We narrowed to 5,861 results for: SPI
-
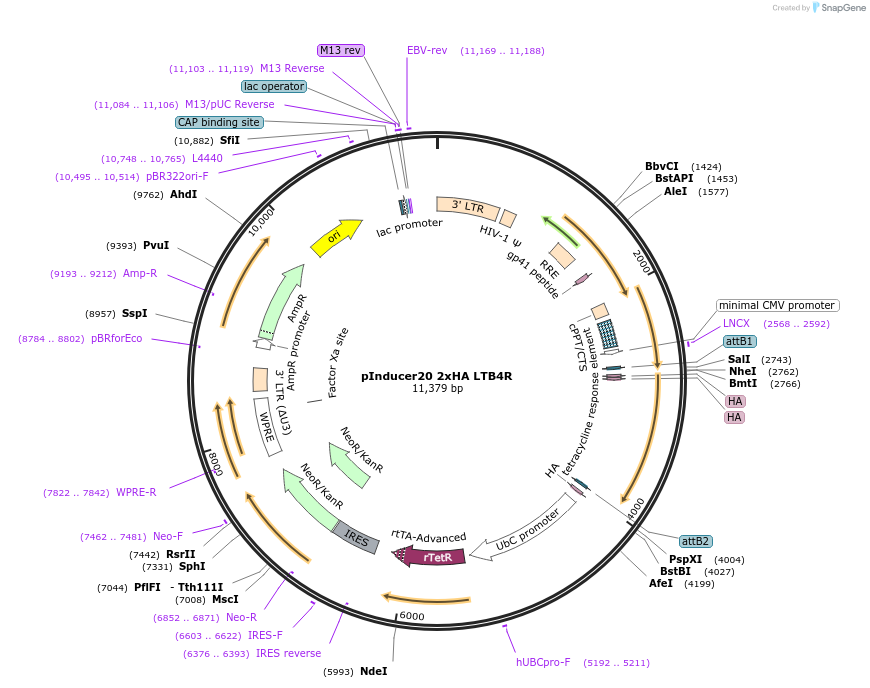
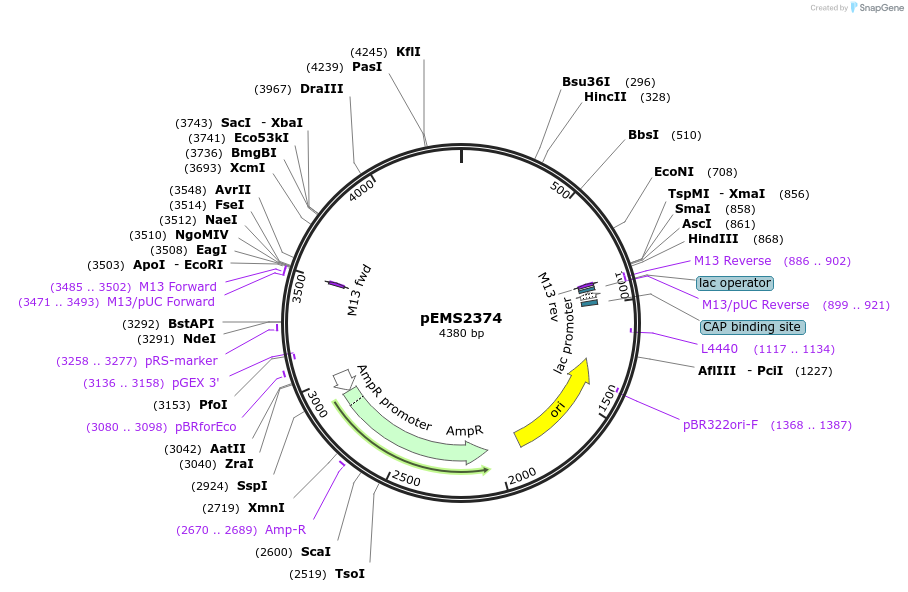
Plasmid#226810PurposeTet-inducible lentiviral vector for 2xHA LTB4R expressionDepositorInsertLTB4R (LTB4R Human)
UseLentiviralTags2xHA tag followed by a GDPPVAT linkerMutationSequence optimized from NP_858043.1Available SinceJan. 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
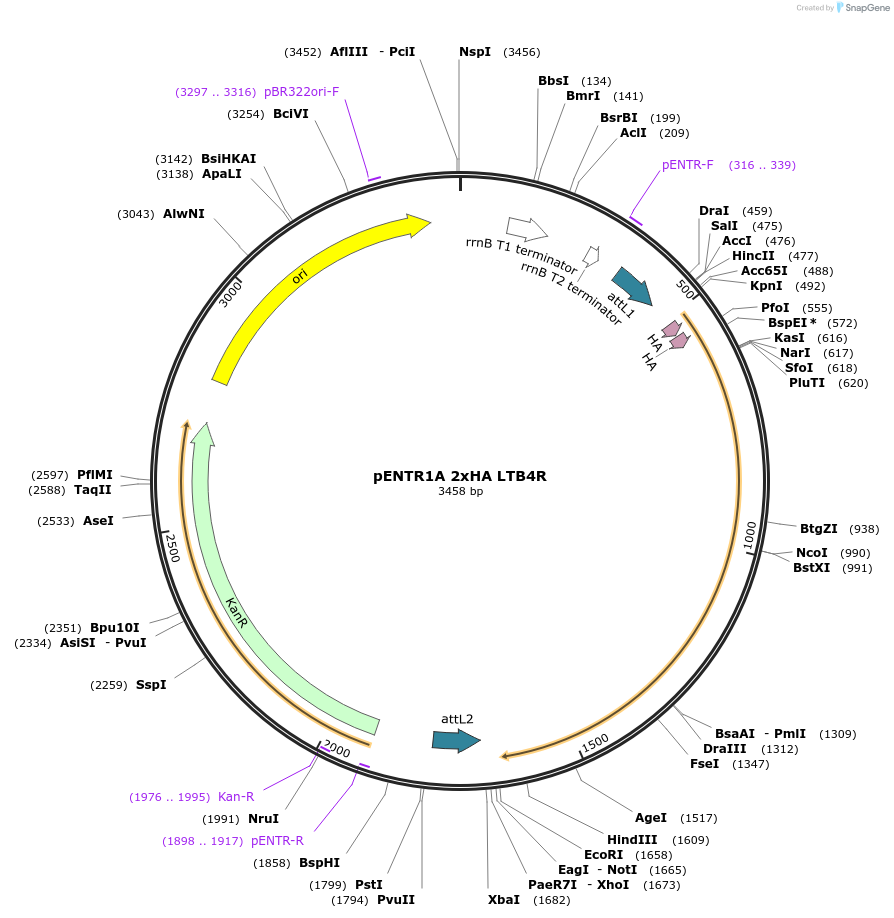
pENTR1A 2xHA GPR153
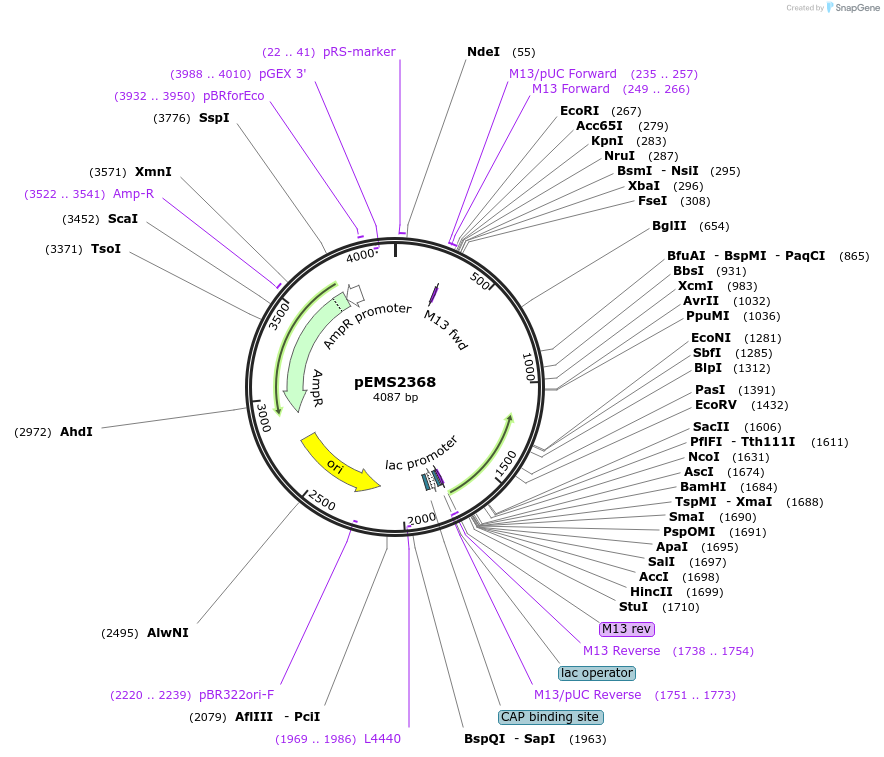
Plasmid#226808PurposeGateway entry vector for 2xHA GPR153DepositorInsertGPR153 (GPR153 Human)
UseGateway entryTags2xHA tag followed by a GDPPVAT linkerMutationSequence optimized from NP_997253.2Available SinceJan. 7, 2025AvailabilityAcademic Institutions and Nonprofits only -
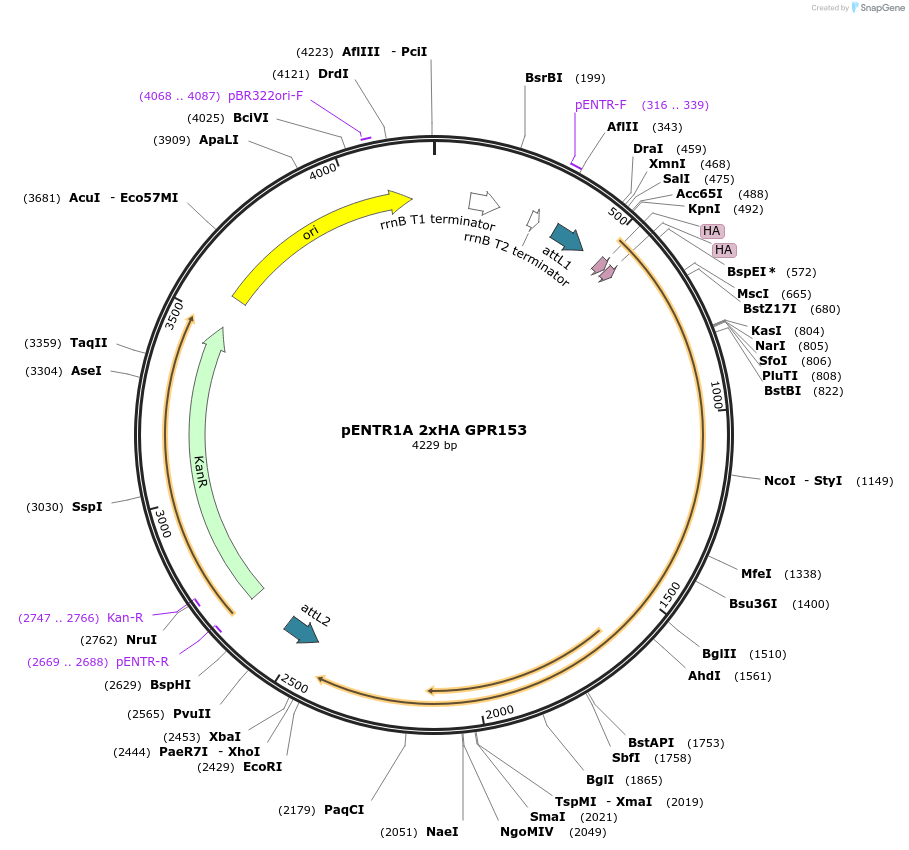
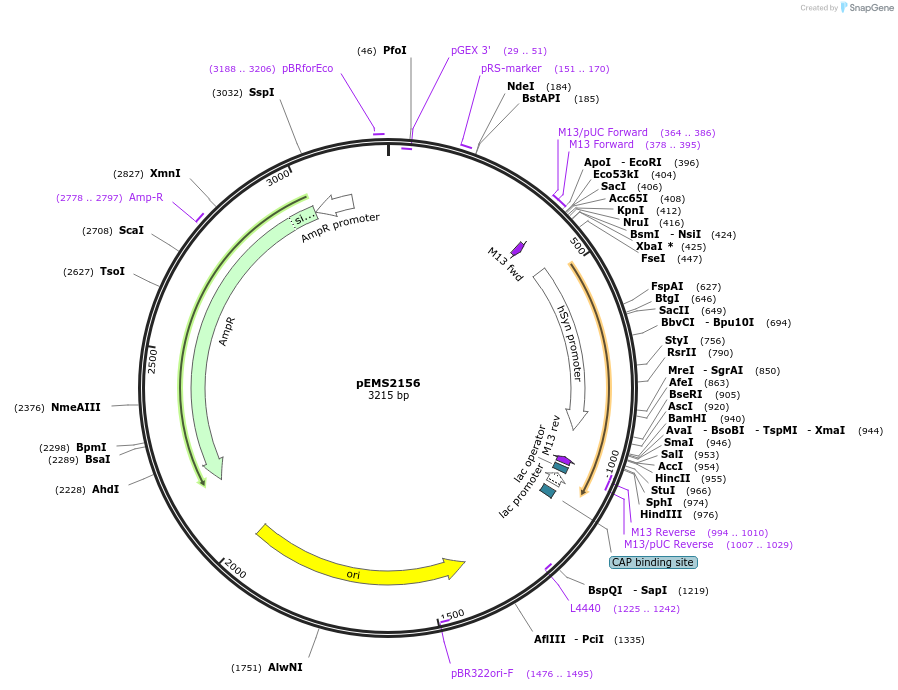
