We narrowed to 5,940 results for: Chia
-
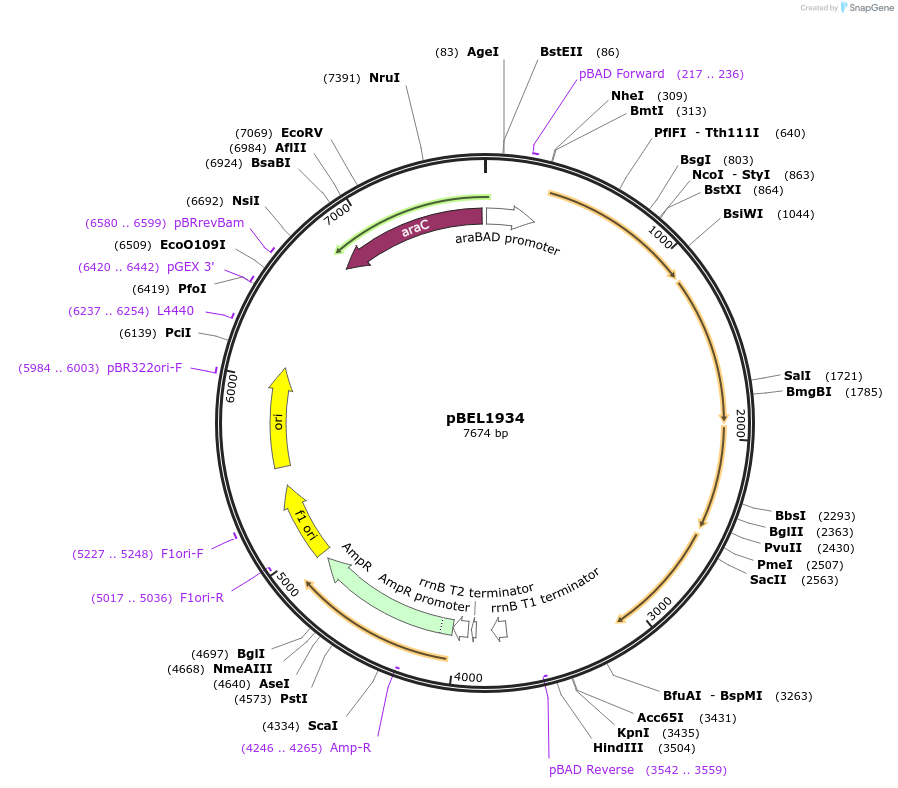
Plasmid#195696PurposeComplementation of MlaF-MlaE-MlaD(L123A)-MlaC-MlaBDepositorInsertMlaFEDCB
ExpressionBacterialMutationMlaF-MlaE-MlaD(L123A)-MlaC-MlaBAvailable SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
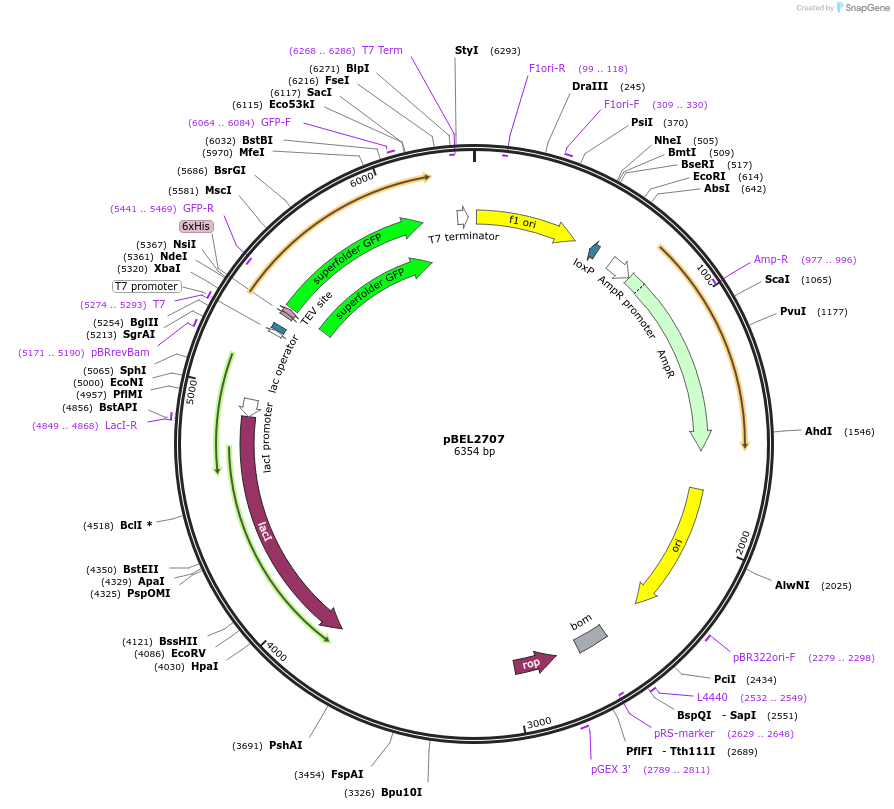
pBEL2707
Plasmid#195636PurposeExpression/purification of 6xHis-TEV-sfGFP-MlaA (227-251)DepositorInsertsfGFP-MlaA
ExpressionBacterialMutation6xHis-TEV-sfGFP-MlaA (227-251)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
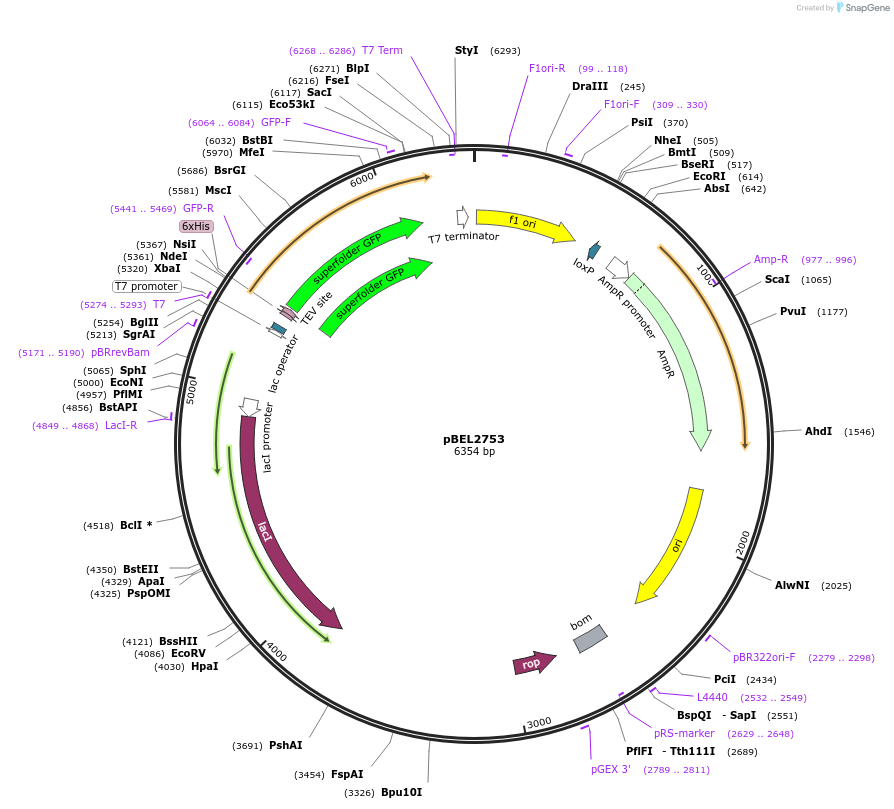
pBEL2753
Plasmid#195637PurposeExpression/purification of 6xHis-TEV-sfGFP-MlaA (227-251; I241N)DepositorInsertsfGFP-MlaA
