We narrowed to 15,933 results for: NOL
-
-
Zebrafish-gRNA-0009
Plasmid#42249PurposegRNA targeted to zebrafish gene tph1aDepositorAvailable SinceJan. 31, 2013AvailabilityAcademic Institutions and Nonprofits only -
-
-
TAL3072
Plasmid#41246DepositorInsertZebrafishCommunity-fan1-Left (si:ch211-201b11.2 Zebrafish)
Uset7Tags3X Flag and WT FOKIExpressionMammalianPromoterCMVAvailable SinceNov. 28, 2012AvailabilityAcademic Institutions and Nonprofits only -
TAL3073
Plasmid#41247DepositorInsertZebrafishCommunity-fan1-Right (si:ch211-201b11.2 Zebrafish)
Uset7Tags3X Flag and WT FOKIExpressionMammalianPromoterCMVAvailable SinceNov. 28, 2012AvailabilityAcademic Institutions and Nonprofits only -
-
PsiB1
Plasmid#21959DepositorAvailable SinceJan. 28, 2010AvailabilityAcademic Institutions and Nonprofits only -
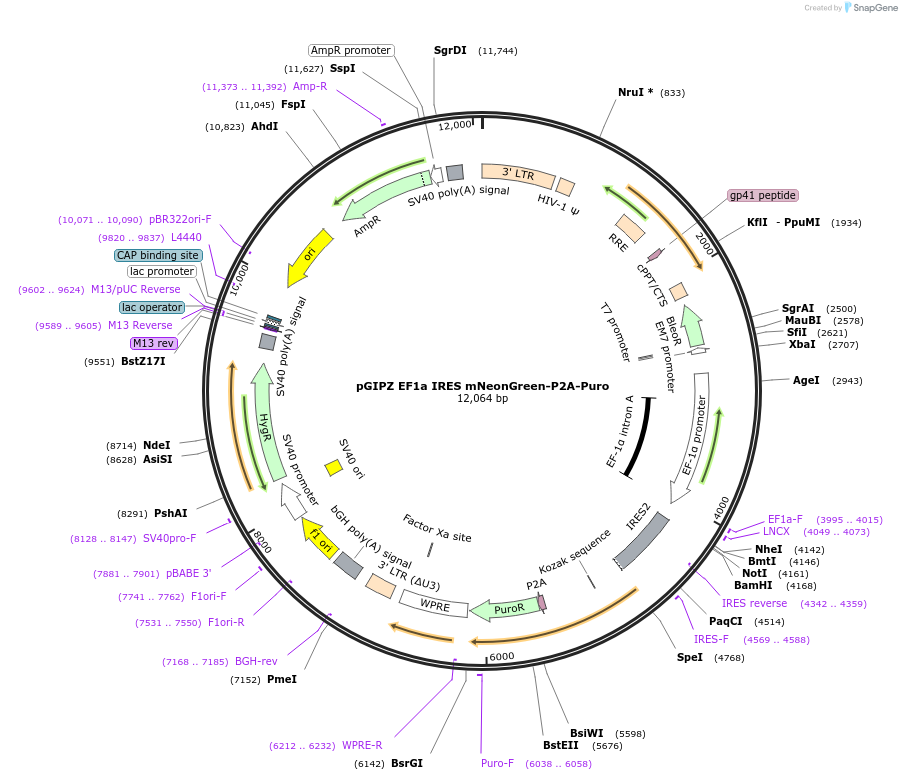
pGIPZ EF1a IRES mNeonGreen-P2A-Puro
Plasmid#248030PurposeBackbone that constitutively expresses mNeonGreen and a puromycin resistance gene in human cellsDepositorTypeEmpty backboneUseLentiviralExpressionMammalianAvailable SinceJan. 16, 2026AvailabilityAcademic Institutions and Nonprofits only -