pENTR1A 2xHA LTB4R
Plasmid#226806PurposeGateway entry vector for 2xHA LTB4RDepositorInsertLTB4R (LTB4R Human)
UseGateway entryTags2xHA tag followed by a GDPPVAT linkerMutationSequence optimized from NP_858043.1Available SinceJan. 7, 2025AvailabilityAcademic Institutions and Nonprofits only -
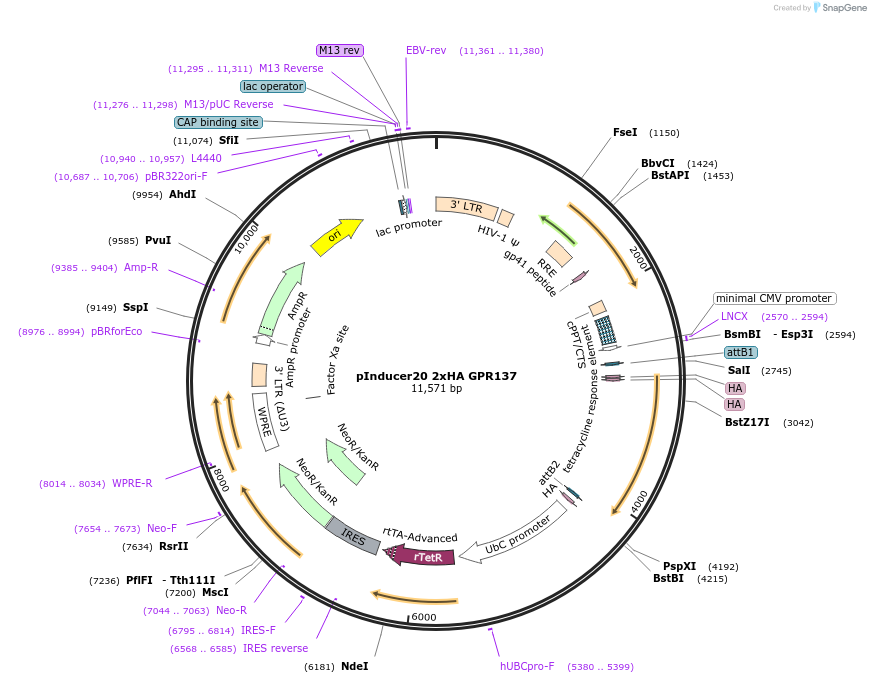
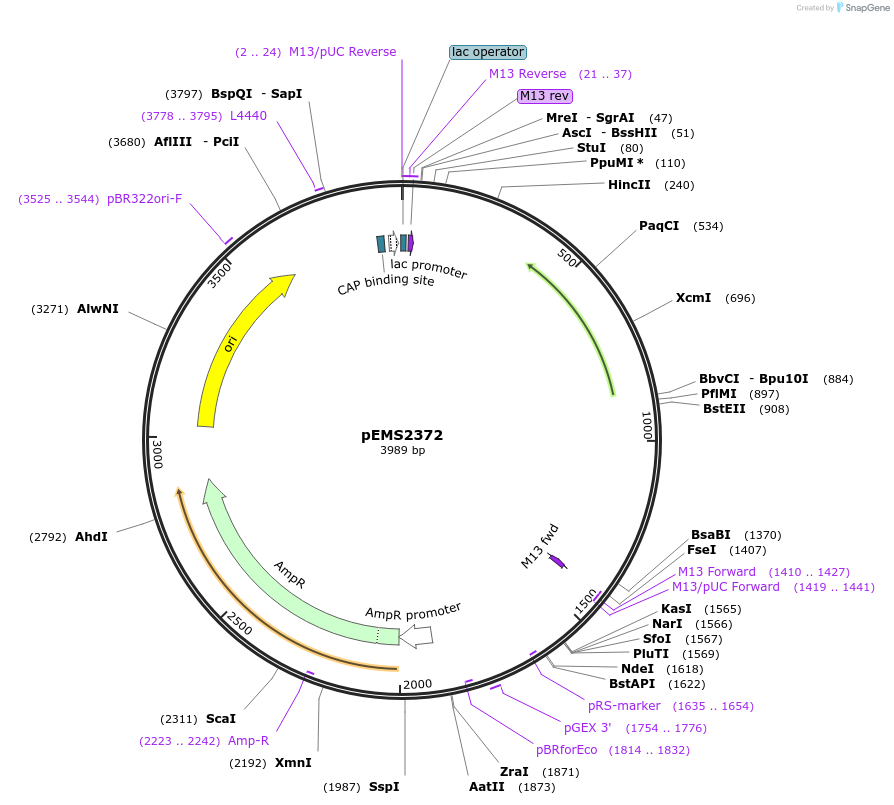
pInducer20 2xHA GPR137
Plasmid#226813PurposeTet-inducible lentiviral vector for 2xHA GPR137 expressionDepositorInsertGPR137 (GPR137 Human)
UseLentiviralTags2xHA tag followed by a GDPPVAT linkerMutationSequence optimized from NP_001164351Available SinceJan. 7, 2025AvailabilityAcademic Institutions and Nonprofits only -
-
-
-
-
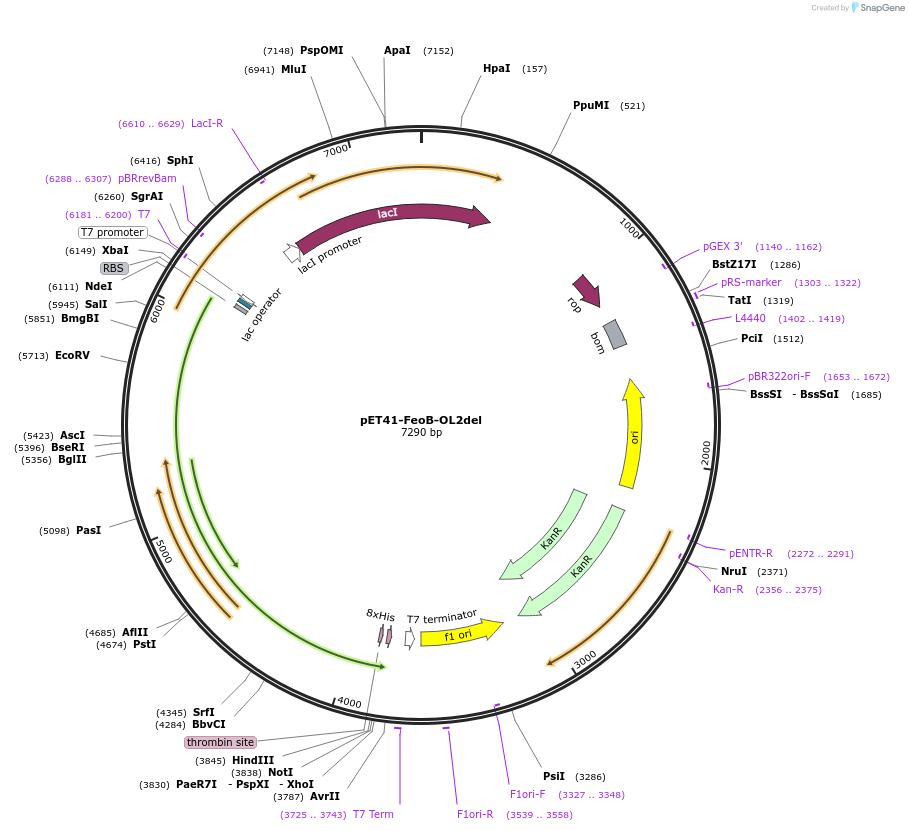
pET41-FeoB-OL2del