ExpressionBacterialMutation6xHis-TEV-sfGFP-MlaA (227-251; I241N)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
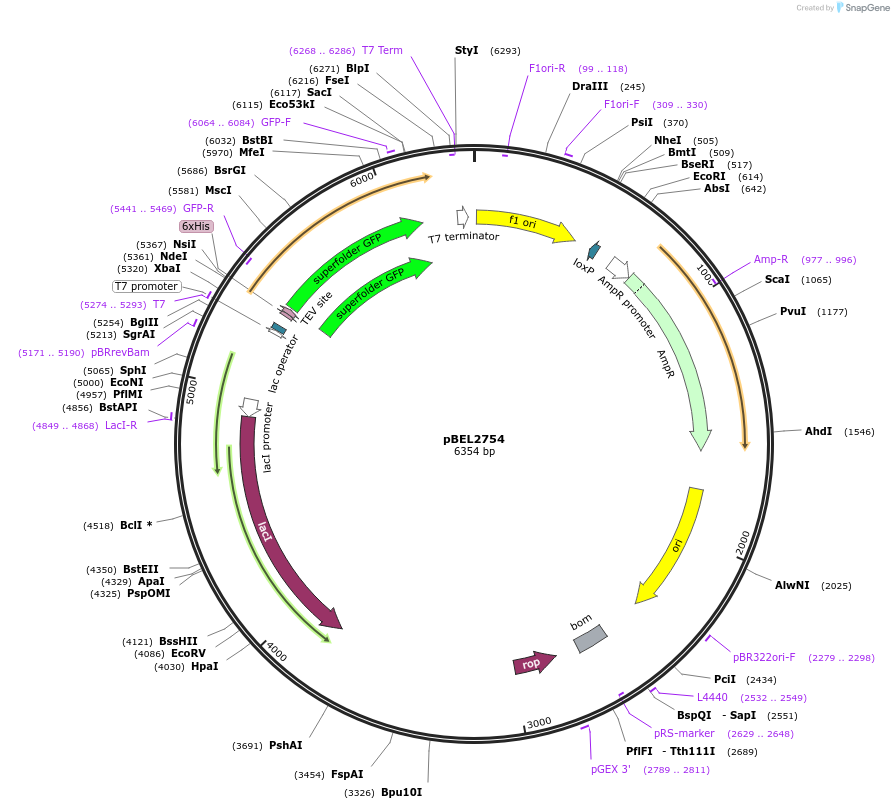
pBEL2754
Plasmid#195638PurposeExpression/purification of 6xHis-TEV-sfGFP-MlaA (227-251; L245N)DepositorInsertsfGFP-MlaA
ExpressionBacterialMutation6xHis-TEV-sfGFP-MlaA (227-251; L245N)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
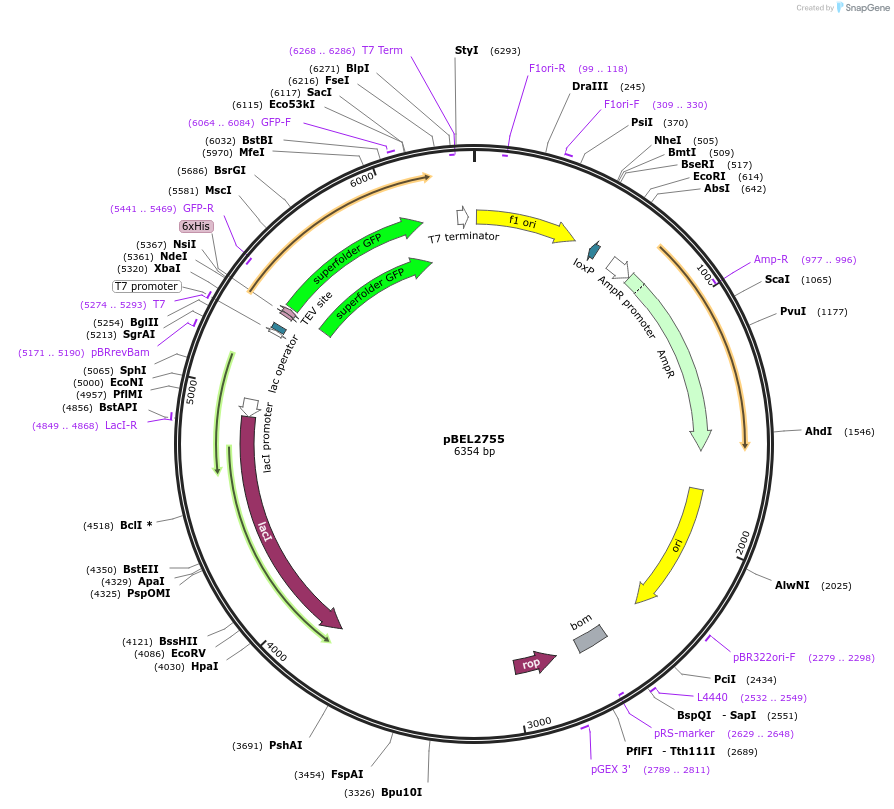
pBEL2755
Plasmid#195639PurposeExpression/purification of 6xHis-TEV-sfGFP-MlaA (227-251; I248N)DepositorInsertsfGFP-MlaA
ExpressionBacterialMutation6xHis-TEV-sfGFP-MlaA (227-251; I248N)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
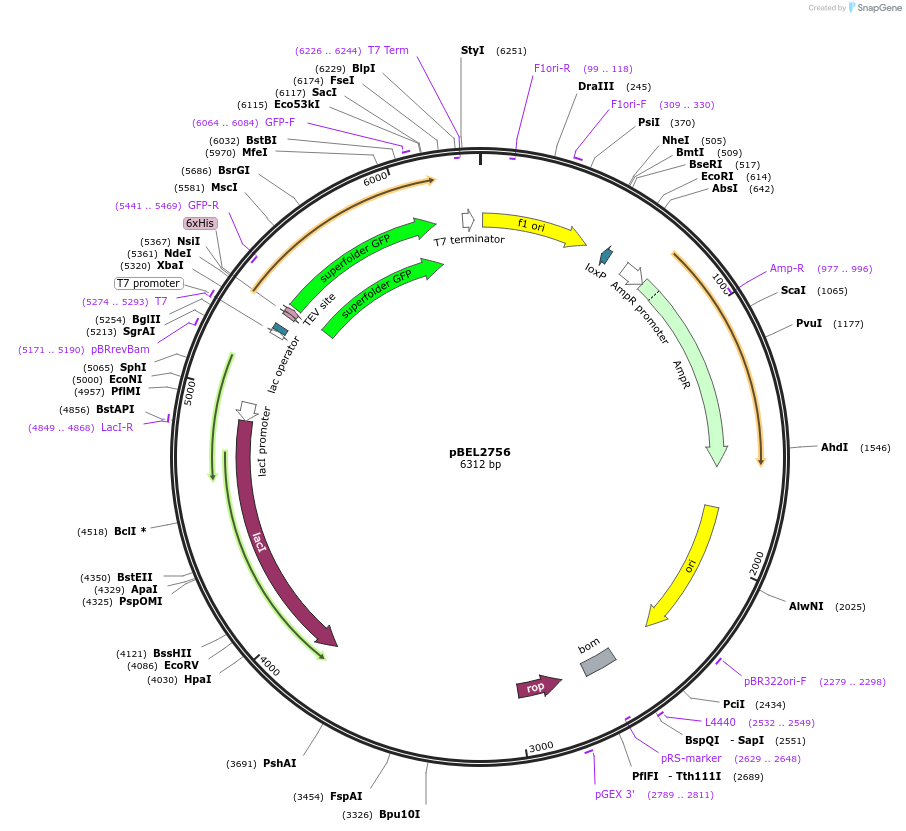
pBEL2756