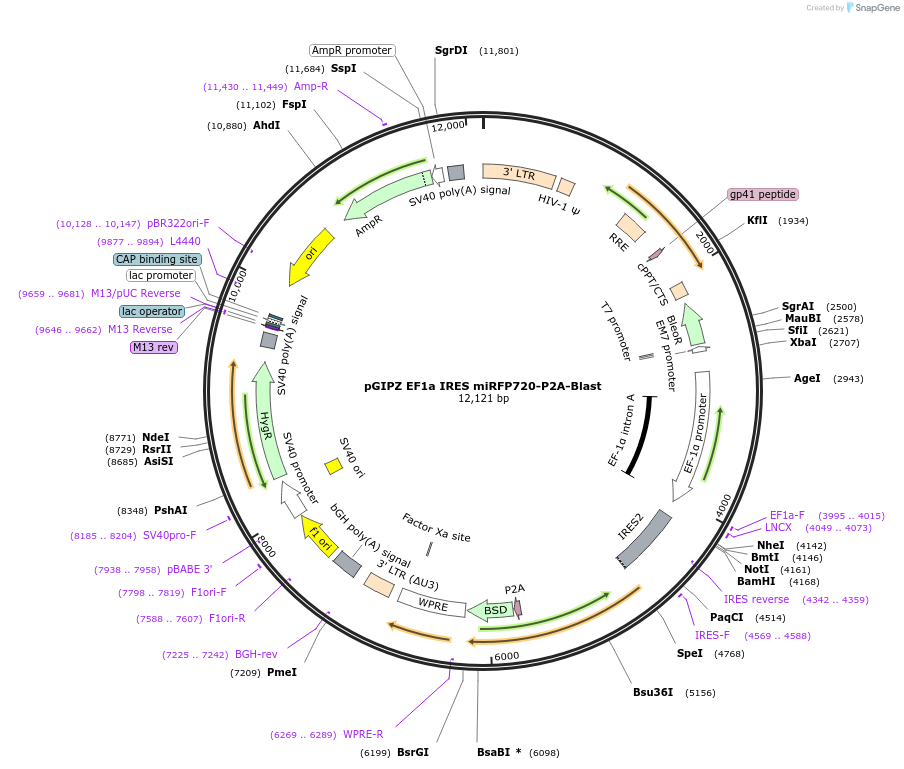
pGIPZ EF1a IRES miRFP720-P2A-Blast
Plasmid#248029PurposeBackbone that constitutively expresses miRFP720 and a blasticidin resistance gene in human cellsDepositorTypeEmpty backboneUseLentiviralExpressionMammalianAvailable SinceJan. 16, 2026AvailabilityAcademic Institutions and Nonprofits only -
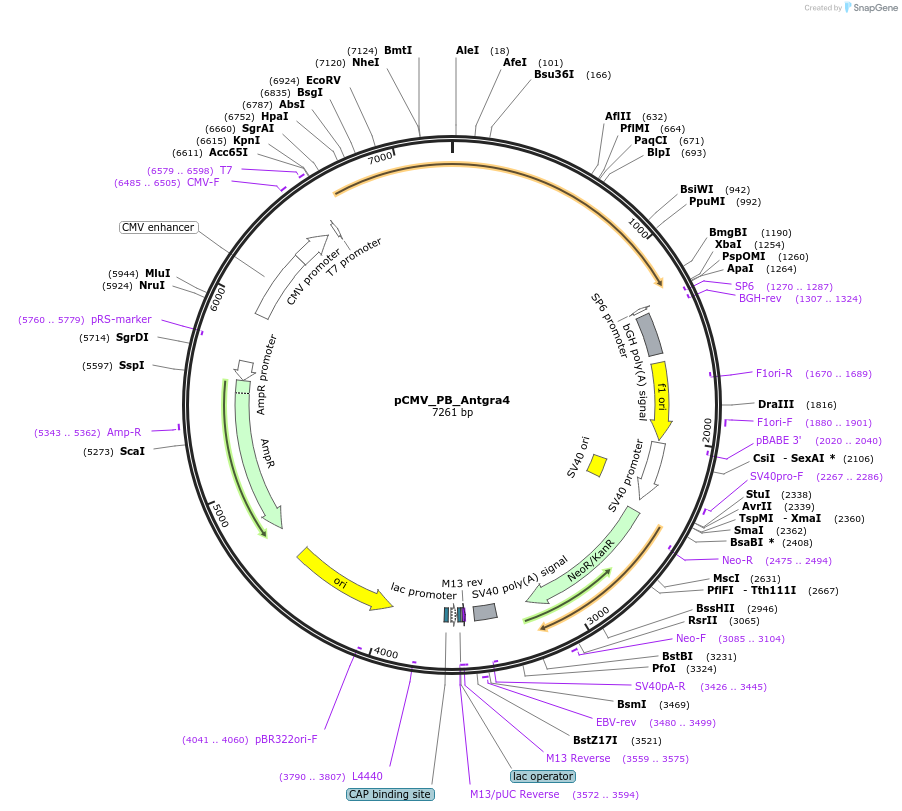
pCMV_PB_Antgra4
Plasmid#248199PurposeAntgra4 transposase expressing plasmidDepositorInsertAntgra4 hyPB
