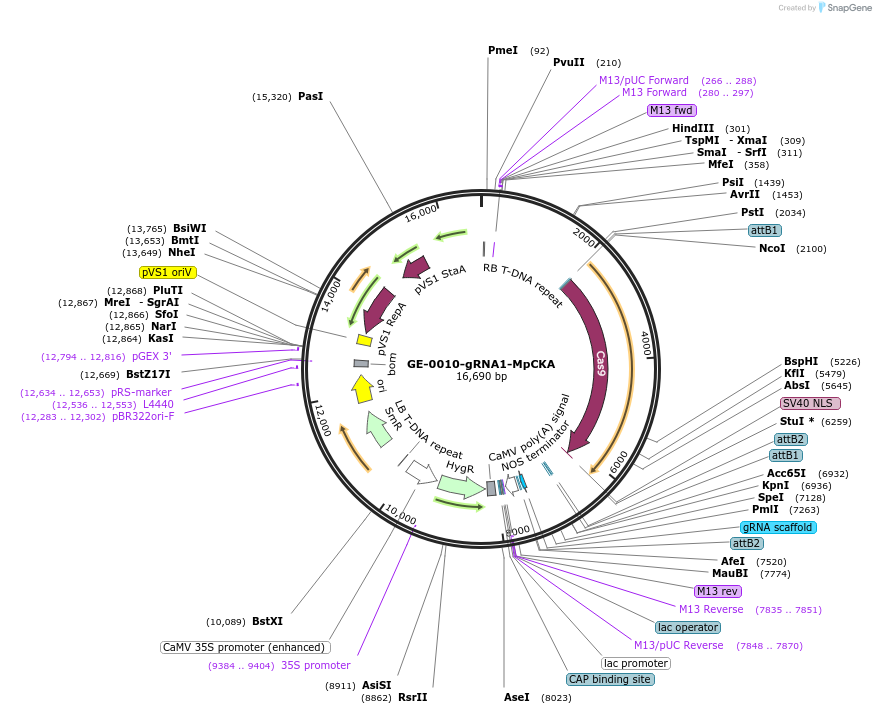
GE-0010-gRNA1-MpCKA
Plasmid#238526PurposePlant expression of Cas9 and gRNA against M. polymorpha CK2 alphaDepositorInsertMpCKA
UseCRISPRExpressionPlantAvailable SinceJune 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
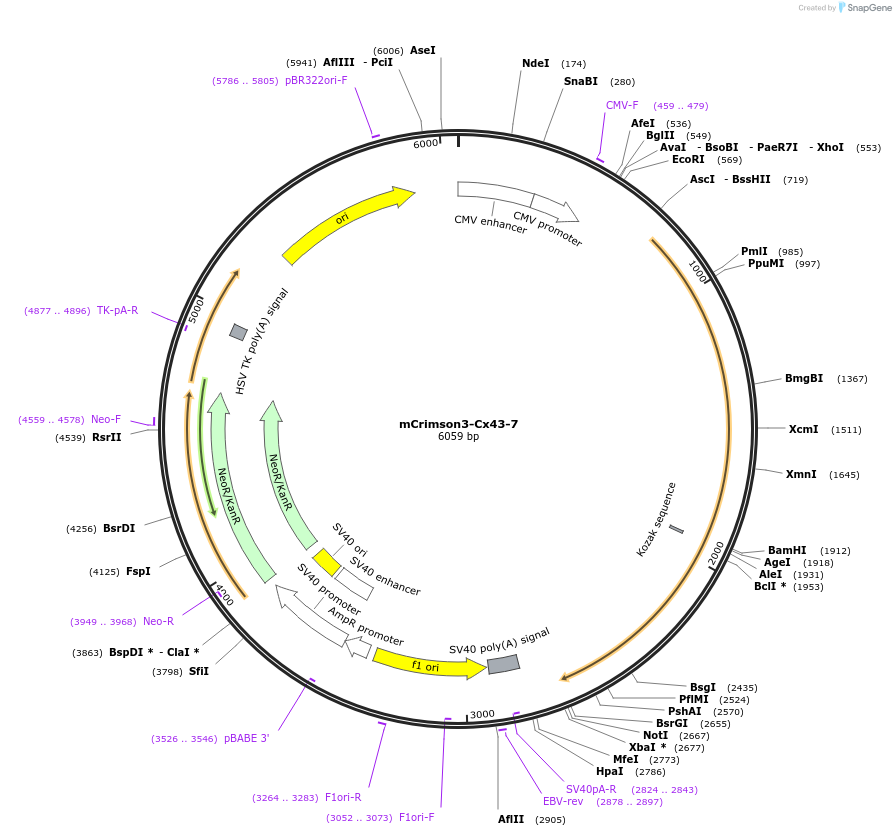
mCrimson3-Cx43-7
Plasmid#56193PurposeLocalization: Gap Junctions, Excitation: 588, Emission: 615DepositorInsertCx43
TagsmCrimson3ExpressionMammalianPromoterCMVAvailable SinceMarch 12, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
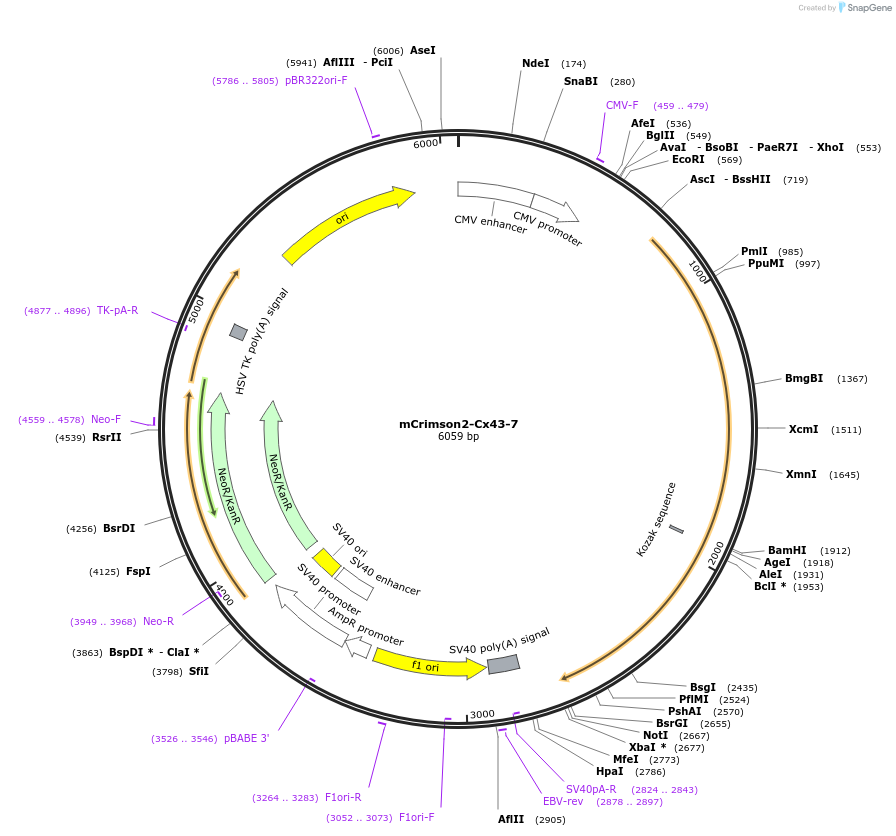
mCrimson2-Cx43-7
Plasmid#56186PurposeLocalization: Gap Junctions, Excitation: 588, Emission: 615DepositorInsertCx43
TagsmCrimson2ExpressionMammalianPromoterCMVAvailable SinceMarch 12, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
mCrimson-Cx43-7
Plasmid#56180PurposeLocalization: Gap Junctions, Excitation: 588, Emission: 615DepositorInsertCx43
TagsmCrimsonExpressionMammalianPromoterCMVAvailable SinceMarch 12, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
DsRed2-Cx43-7
Plasmid#55834PurposeLocalization: Gap Junctions, Excitation: 558, Emission: 583DepositorInsertCx43
TagsDsRed2ExpressionMammalianPromoterCMVAvailable SinceMarch 12, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
mRFP1-Cx43-7
Plasmid#55919PurposeLocalization: Gap Junctions, Excitation: 584, Emission: 607DepositorInsertCx43
TagsmRFP1ExpressionMammalianPromoterCMVAvailable SinceMarch 10, 2015AvailabilityAcademic Institutions and Nonprofits only -
YPet-Cx43-7
Plasmid#56631PurposeLocalization: Gap Junctions, Excitation: 513, Emission: 527DepositorInsertCx43
TagsYPetExpressionMammalianPromoterCMVAvailable SinceDec. 18, 2014AvailabilityIndustry, Academic Institutions, and Nonprofits