We narrowed to 32,559 results for: STO;
-
Plasmid#66296PurposeExpression of G protein-coupled receptors for PRESTO-Tango: parallel receptorome expression and screening via transcriptional output, with transcriptional activation following arrestin translocationDepositorAvailable SinceSept. 15, 2015AvailabilityAcademic Institutions and Nonprofits only
-
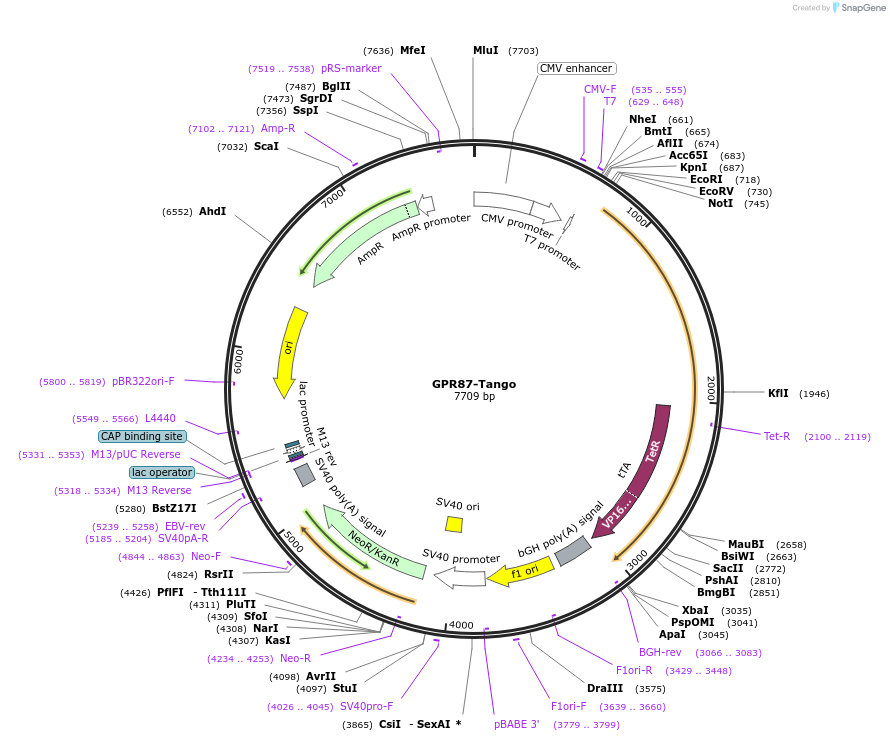
GPR87-Tango
Plasmid#66379PurposeExpression of G protein-coupled receptors for PRESTO-Tango: parallel receptorome expression and screening via transcriptional output, with transcriptional activation following arrestin translocationDepositorAvailable SinceSept. 15, 2015AvailabilityAcademic Institutions and Nonprofits only -
CHRM3-Tango
Plasmid#66250PurposeExpression of G protein-coupled receptors for PRESTO-Tango: parallel receptorome expression and screening via transcriptional output, with transcriptional activation following arrestin translocationDepositorAvailable SinceSept. 15, 2015AvailabilityAcademic Institutions and Nonprofits only -
BDKBR1-Tango
Plasmid#66229PurposeExpression of G protein-coupled receptors for PRESTO-Tango: parallel receptorome expression and screening via transcriptional output, with transcriptional activation following arrestin translocationDepositorAvailable SinceSept. 15, 2015AvailabilityAcademic Institutions and Nonprofits only -
HCTR1-Tango
Plasmid#66398PurposeExpression of G protein-coupled receptors for PRESTO-Tango: parallel receptorome expression and screening via transcriptional output, with transcriptional activation following arrestin translocationDepositorAvailable SinceSept. 15, 2015AvailabilityAcademic Institutions and Nonprofits only -