ExpressionMammalianAvailable SinceJan. 12, 2026AvailabilityAcademic Institutions and Nonprofits only -
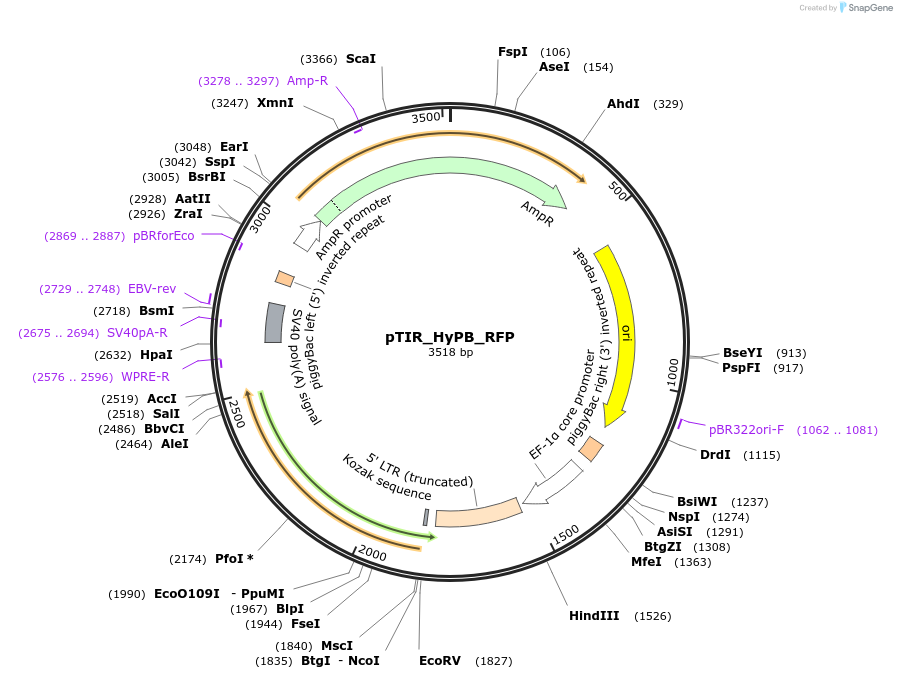
pTIR_HyPB_RFP
Plasmid#248201PurposeHyPB RFP cargo plasmidDepositorInsertRFP
ExpressionMammalianAvailable SinceJan. 12, 2026AvailabilityAcademic Institutions and Nonprofits only -
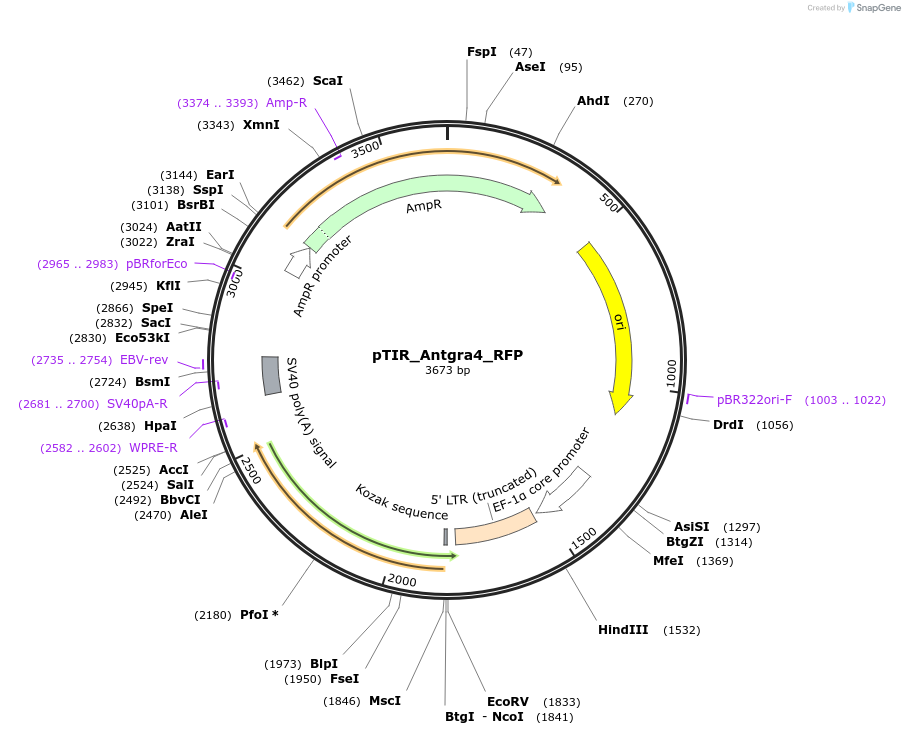
pTIR_Antgra4_RFP
Plasmid#248203PurposeAntgra4 RFP cargo plasmidDepositorInsertRFP
ExpressionMammalianAvailable SinceJan. 12, 2026AvailabilityAcademic Institutions and Nonprofits only -
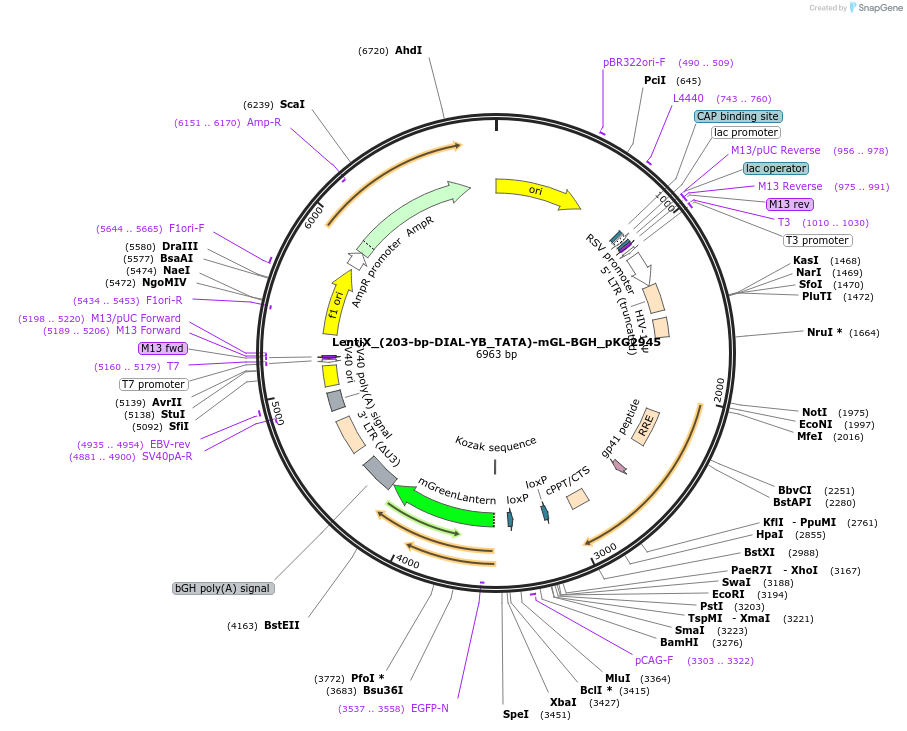