Plasmid#216742PurposeStudy the interaction of FeoB in P. aeruginosa, specifically focusing on the role of the outer loop 2 (OL2) by examining the variant lacking OL2 in its interaction with exogenous peptides.DepositorInsertfeoB (PA4358 P. aeruginosa (Bacteria))
Tags8x HisExpressionBacterialMutationOuter loop 2 (OL2) with the sequence of [LEDSGYMA…Available SinceSept. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
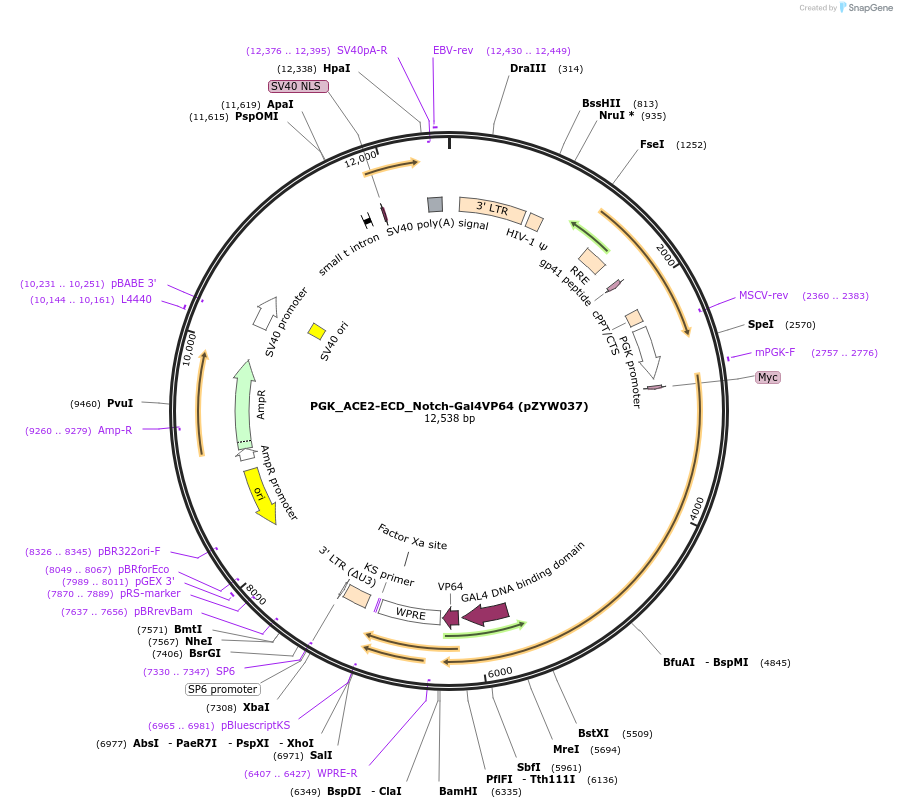
PGK_ACE2-ECD_Notch-Gal4VP64 (pZYW037)
Plasmid#194226PurposeLentiviral expression vector expressing ACE2-ECD SynNotch and mCherry transduction markerDepositorAvailable SinceSept. 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
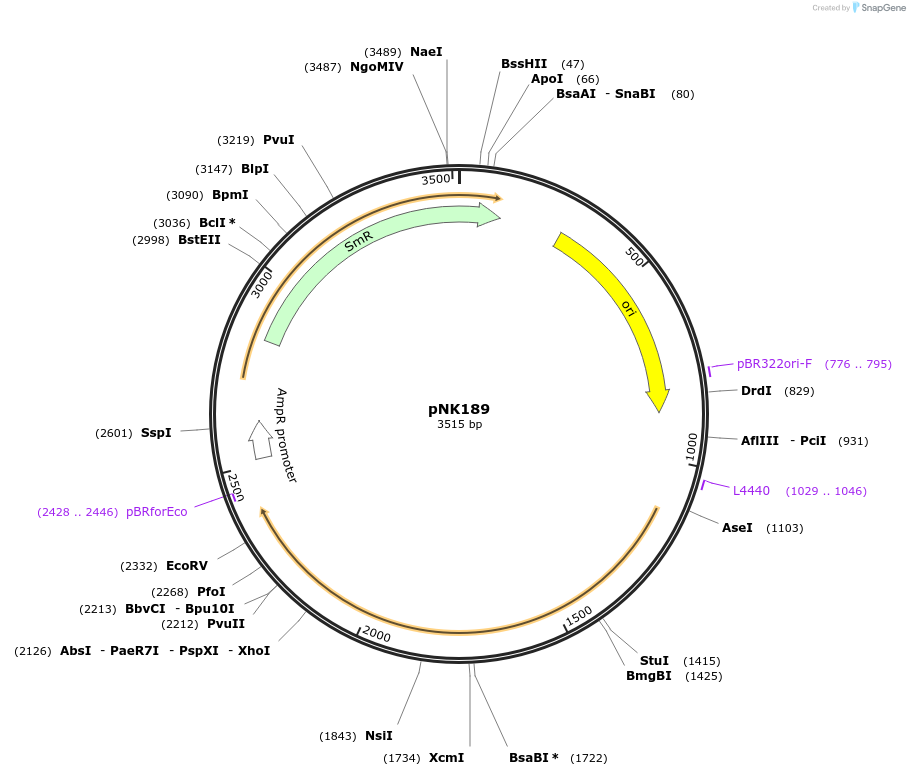
pNK189
Plasmid#219743PurposeMoClo-compatible Level 0 promoterless vector encoding mutant of Neonothopanus nambi hispidin-3-hydroxylase nnH3H_v2 codon-optimised for expression in Homo sapiensDepositorInsertmutant of fungal hispidin-3-hydroxylase
