We narrowed to 24,781 results for: CHI
-
Plasmid#26329DepositorInsertmmu-mir-139 (Mir139 Mouse)
ExpressionMammalianAvailable SinceSept. 29, 2010AvailabilityAcademic Institutions and Nonprofits only -
Tmem194 pmRFP-N2 (1389)
Plasmid#62048Purposemammalian expression of nuclear envelope transmembrane proteinDepositorAvailable SinceJan. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3.2/V5 mmu-mir-1224
Plasmid#26337DepositorInsertmmu-mir-1224 (Mir1224 Mouse)
ExpressionMammalianAvailable SinceOct. 19, 2010AvailabilityAcademic Institutions and Nonprofits only -
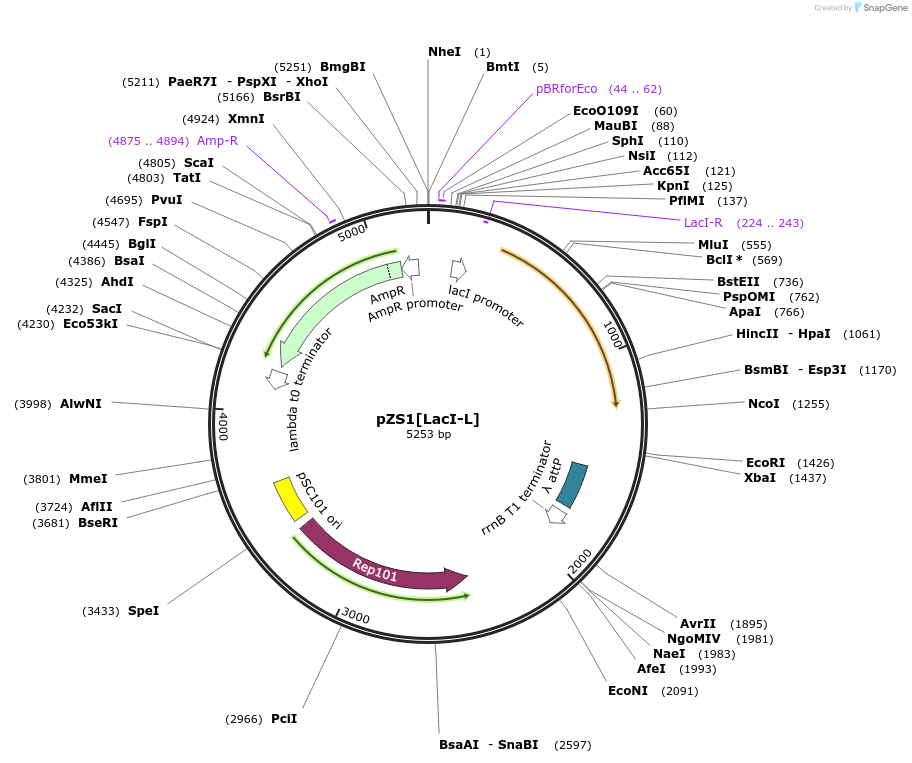
pZS1[LacI-L]
Plasmid#60745PurposeContains PI driving expression dimeric LacI, the Lactose inducible repressor.DepositorInsertLacI