LentiX_(203-bp-DIAL-YB_TATA)-mGL-BGH_pKG2945
Plasmid#246342Purpose203-bp DIAL Reporter Lentivirus expressing mGreenLantern in the presence of ZFa and editable by Cre recombinaseDepositorInsertmGreenLantern
UseLentiviral and Synthetic BiologyAvailable SinceJan. 7, 2026AvailabilityAcademic Institutions and Nonprofits only -
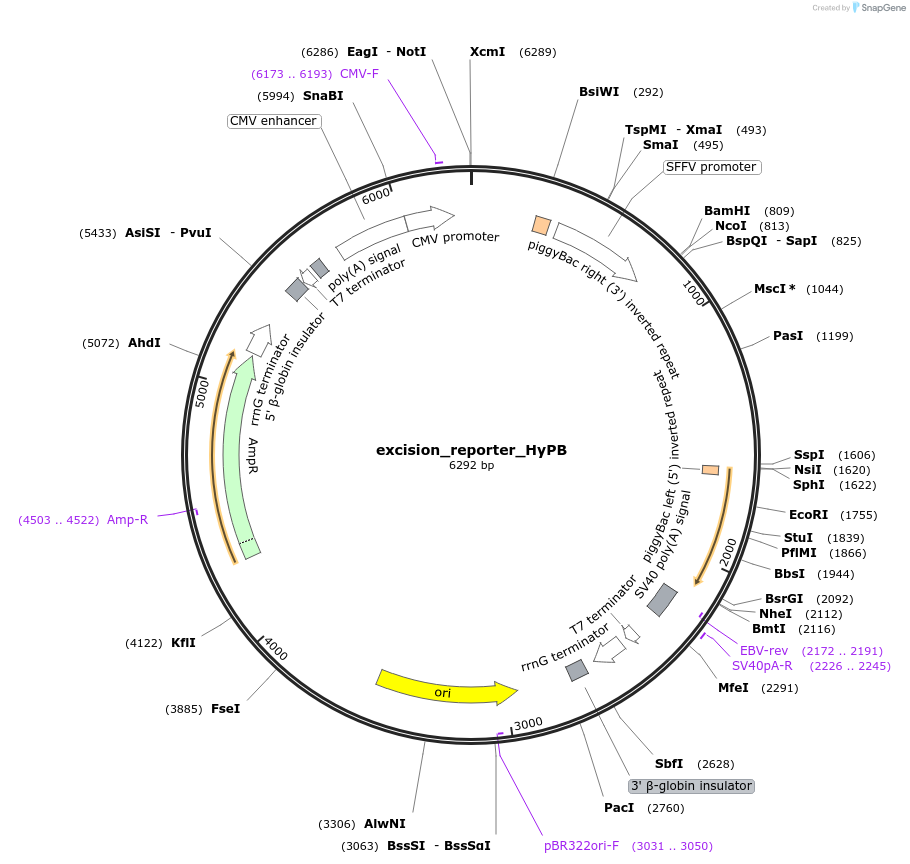
excision_reporter_HyPB
Plasmid#248200PurposeHyPB and synthetic transposase excision reporter and targeted integration reporterDepositorInsertExcision RFP reporter