UseSynthetic BiologyMutationD37E, V181I, S323M, M385KAvailable SinceJuly 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
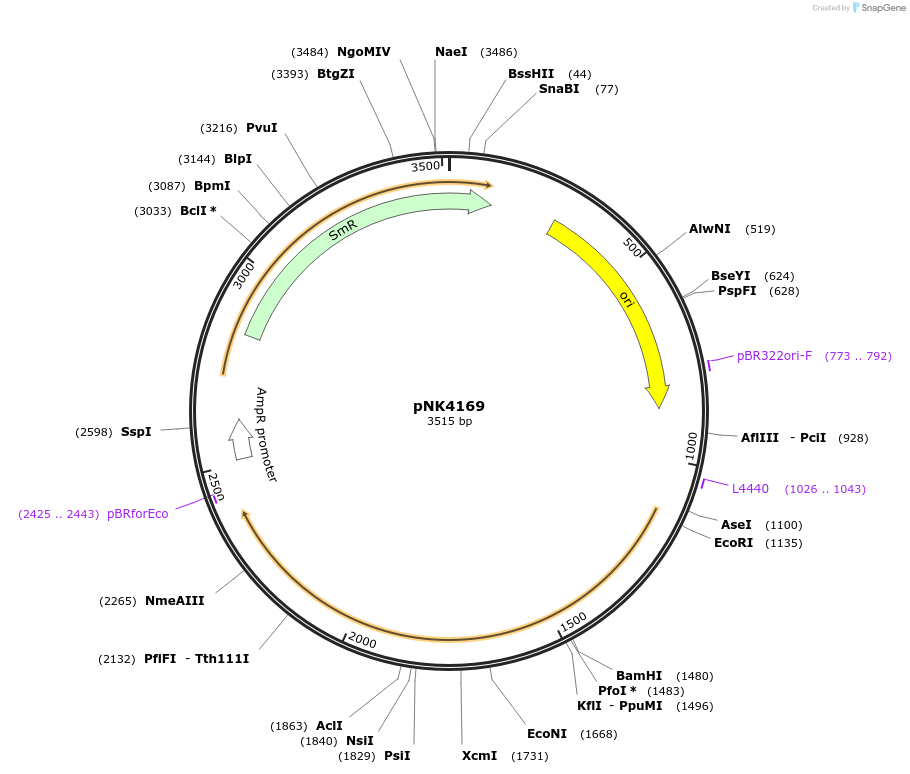
pNK4169
Plasmid#219744PurposeMoClo-compatible Level 0 promoterless vector encoding mutant of Neonothopanus nambi hispidin-3-hydroxylase nnH3H_v2 codon-optimised for expression in Pichia pastorisDepositorInsertmutant of fungal hispidin-3-hydroxylase
UseSynthetic BiologyMutationD37E, V181I, S323M, M385KAvailable SinceJuly 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
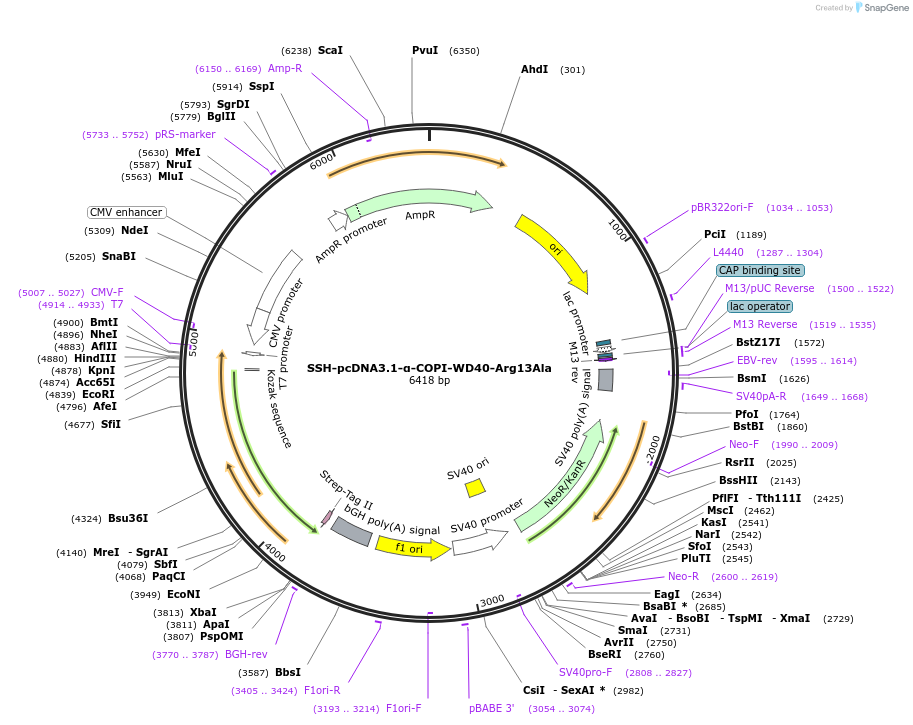
SSH-pcDNA3.1-α-COPI-WD40-Arg13Ala
Plasmid#208172PurposeMammalian expression plasmid for Schizosaccharomyces pombe α-COPI-WD40 Arg13Ala mutant (residues 1-327)DepositorInsertN-terminal WD40 domain of yeast α-COPI(1-327);six substitutions(Arg13Ala, Leu181Lys, Leu185Lys, Ile192Lys, Leu196Lys, Phe197Lys)