UseSynthetic BiologyExpressionBacterialMutationwt LacI without the tetramerization domainAvailable SinceAug. 6, 2015AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3.2/V5 mmu-mir-217
Plasmid#26333DepositorInsertmmu-mir-217 (Mir217 Mouse)
ExpressionMammalianAvailable SinceSept. 29, 2010AvailabilityAcademic Institutions and Nonprofits only -
Tmem194 pEGFP-N2 (1074)
Plasmid#62027Purposemammalian expression of nuclear envelope transmembrane proteinDepositorAvailable SinceJan. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
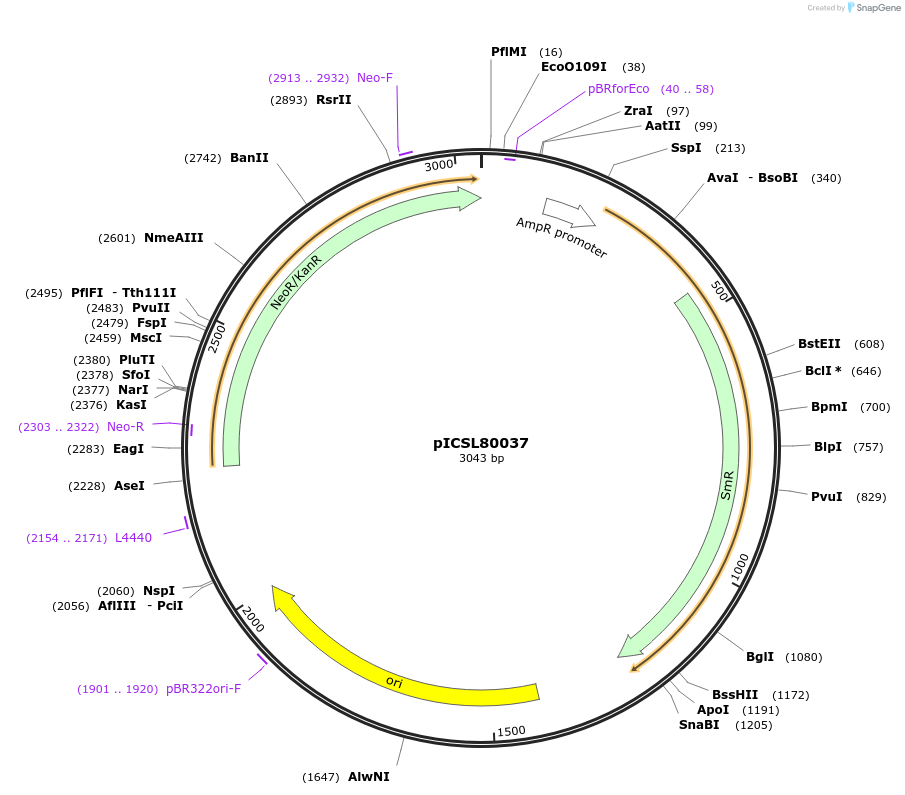
pICSL80037
Plasmid#68260PurposeLevel 0 Golden Gate Part: Coding sequence, neomycin phophotransferase II (Escherichia coli)DepositorInsertCoding sequence, neomycin phophotransferase II (Escherichia coli)