Plasmid#195640PurposeExpression/purification of 6xHis-TEV-sfGFP-MlaA(227-251; delta238-251)DepositorInsertsfGFP-MlaA
ExpressionBacterialMutation6xHis-TEV-sfGFP-MlaA(227-251; delta238-251)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
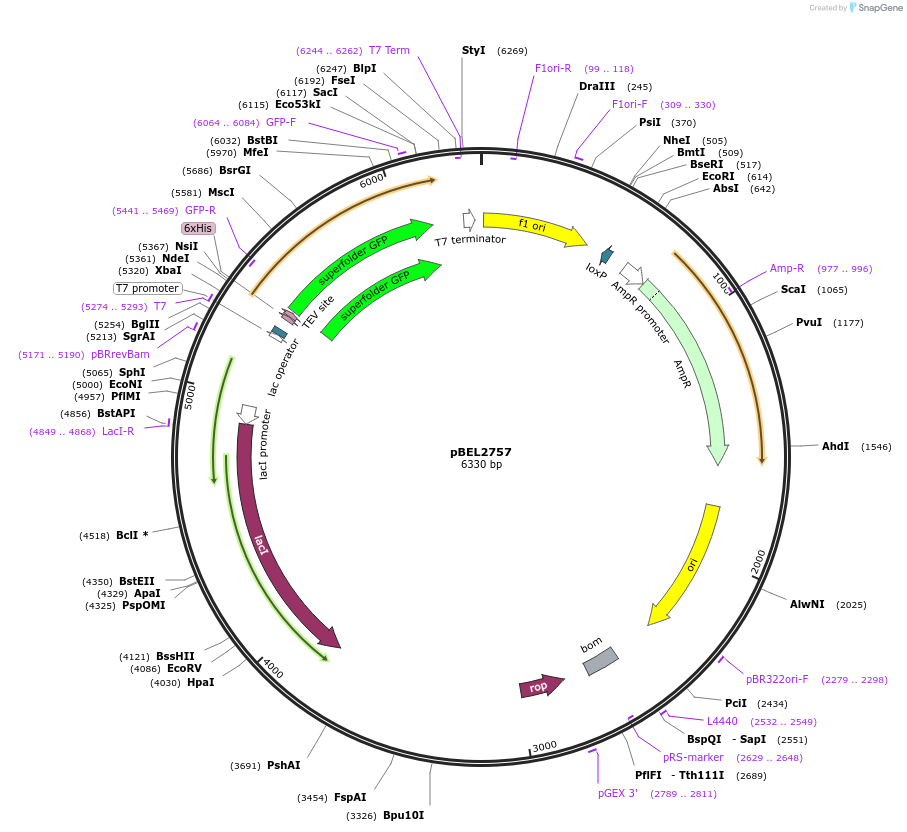
pBEL2757
Plasmid#195641PurposeExpression/purification of 6xHis-TEV-sfGFP-MlaA(227-251; delta244-251)DepositorInsertsfGFP-MlaA
ExpressionBacterialMutation6xHis-TEV-sfGFP-MlaA(227-251; delta244-251)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
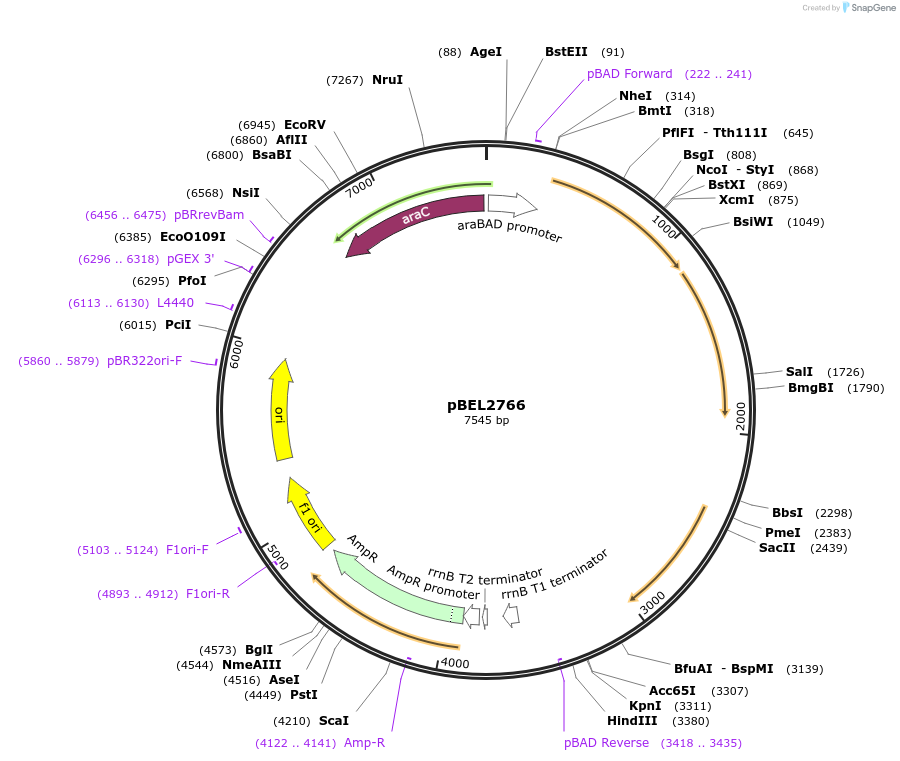
pBEL2766
Plasmid#196028PurposeComplementation of MlaF-MlaE-MlaDdelta141-183-MlaC-MlaBDepositorInsertMlaFEDCB
ExpressionBacterialMutationMlaF-MlaE-MlaDdelta141-183-MlaC-MlaBAvailable SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