TagsStrep-tagExpressionMammalianMutationArg13Ala, Leu181Lys, Leu185Lys, Ile192Lys, Leu196…Available SinceJune 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
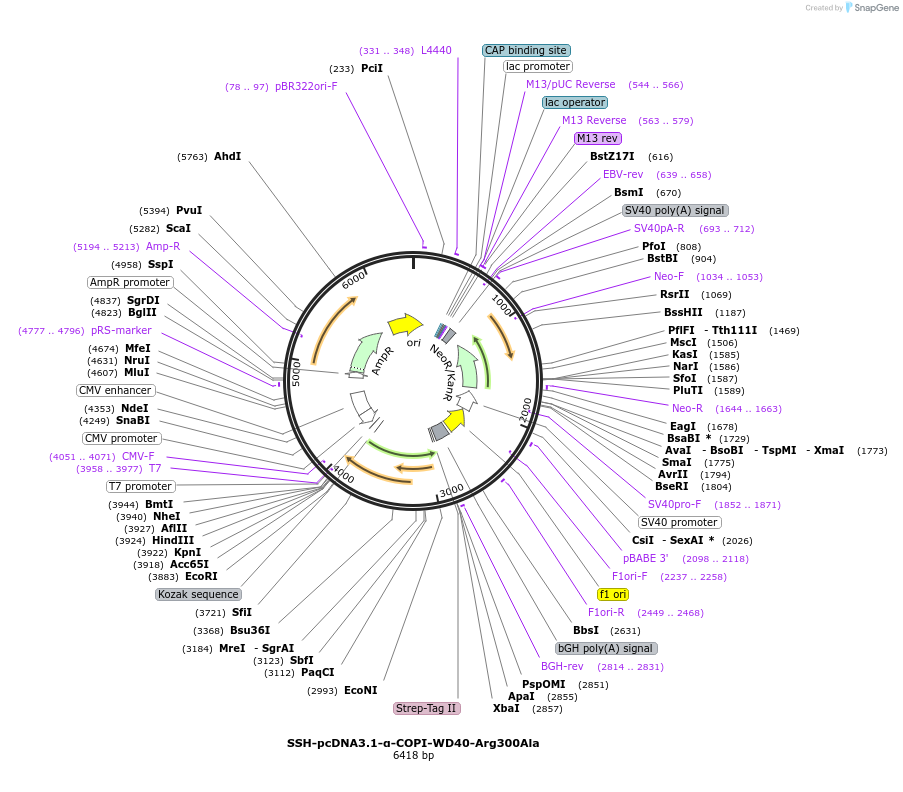
SSH-pcDNA3.1-α-COPI-WD40-Arg300Ala
Plasmid#208173PurposeMammalian expression plasmid for Schizosaccharomyces pombe α-COPI-WD40 mutant Arg300Ala (residues 1-327)DepositorInsertN-terminal WD40 domain of yeast α-COPI(1-327);six substitutions(Leu181Lys, Leu185Lys, Ile192Lys, Leu196Lys, Phe197Lys,Arg300Ala)
TagsStrep-tagExpressionMammalianMutationLeu181Lys, Leu185Lys, Ile192Lys, Leu196Lys, Phe19…Available SinceJune 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
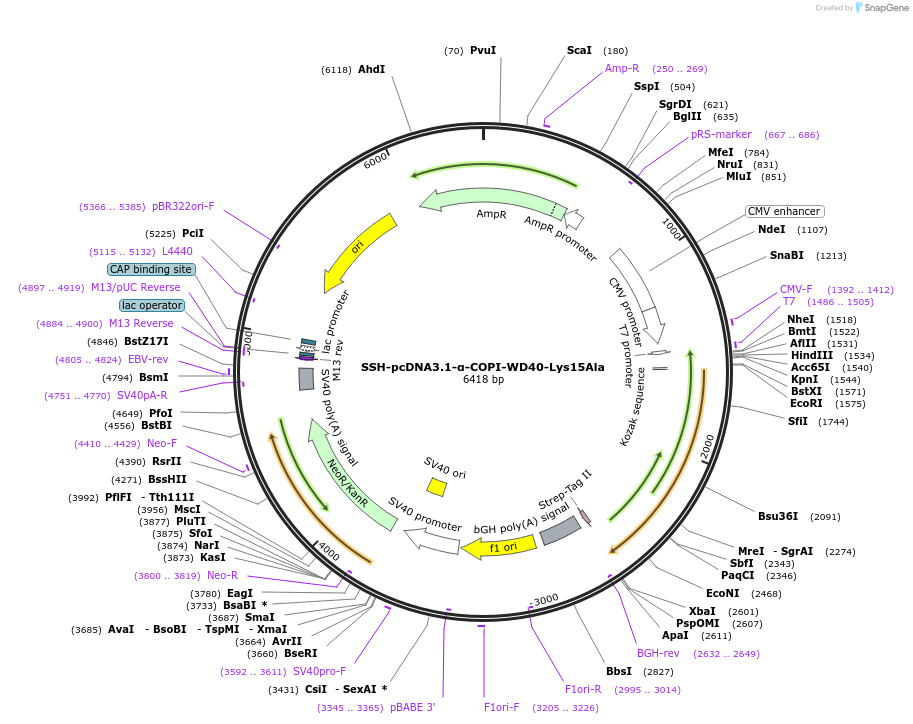
SSH-pcDNA3.1-α-COPI-WD40-Lys15Ala