UseSynthetic BiologyAvailable SinceAug. 20, 2015AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3.2/V5 mmu-mir-1197
Plasmid#26339DepositorInsertmmu-mir-1197 (Mir1197 Mouse)
ExpressionMammalianAvailable SinceSept. 29, 2010AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3.2/V5 mmu-mir-216a
Plasmid#26332DepositorInsertmmu-mir-216a (Mir216a Mouse)
ExpressionMammalianAvailable SinceSept. 29, 2010AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3.2/V5 mmu-mir-1964
Plasmid#26343DepositorInsertmmu-mir-1964 (Mir1964 Mouse)
ExpressionMammalianAvailable SinceSept. 29, 2010AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3.2/V5 mmu-mir-224
Plasmid#26335DepositorInsertmmu-mir-224 (Mir224 Mouse)
ExpressionMammalianAvailable SinceSept. 29, 2010AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3.2/V5 hsa-mir-888
Plasmid#26316DepositorInserthsa-mir-888 (MIR888 Human)
ExpressionMammalianAvailable SinceSept. 29, 2010AvailabilityAcademic Institutions and Nonprofits only -
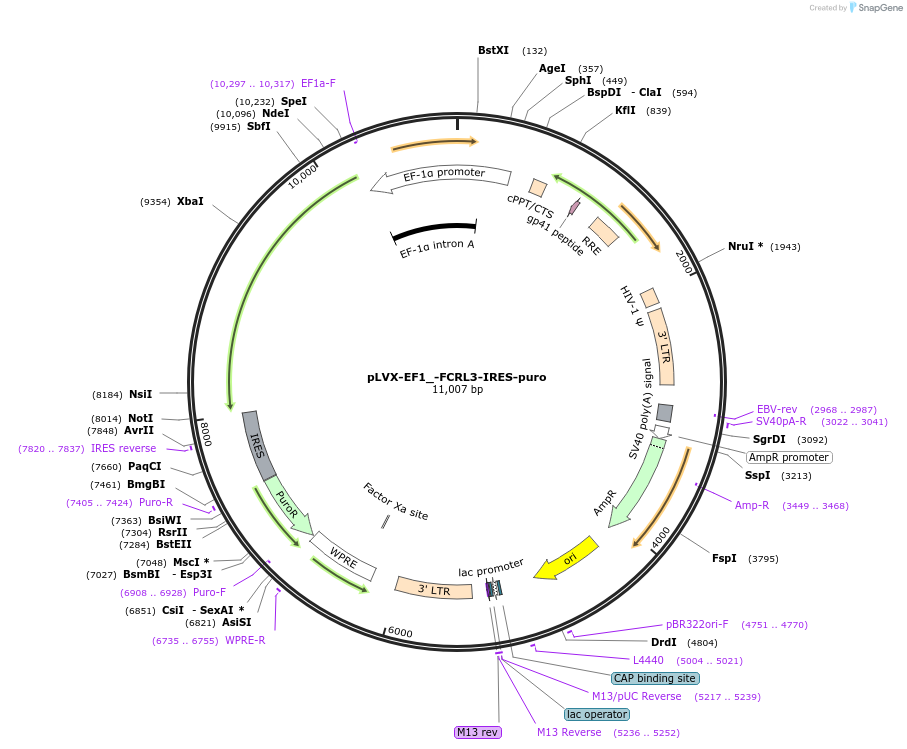
pLVX-EF1_-FCRL3-IRES-puro
Plasmid#248148PurposeLentiviral vector encoding human FCRL3DepositorAvailable SinceDec. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
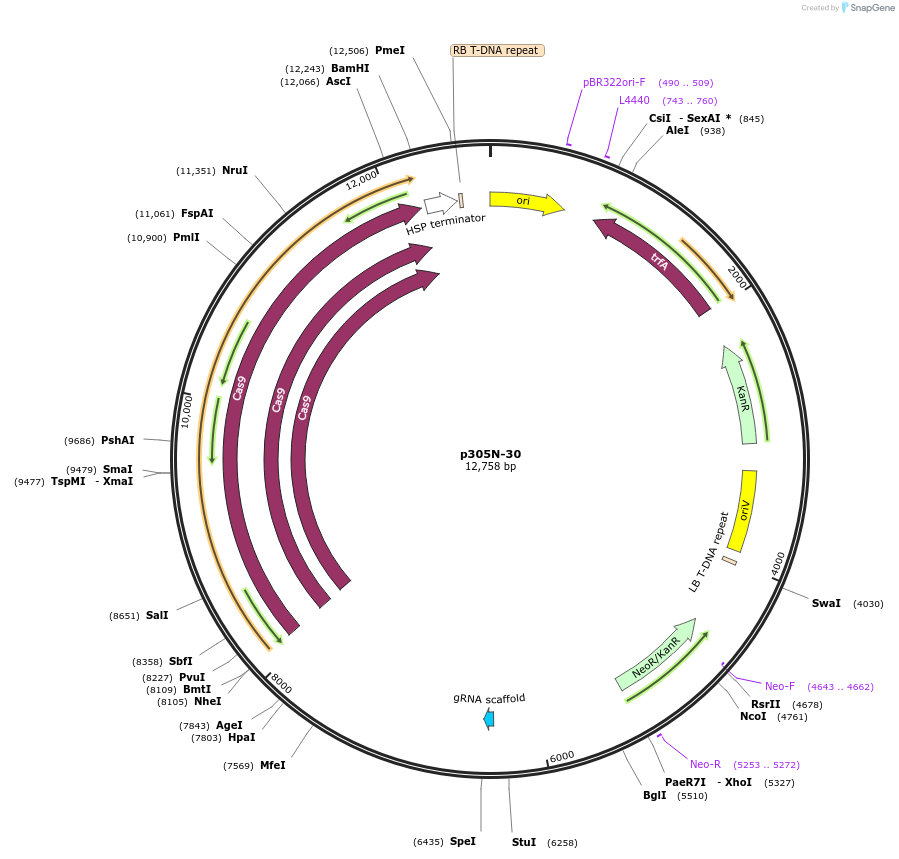
p305N-30
Plasmid#246313PurposeEvaluation of PtU6.2c3 promoter (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertPtU6.2c3 promoter