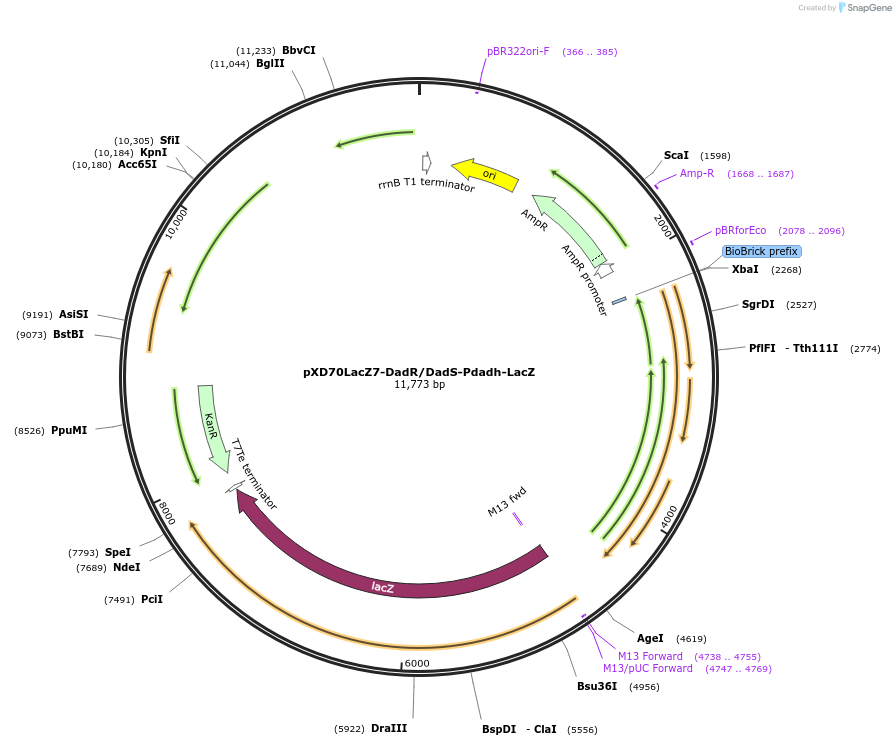
pXD70LacZ7-DadR/DadS-Pdadh-LacZ
Plasmid#191622PurposeE. coli - Eggerthella lenta shuttle plasmid (KanR), Pdadh dopamine dehydroxylase promoter-lacZ with transcriptional regulators DadR/DadSDepositorInsertlacZ
ExpressionBacterialAvailable SinceJan. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
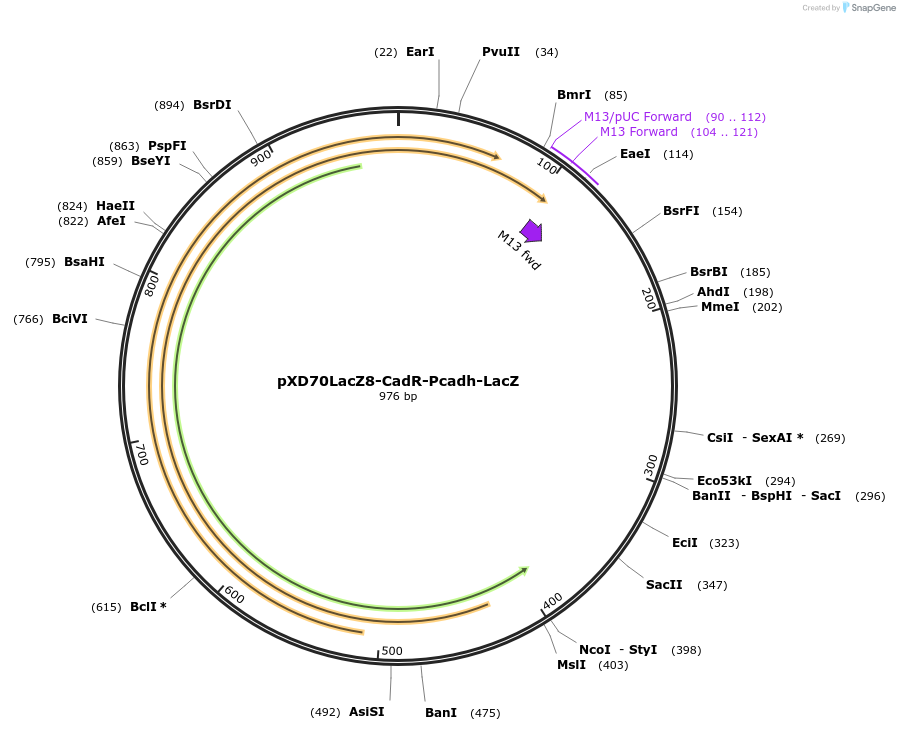
pXD70LacZ8-CadR-Pcadh-LacZ
Plasmid#191623PurposeE. coli - Eggerthella lenta shuttle plasmid (KanR), Pcadh catechin dehydroxylase promoter-lacZ with transcriptional regulator CadRDepositorInsertlacZ
ExpressionBacterialAvailable SinceJan. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
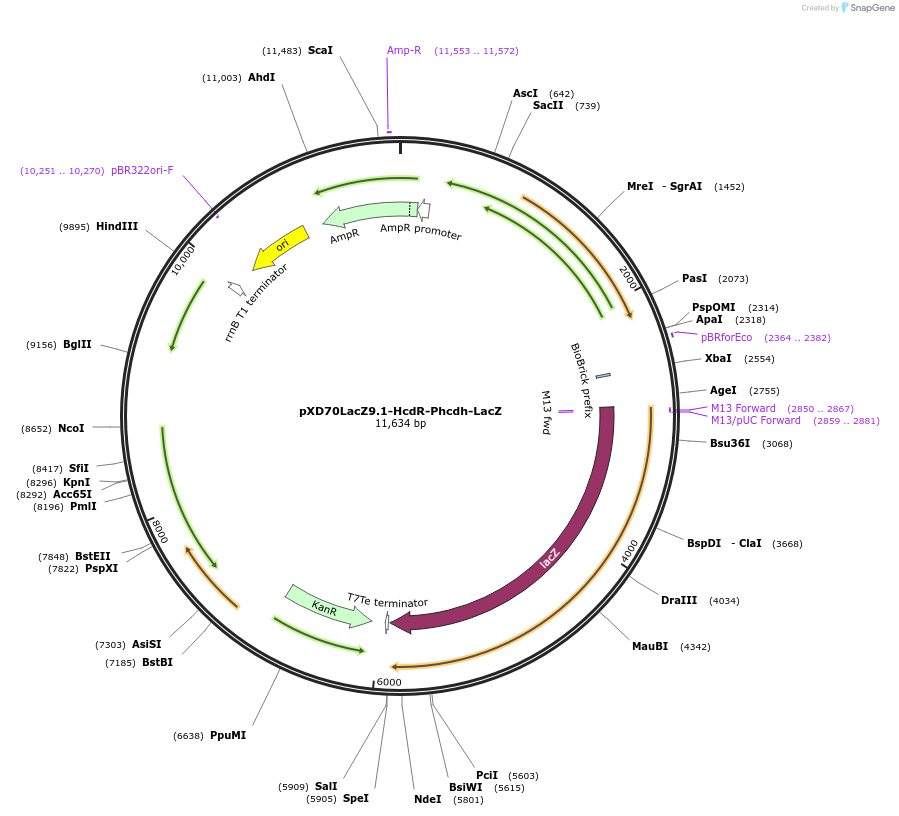
pXD70LacZ9.1-HcdR-Phcdh-LacZ