ExpressionMammalianAvailable SinceJan. 7, 2026AvailabilityAcademic Institutions and Nonprofits only -
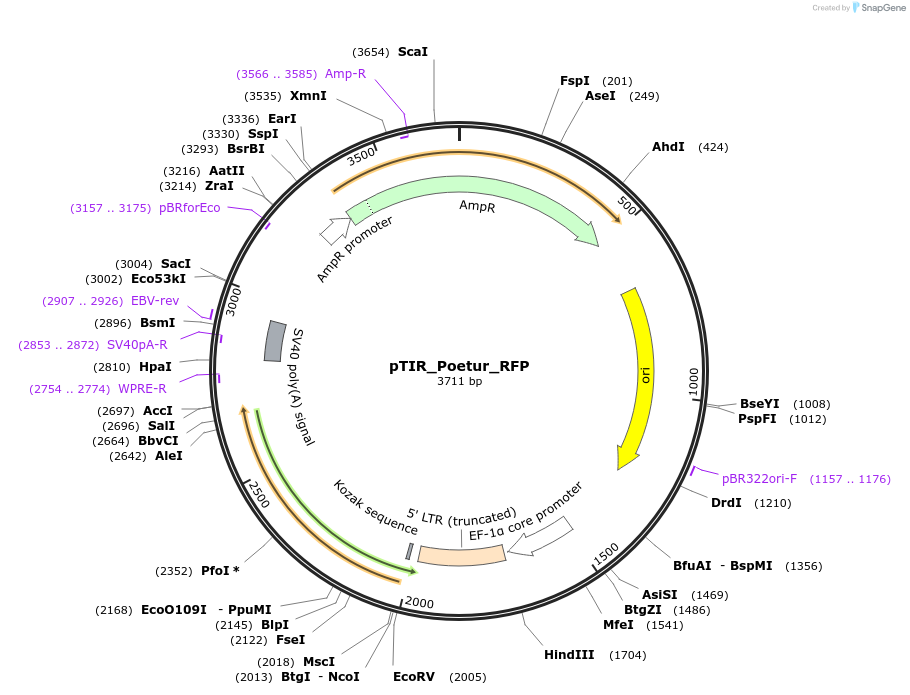
pTIR_Poetur_RFP
Plasmid#248202PurposePoetur RFP cargo plasmidDepositorInsertRFP
ExpressionMammalianAvailable SinceJan. 7, 2026AvailabilityAcademic Institutions and Nonprofits only -
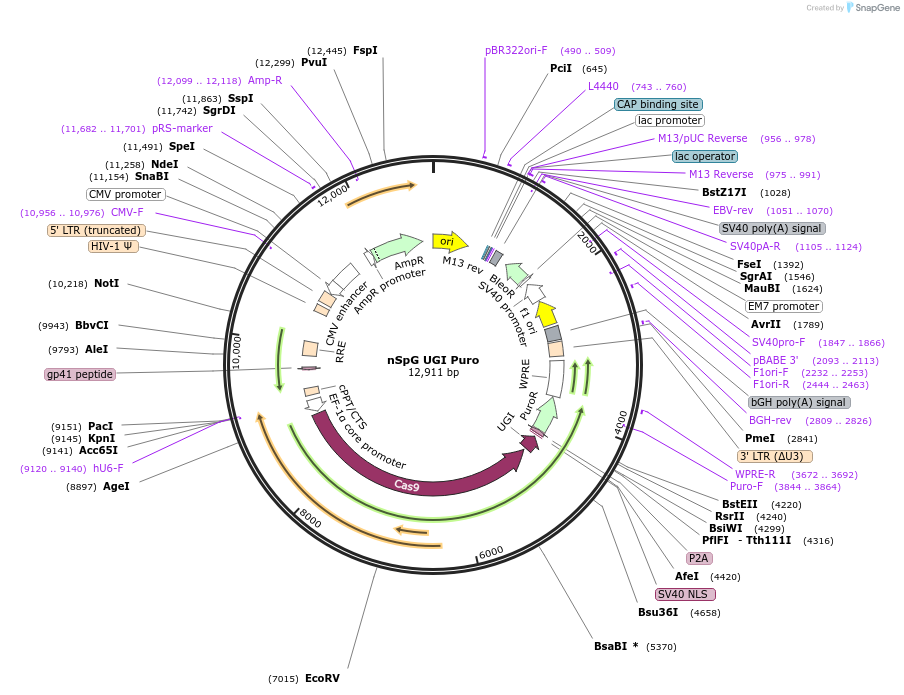
nSpG UGI Puro
Plasmid#235046PurposeControl nickase Cas9DepositorInsertnCas9
UseCRISPR and LentiviralTagsP2A-PuroRPromoterEF1a coreAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