UseCRISPRExpressionPlantMutationdeletion: -T (-29 from TSS) within TATAPromoterPtU6.2c3 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
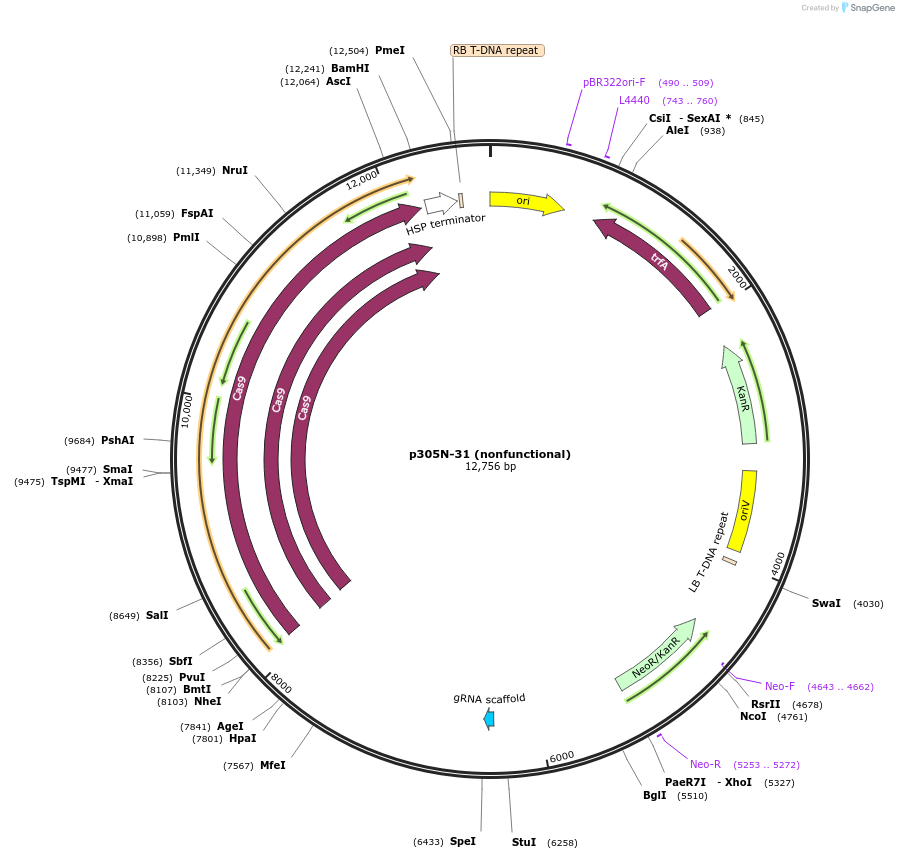
p305N-31 (nonfunctional)
Plasmid#246314PurposeEvaluation of PtU6.2c4 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertPtU6.2c4 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationdeletions: -A (-41 from TSS), -T (-30), -T (-29);…PromoterPtU6.2c4 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
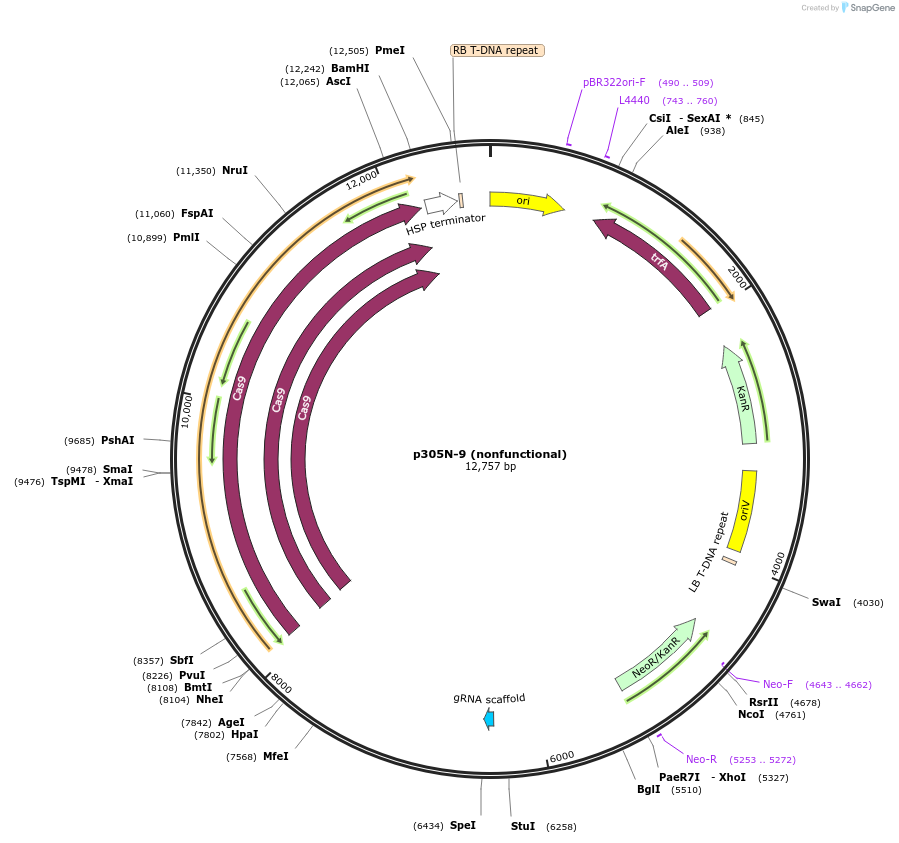
p305N-9 (nonfunctional)
Plasmid#246292PurposeEvaluation of AtU6.29c13 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertAtU6.29c13 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationdeletions: -A (-58 from TSS), -T (-57) within USEPromoterAtU6.29c13 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
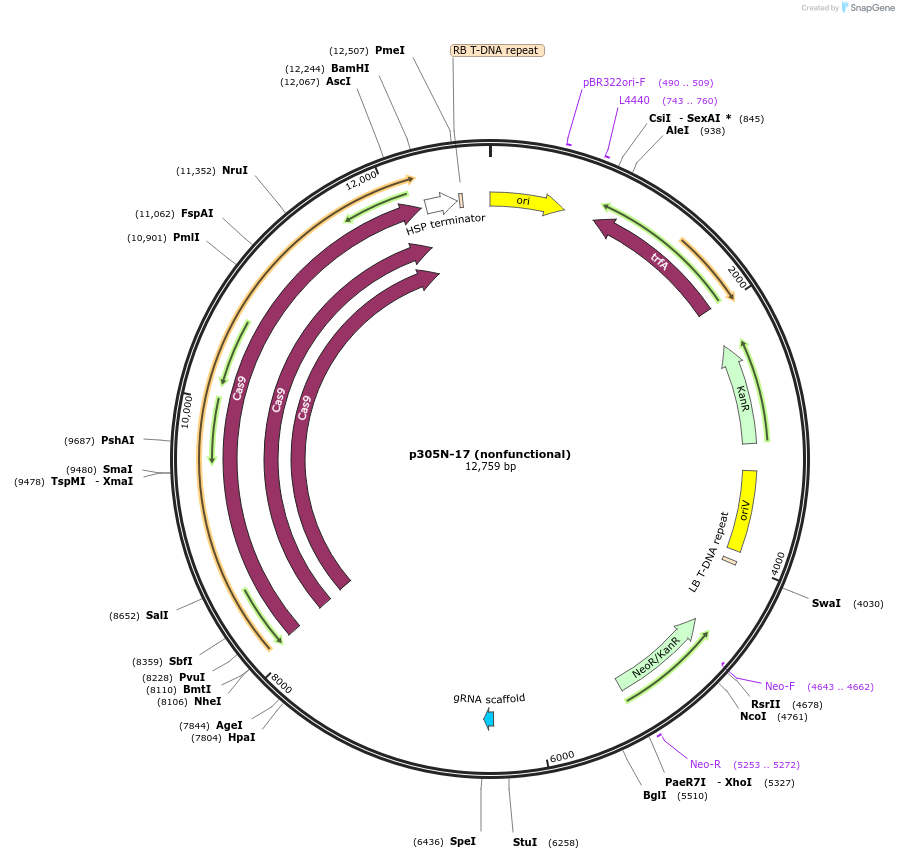
p305N-17 (nonfunctional)
Plasmid#246300PurposeEvaluation of HbU6.2m3 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertHbU6.2m3 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationC-to-T (-29 from TSS), G-to-A (-24)PromoterHbU6.2m3 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
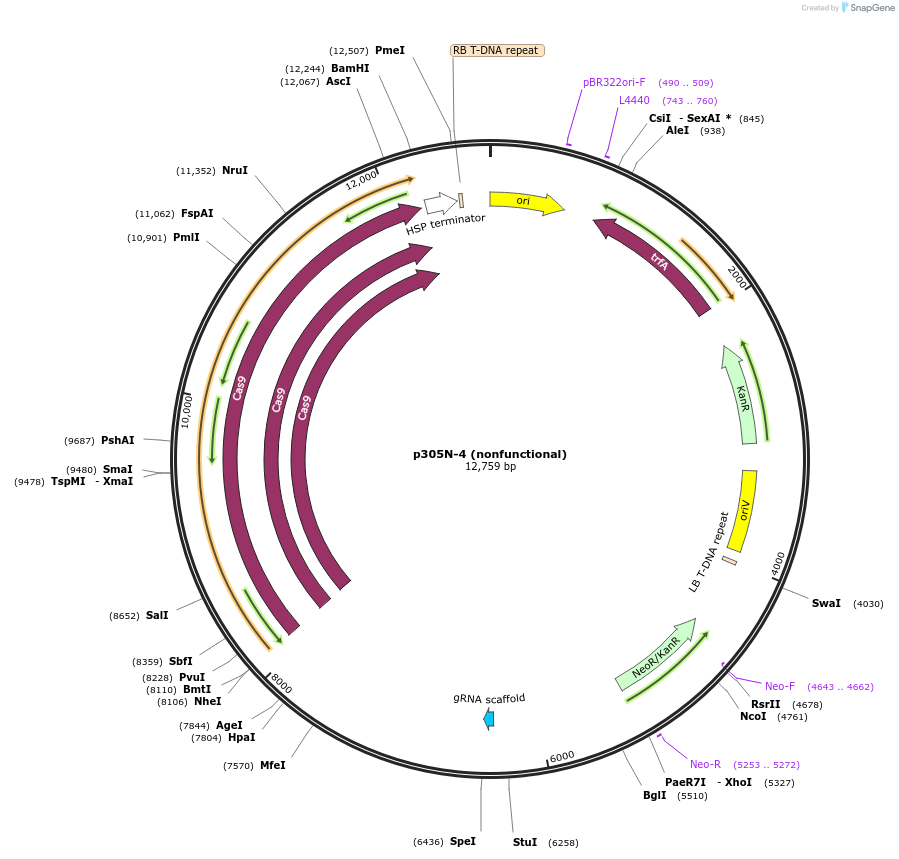
p305N-4 (nonfunctional)
Plasmid#246287PurposeEvaluation of AtU6.1c1 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertAtU6.1c1 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationG-to-C at -15 from TSSPromoterAtU6.1c1 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