F2R-Tango
Plasmid#66276PurposeExpression of G protein-coupled receptors for PRESTO-Tango: parallel receptorome expression and screening via transcriptional output, with transcriptional activation following arrestin translocationDepositorAvailable SinceSept. 15, 2015AvailabilityAcademic Institutions and Nonprofits only -
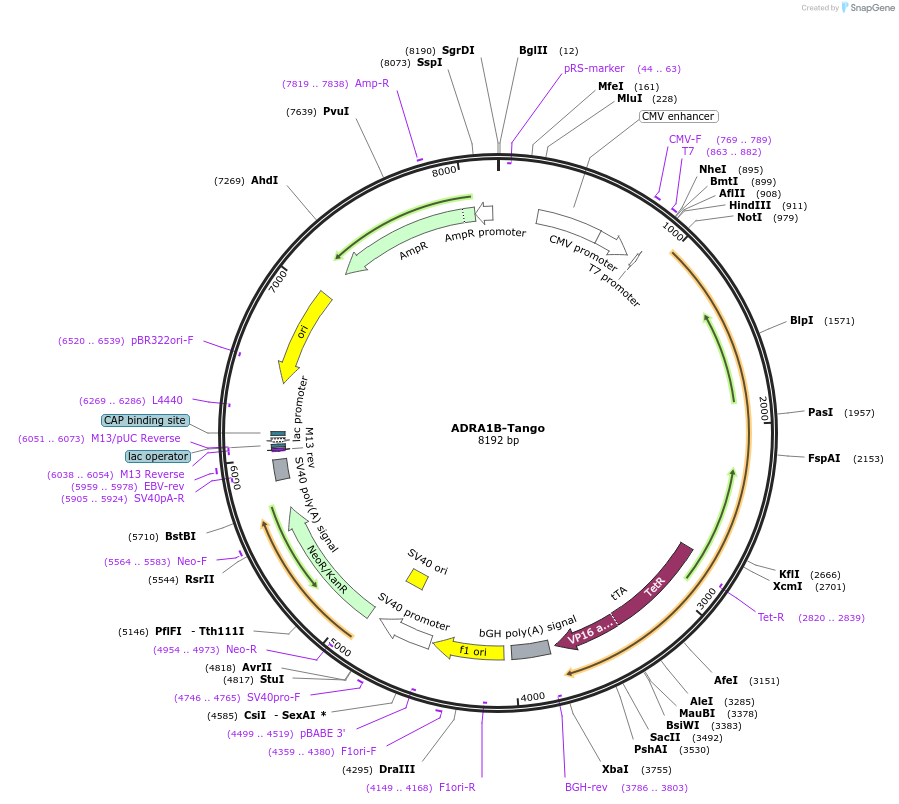
ADRA1B-Tango
Plasmid#66214PurposeExpression of G protein-coupled receptors for PRESTO-Tango: parallel receptorome expression and screening via transcriptional output, with transcriptional activation following arrestin translocationDepositorAvailable SinceSept. 15, 2015AvailabilityAcademic Institutions and Nonprofits only -
GPR37L1-Tango
Plasmid#66356PurposeExpression of G protein-coupled receptors for PRESTO-Tango: parallel receptorome expression and screening via transcriptional output, with transcriptional activation following arrestin translocationDepositorAvailable SinceSept. 15, 2015AvailabilityAcademic Institutions and Nonprofits only -
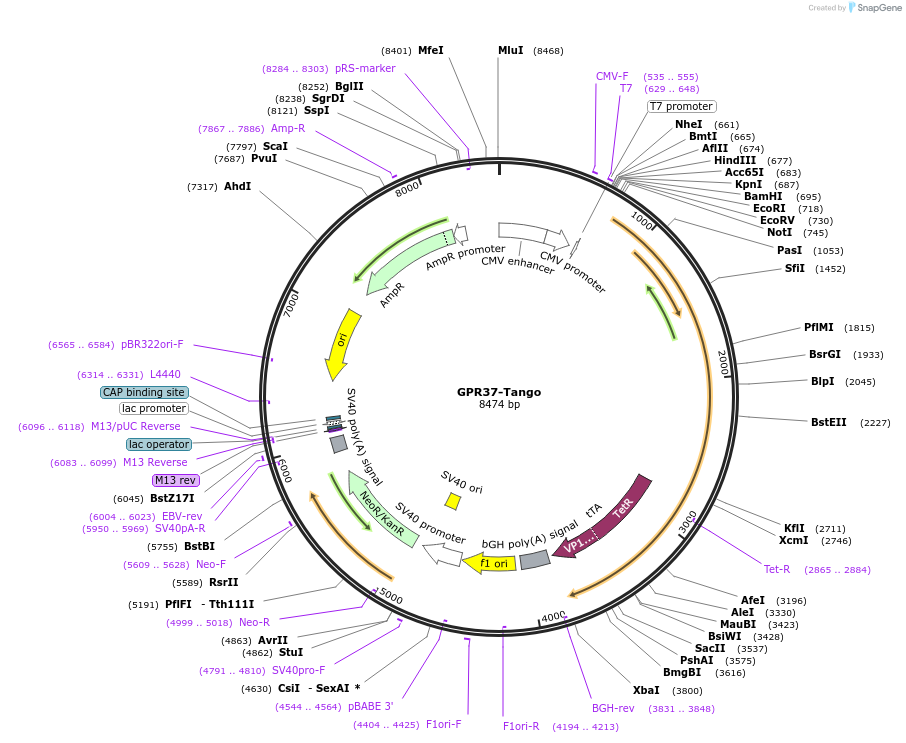
GPR37-Tango
Plasmid#66355PurposeExpression of G protein-coupled receptors for PRESTO-Tango: parallel receptorome expression and screening via transcriptional output, with transcriptional activation following arrestin translocationDepositorAvailable SinceSept. 17, 2015AvailabilityAcademic Institutions and Nonprofits only -
GPR119-Tango
Plasmid#66308PurposeExpression of G protein-coupled receptors for PRESTO-Tango: parallel receptorome expression and screening via transcriptional output, with transcriptional activation following arrestin translocationDepositorAvailable SinceSept. 15, 2015AvailabilityAcademic Institutions and Nonprofits only -