Plasmid#191629PurposeE. coli - Eggerthella lenta shuttle plasmid (KanR), Phcdh hydrocaffeic acid dehydroxylase promoter-lacZ with transcriptional regulator HcdRDepositorInsertlacZ
ExpressionBacterialAvailable SinceJan. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
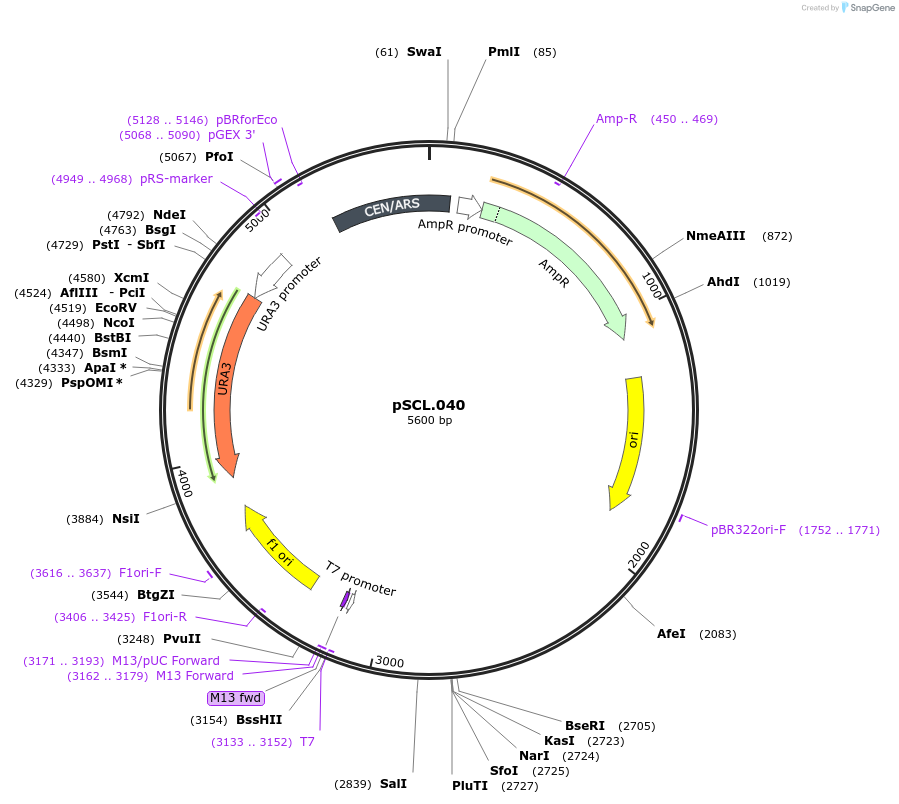
pSCL.040
Plasmid#184974PurposeTest effect of extending a1/a2 on ADE2 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, ADE2 P272X, a1/a2 length: 27 v2
ExpressionYeastMutationADE2 donor P272stop, a1/a2 length extended to 27 …PromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
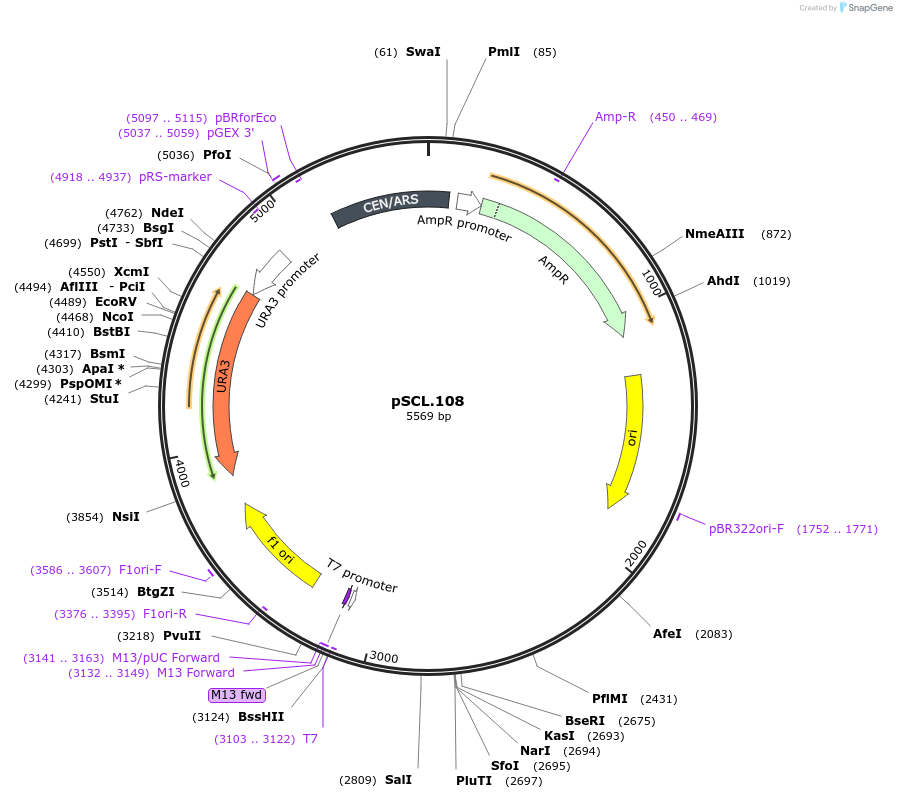
pSCL.108
Plasmid#184977PurposeExpress -Eco1 LYP1 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, LYP1 E27X, a1/a2 length: 12