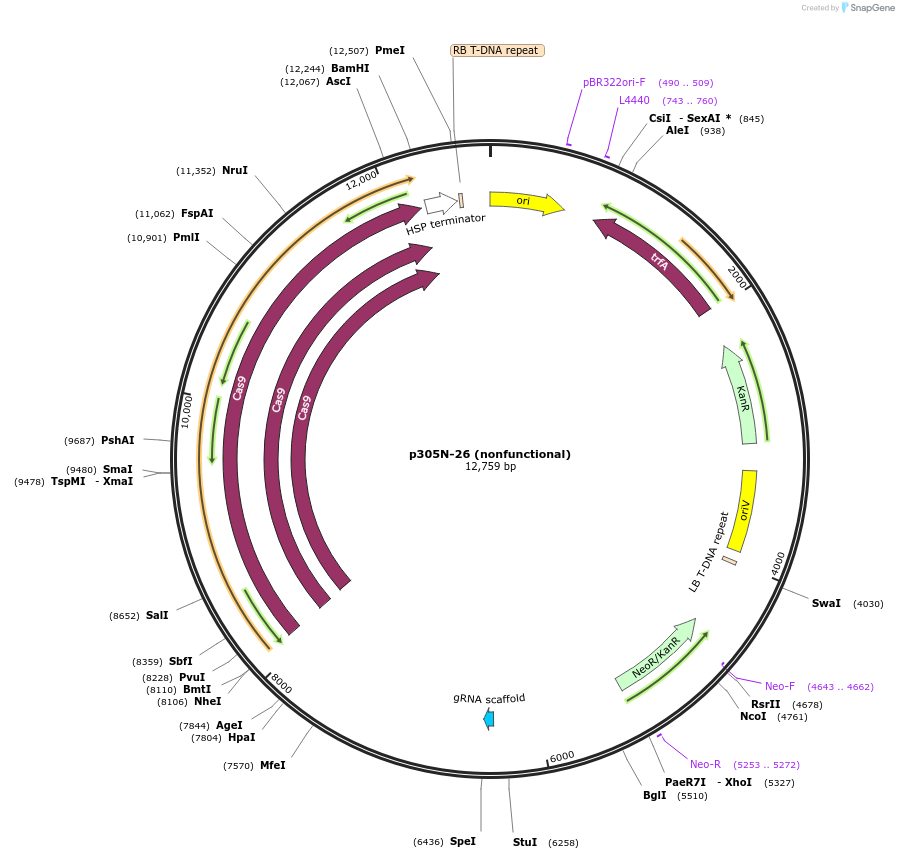
p305N-26 (nonfunctional)
Plasmid#246309PurposeEvaluation of MtU6.6m7 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertMtU6.6m7 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationC-to-A (-63 from TSS), T-to-G (-57)PromoterMtU6.6m7 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
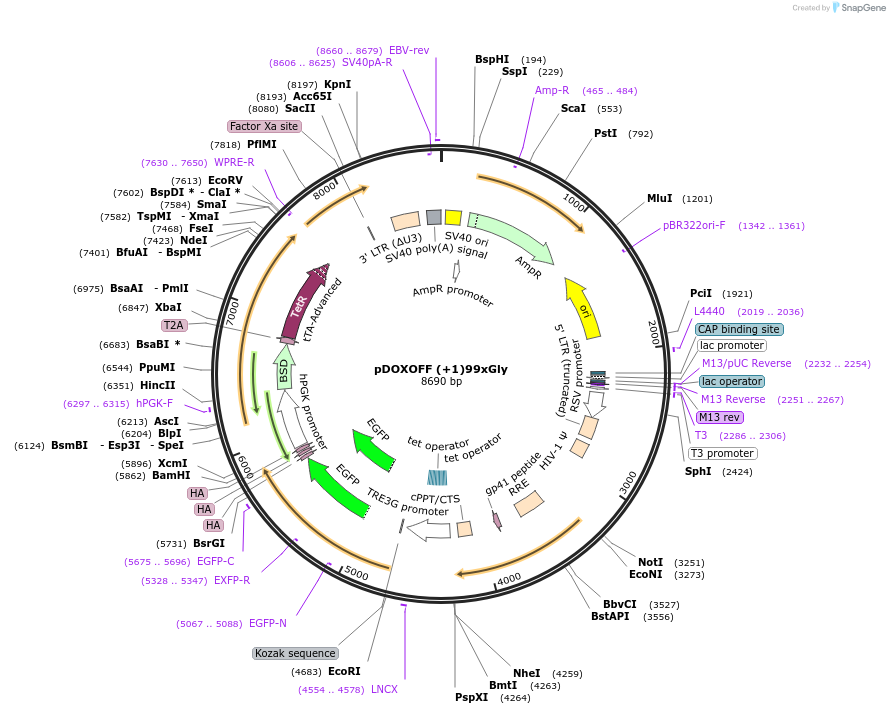
pDOXOFF (+1)99xGly
Plasmid#211347PurposeDOX-regulated lentiviral expression of +1 frameshifted 99xGly-EGFP-3xHADepositorInsert(+1)99xGly-EGFP-3xHA
UseLentiviralTagsEGFP-3xHAMutation+1 frameshift to produce polyR frameAvailable SinceOct. 28, 2025AvailabilityAcademic Institutions and Nonprofits only