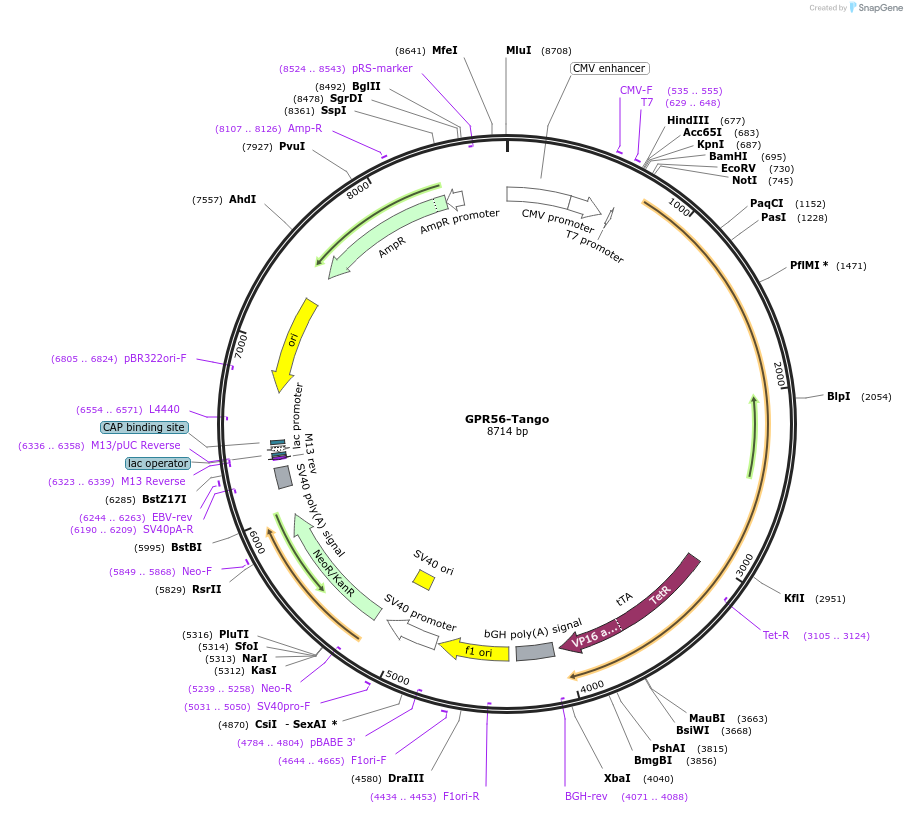
GPR56-Tango
Plasmid#66364PurposeExpression of G protein-coupled receptors for PRESTO-Tango: parallel receptorome expression and screening via transcriptional output, with transcriptional activation following arrestin translocationDepositorAvailable SinceSept. 15, 2015AvailabilityAcademic Institutions and Nonprofits only -
GPR125-Tango
Plasmid#66313PurposeExpression of G protein-coupled receptors for PRESTO-Tango: parallel receptorome expression and screening via transcriptional output, with transcriptional activation following arrestin translocationDepositorAvailable SinceSept. 15, 2015AvailabilityAcademic Institutions and Nonprofits only -
GPR55-Tango
Plasmid#66363PurposeExpression of G protein-coupled receptors for PRESTO-Tango: parallel receptorome expression and screening via transcriptional output, with transcriptional activation following arrestin translocationDepositorAvailable SinceSept. 15, 2015AvailabilityAcademic Institutions and Nonprofits only -
F2RL3-Tango
Plasmid#66279PurposeExpression of G protein-coupled receptors for PRESTO-Tango: parallel receptorome expression and screening via transcriptional output, with transcriptional activation following arrestin translocationDepositorAvailable SinceSept. 15, 2015AvailabilityAcademic Institutions and Nonprofits only -
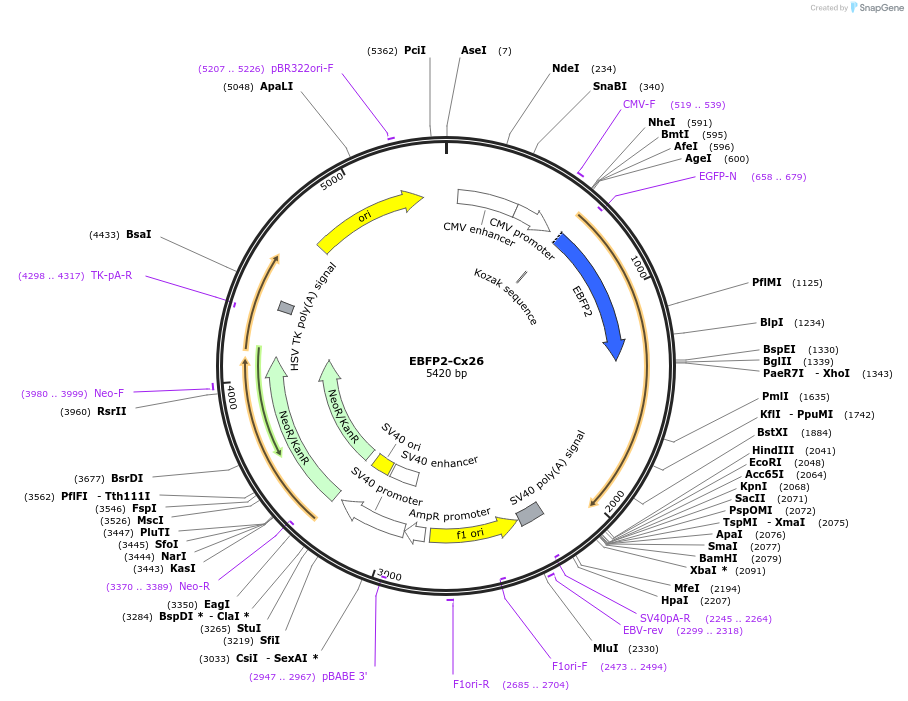
EBFP2-Cx26
Plasmid#69021PurposeExpresses rat Cx26 (Gjb2 CDS) with an 8 amino acid linker on the N-terminus of the Connexin linking to Enhanced Blue Fluorescent Protein 2 (FP fused to NT of connexin). CMV promoter.DepositorAvailable SinceOct. 20, 2015AvailabilityAcademic Institutions and Nonprofits only -