Plasmid#208174PurposeMammalian expression plasmid for Schizosaccharomyces pombe α-COPI-WD40 mutant Lys15Ala (residues 1-327)DepositorInsertN-terminal WD40 domain of yeast α-COPI(1-327);six substitutions(Lys15Ala, Leu181Lys, Leu185Lys, Ile192Lys, Leu196Lys, Phe197Lys)
TagsStrep-tagExpressionMammalianMutationLys15Ala, Leu181Lys, Leu185Lys, Ile192Lys, Leu196…Available SinceJune 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
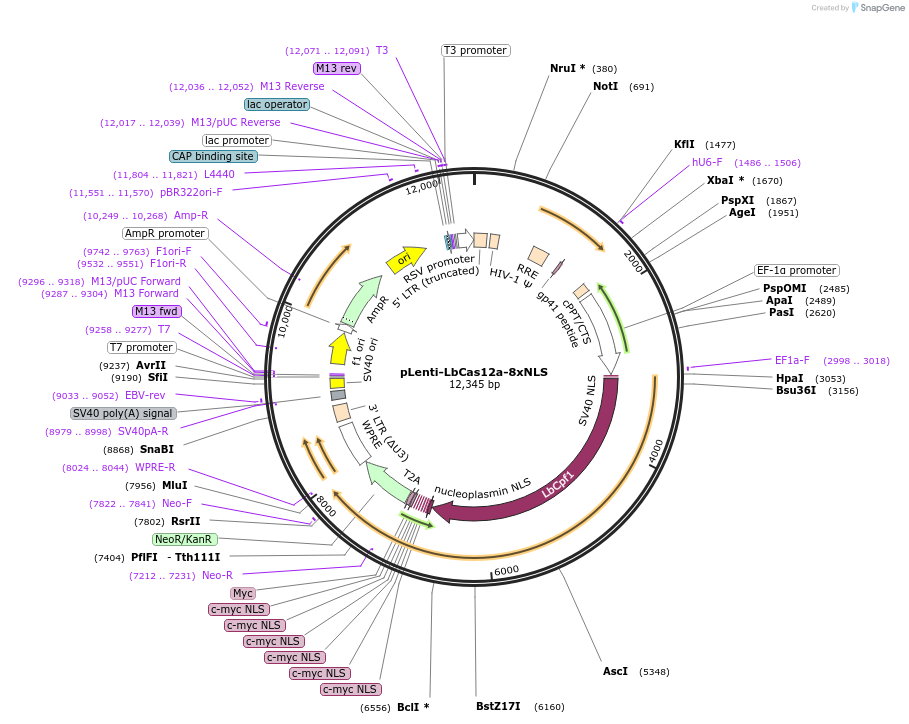
pLenti-LbCas12a-8xNLS
Plasmid#209020PurposeLentiviral vector expressing human codon-optimized LbCas12a with 8xNLSDepositorInsertLbCas12a-8xNLS-Myc-NeoR
UseCRISPR and LentiviralTagsMYCExpressionMammalianPromoterEF-1aAvailable SinceJune 20, 2024AvailabilityAcademic Institutions and Nonprofits only -
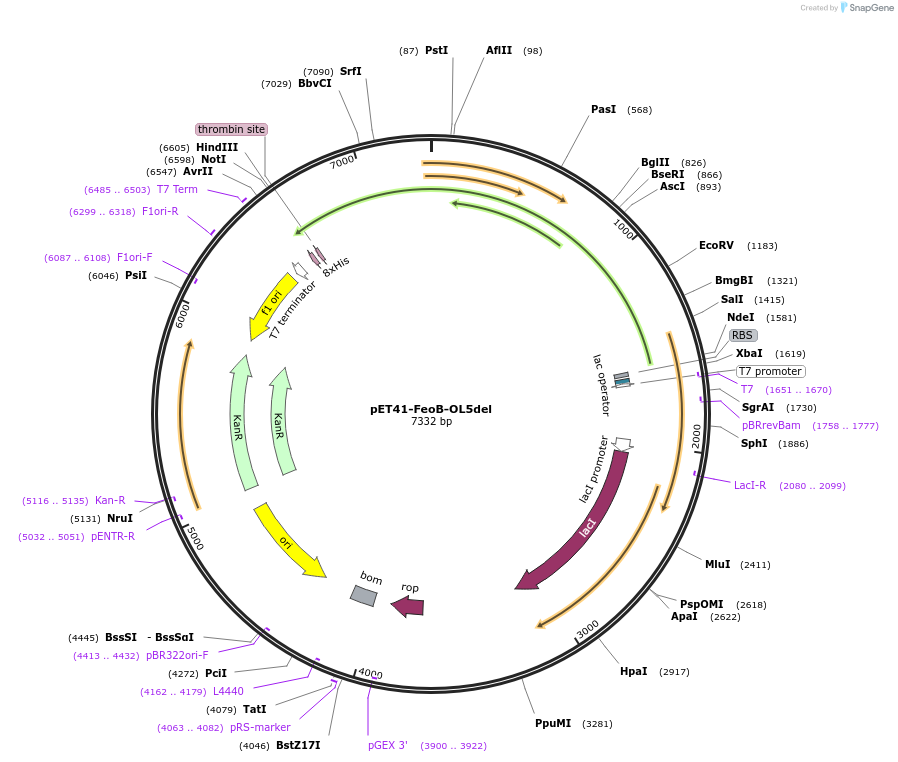
pET41-FeoB-OL5del