ExpressionYeastMutationLYP1 donor E27stopPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
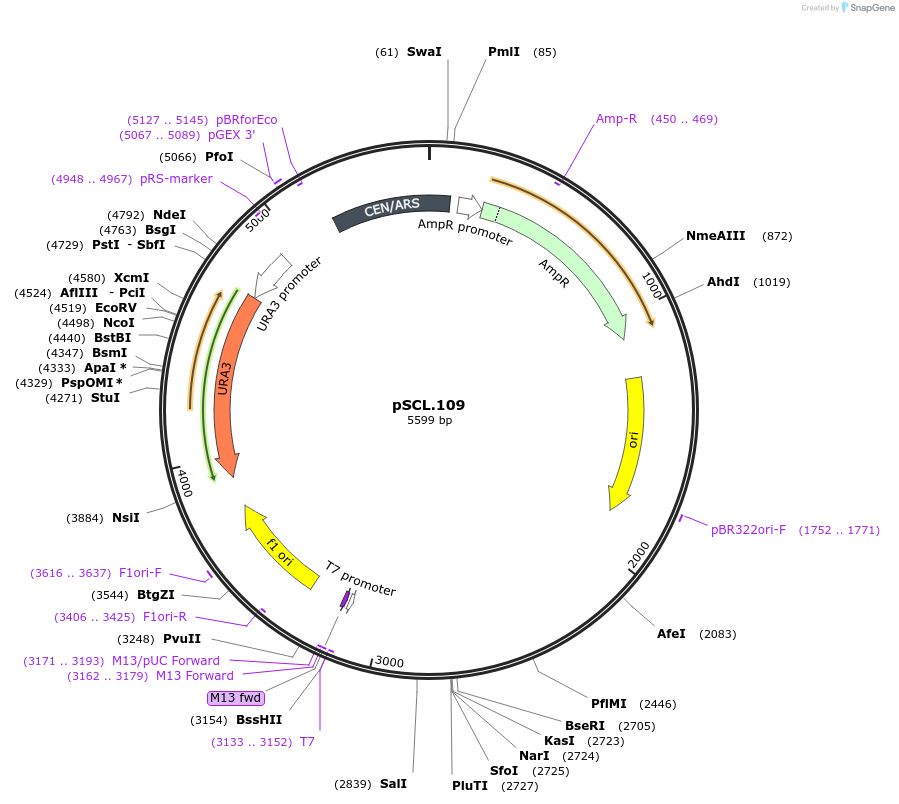
pSCL.109
Plasmid#184978PurposeTest effect of extending a1/a2 on LYP1 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, LYP1 E27X, a1/a2 length: 27 v1
ExpressionYeastMutationLYP1 donor E27stop, a1/a2 length extended to 27 bpPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
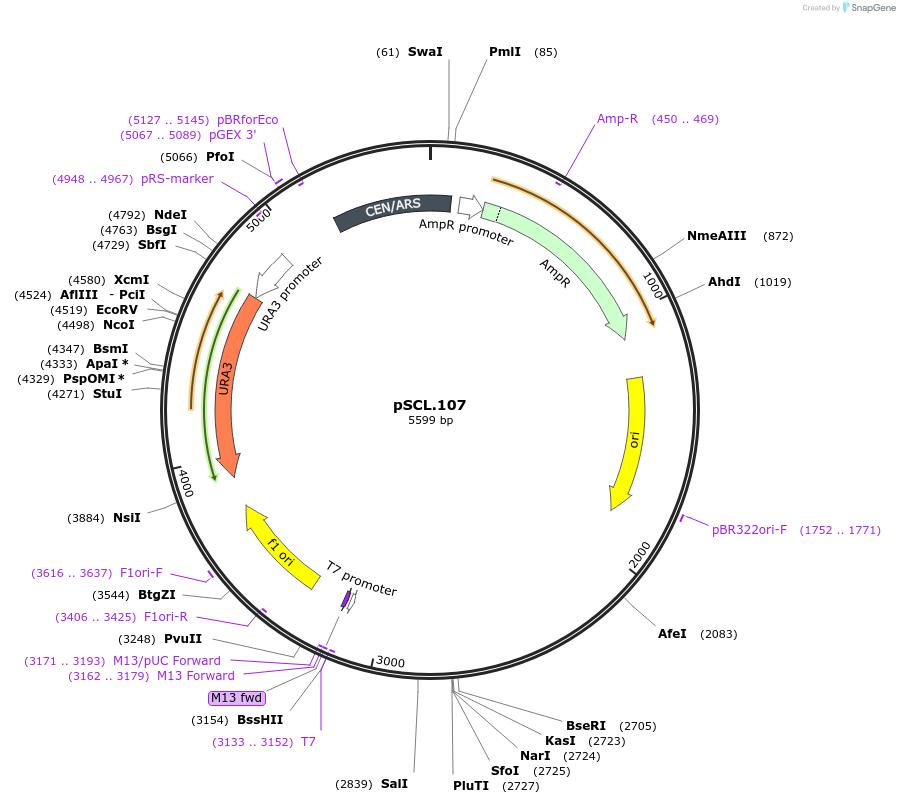
pSCL.107
Plasmid#184976PurposeTest effect of extending a1/a2 on CAN1 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, CAN1 G444X, a1/a2 length: 27 v1
ExpressionYeastMutationCAN1 donor G444stop, a1/a2 length extended to 27 …PromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