MAS1-Tango
Plasmid#66425PurposeExpression of G protein-coupled receptors for PRESTO-Tango: parallel receptorome expression and screening via transcriptional output, with transcriptional activation following arrestin translocationDepositorAvailable SinceSept. 15, 2015AvailabilityAcademic Institutions and Nonprofits only -
GPR171-Tango
Plasmid#66337PurposeExpression of G protein-coupled receptors for PRESTO-Tango: parallel receptorome expression and screening via transcriptional output, with transcriptional activation following arrestin translocationDepositorAvailable SinceSept. 15, 2015AvailabilityAcademic Institutions and Nonprofits only -
GPR84-Tango
Plasmid#66377PurposeExpression of G protein-coupled receptors for PRESTO-Tango: parallel receptorome expression and screening via transcriptional output, with transcriptional activation following arrestin translocationDepositorAvailable SinceSept. 15, 2015AvailabilityAcademic Institutions and Nonprofits only -
GHSR-Tango
Plasmid#66293PurposeExpression of G protein-coupled receptors for PRESTO-Tango: parallel receptorome expression and screening via transcriptional output, with transcriptional activation following arrestin translocationDepositorAvailable SinceSept. 15, 2015AvailabilityAcademic Institutions and Nonprofits only -
HTR1F-Tango
Plasmid#66408PurposeExpression of G protein-coupled receptors for PRESTO-Tango: parallel receptorome expression and screening via transcriptional output, with transcriptional activation following arrestin translocationDepositorAvailable SinceSept. 15, 2015AvailabilityAcademic Institutions and Nonprofits only