Plasmid#216745PurposeStudy the interaction of FeoB in P. aeruginosa, specifically focusing on the role of the outer loop 5 (OL5) by examining the variant lacking OL5 in its interaction with exogenous peptides.DepositorInsertfeoB (PA4358 P. aeruginosa (Bacteria))
Tags8x HisExpressionBacterialMutationOuter loop 5 (OL5) with the sequence of [ATFAA] i…Available SinceMay 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
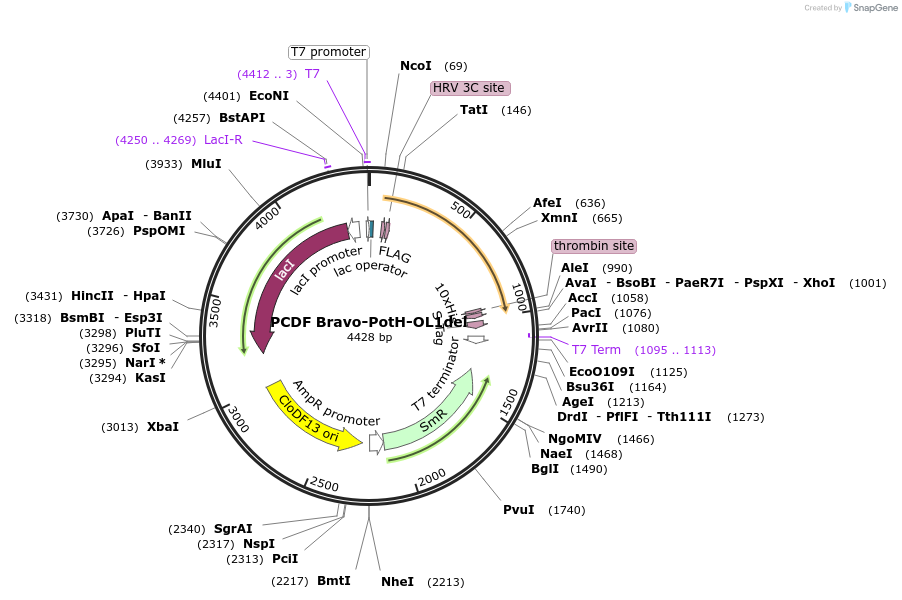
PCDF Bravo-PotH-OL1del
Plasmid#216749PurposeStudy the interaction of PotH in E. coli, specifically focusing on the role of the outer loop 1 (OL1) by examining the variant lacking OL1 in its interaction with exogenous peptides.DepositorInsertpotH (potH E. coli (Bacteria))
Tags10x HisExpressionBacterialMutationOuter loop 1 (OL1) with the sequence of [FKISLAEM…Available SinceMay 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
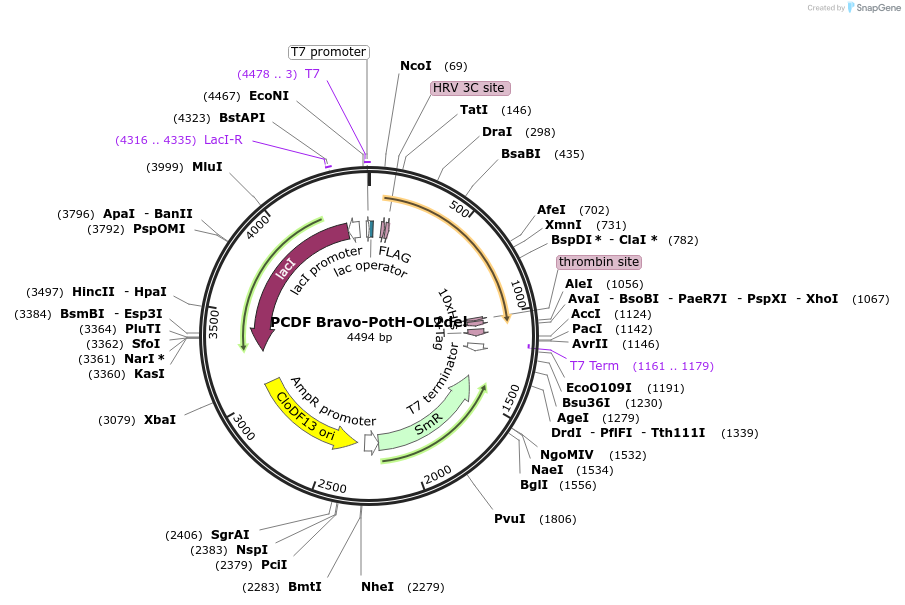
PCDF Bravo-PotH-OL2del