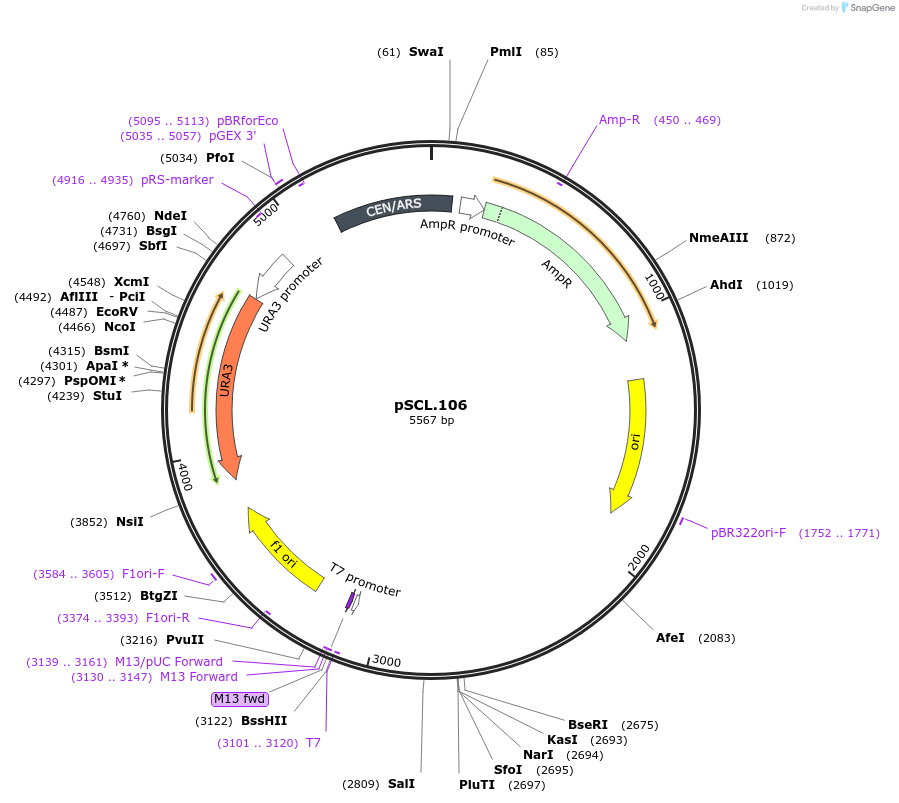
pSCL.106
Plasmid#184975PurposeExpress -Eco1 CAN1 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, CAN1 G444X, a1/a2 length: 12
ExpressionYeastMutationCAN1 donor G444stopPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
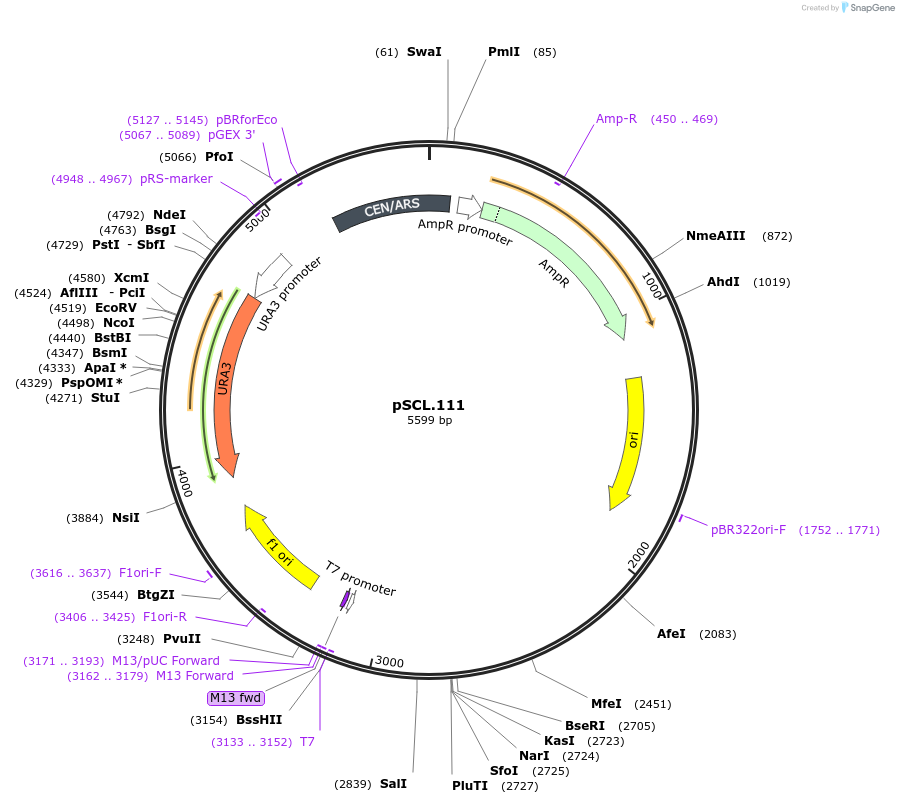
pSCL.111
Plasmid#184980PurposeTest effect of extending a1/a2 on TRP2 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, TRP2 E64X, a1/a2 length: 27 v1
ExpressionYeastMutationTRP2 donor E64stop, a1/a2 length extended to 27 bpPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
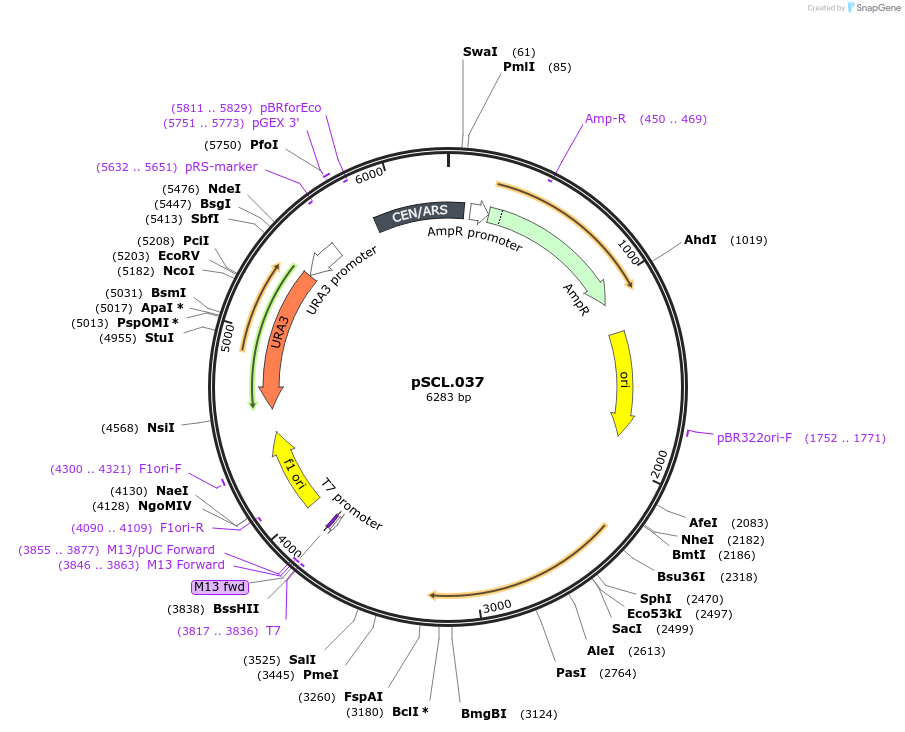
pSCL.037
Plasmid#184966PurposeExpress -Eco1 dead RT and ncRNA in yeasstDepositorInsertEco1: RT and ncRNA(wt), a1/a2 length: 12, dead RT