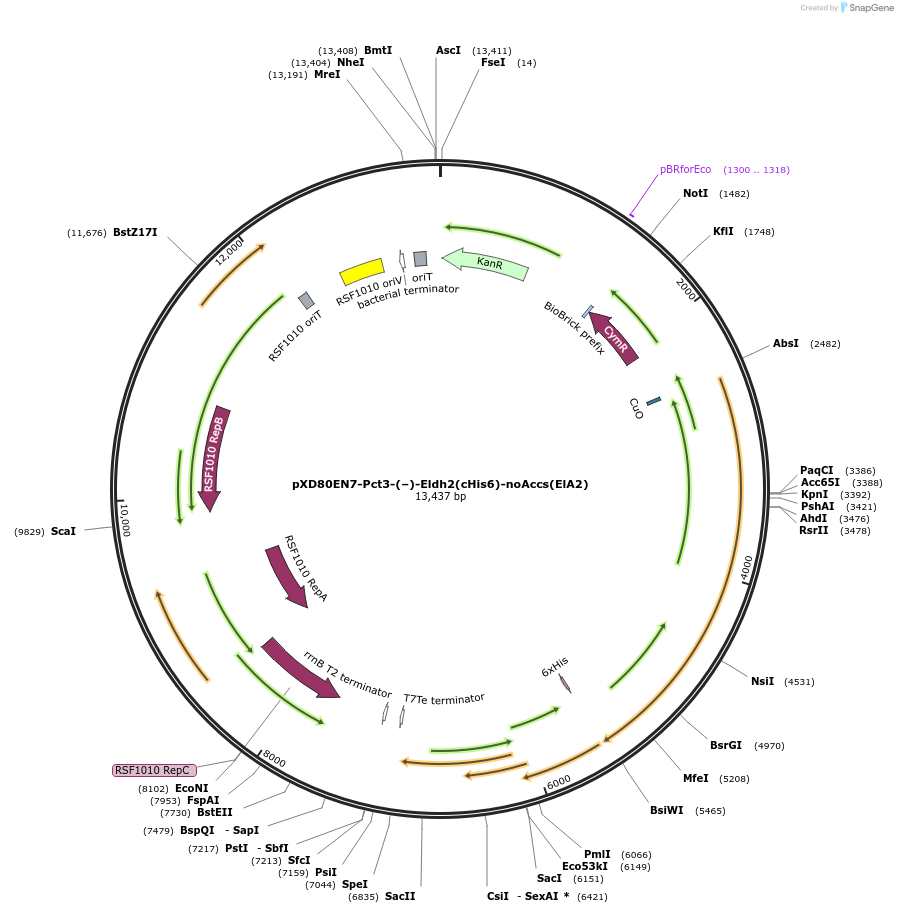
pXD80EN7-Pct3-(–)-Eldh2(cHis6)-noAccs(ElA2)
Plasmid#233878PurposeExpresses His-tagged Eggerthella lenta A2 (–)-Eldh2 with a cumate inducible promoter, without accessory genes (inactive), in Gordonibacter urolithinfaciens DSM 27213DepositorInsert(–)-Eldh2
TagsHisExpressionBacterialAvailable SinceDec. 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
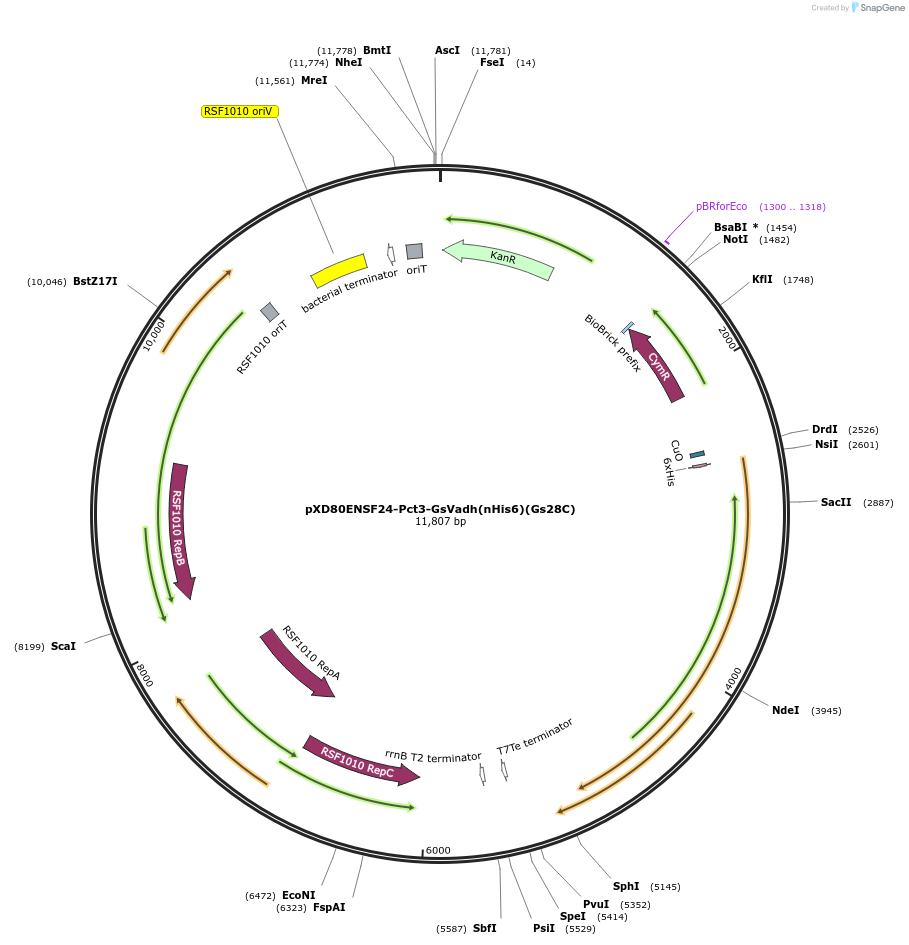
pXD80ENSF24-Pct3-GsVadh(nHis6)(Gs28C)
Plasmid#233872PurposeExpresses His-tagged Gordonibacter sp. 28C Gs Vadh with a cumate