Plasmid#216750PurposeStudy the interaction of PotH in E. coli, specifically focusing on the role of the outer loop 2 (OL2) by examining the variant lacking OL1 in its interaction with exogenous peptides.DepositorInsertpotH (potH E. coli (Bacteria))
Tags10x HisExpressionBacterialMutationOuter loop 2 (OL2) with the sequence of [WMGILKNN…Available SinceMay 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
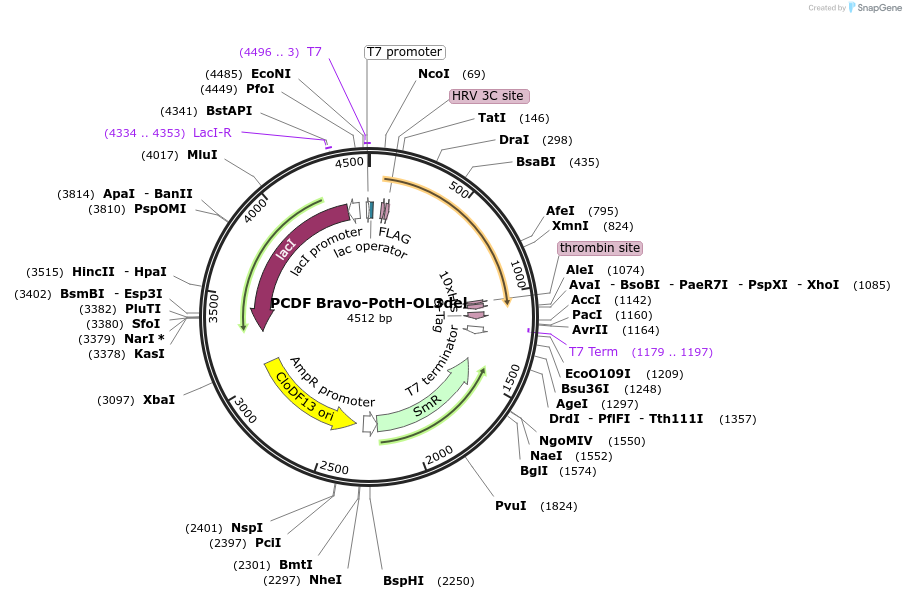
PCDF Bravo-PotH-OL3del
Plasmid#216751PurposeStudy the interaction of PotH in E. coli, specifically focusing on the role of the outer loop 3 (OL3) by examining the variant lacking OL1 in its interaction with exogenous peptides.DepositorInsertpotH (potH E. coli (Bacteria))
Tags10x HisExpressionBacterialMutationOuter loop 3 (OL3) with the sequence of [ELLGGPDS…Available SinceMay 23, 2024AvailabilityAcademic Institutions and Nonprofits only