ExpressionYeastMutationHuman codon optimized RT, YXDD catalytic region m…PromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
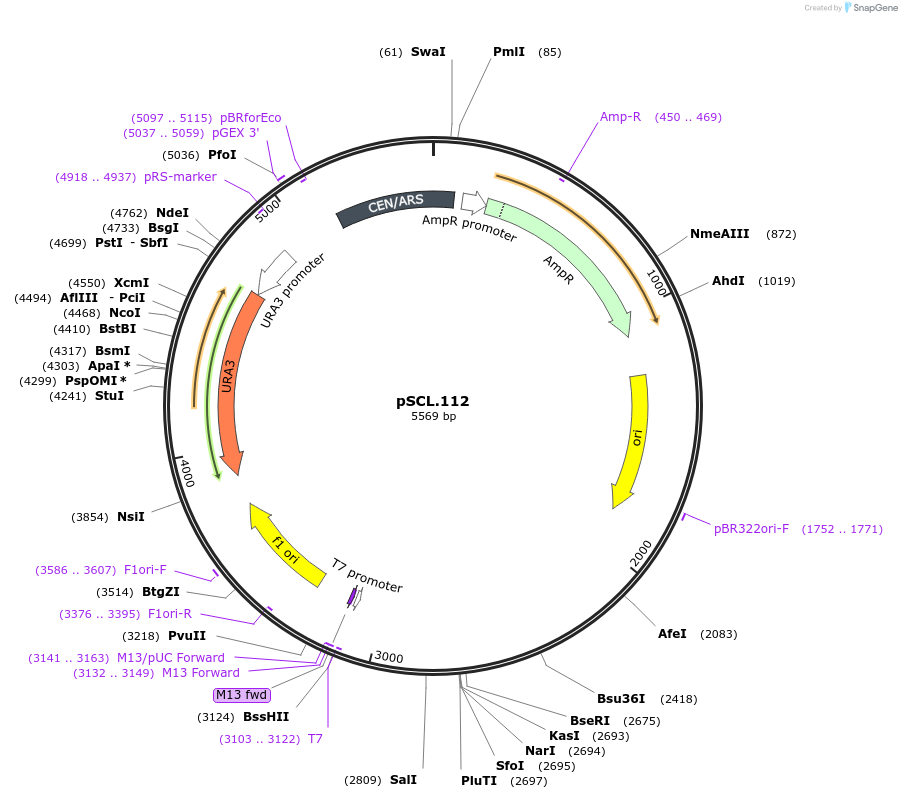
pSCL.112
Plasmid#184981PurposeExpress -Eco1 FAA1 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, FAA1 P233X, a1/a2 length: 12
ExpressionYeastMutationFAA1 donor P233stopPromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
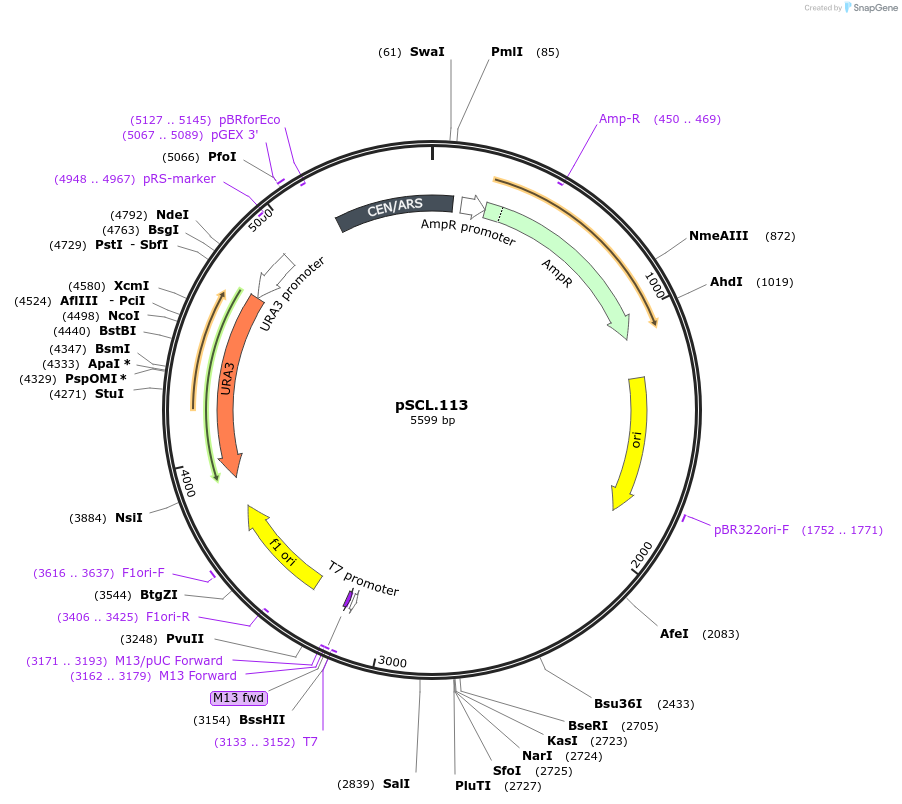
pSCL.113
Plasmid#184982PurposeTest effect of extending a1/a2 on FAA1 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, FAA1 P233X, a1/a2 length: 27 v1
ExpressionYeastMutationFAA1 donor P233stop, a1/a2 length extended to 27 …PromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only