inducible promoter in Gordonibacter urolithinfaciens DSM 27213 for protein purificationDepositorInsertGs Vadh
TagsHisExpressionBacterialAvailable SinceDec. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
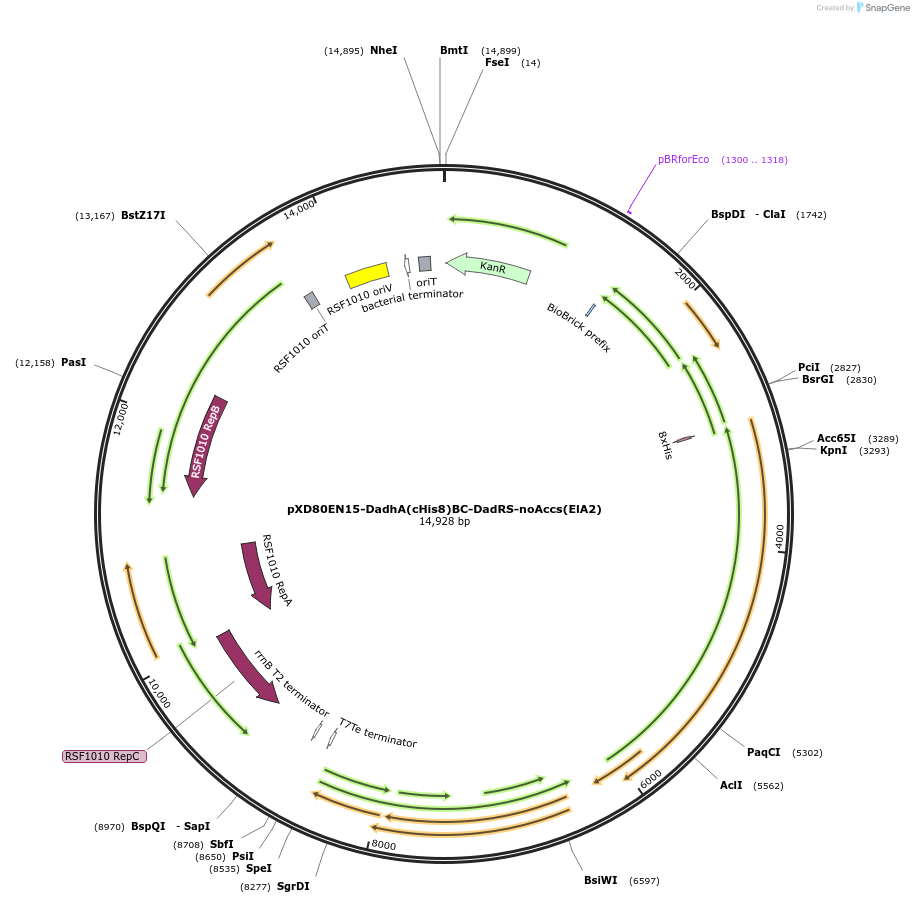
pXD80EN15-DadhA(cHis8)BC-DadRS-noAccs(ElA2)
Plasmid#225327PurposeExpresses Eggerthella lenta A2 Dadh without accessory genes (inactive) in Gordonibacter urolithinfaciens DSM 27213DepositorInsertDadh
TagsHis8ExpressionBacterialAvailable SinceDec. 2, 2025AvailabilityAcademic Institutions and Nonprofits only