We narrowed to 25,428 results for: Nes
-
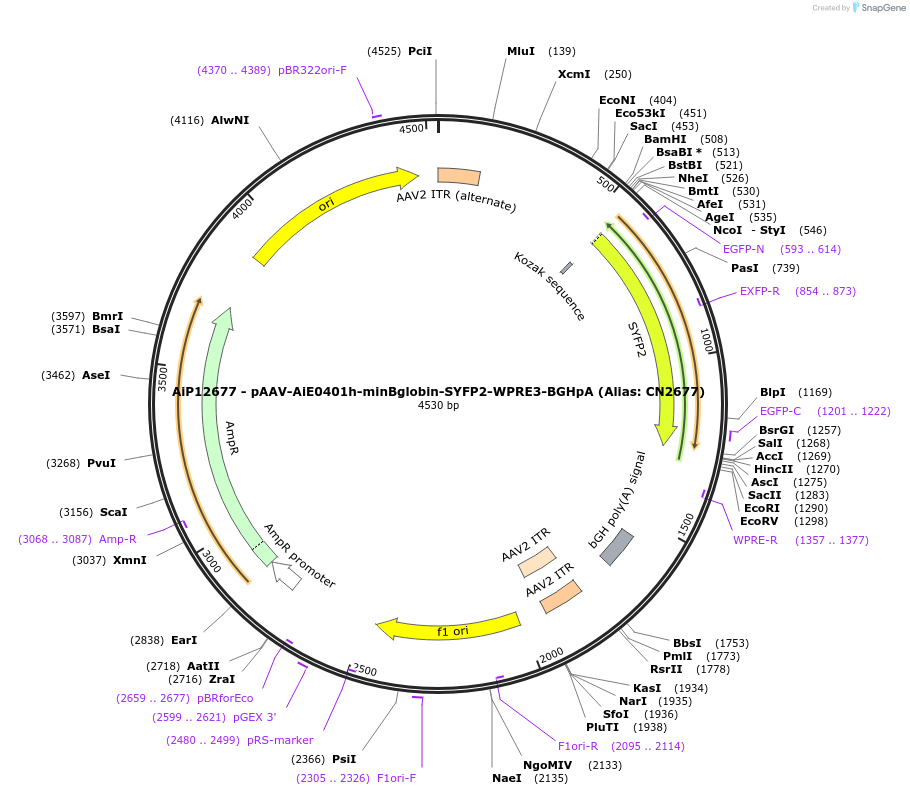
Plasmid#208153PurposeDirect-expressing SYFP2 in oligodendrocytes AAV VirusDepositorInsertSYFP2
UseAAVMutationNAPromoterBeta Globin minimal promoterAvailable SinceOct. 23, 2023AvailabilityAcademic Institutions and Nonprofits only -
AiP12106 - pAAV-AiE0400m-minBglobin-SYFP2-WPRE3-BGHpA (Alias: CN2106)
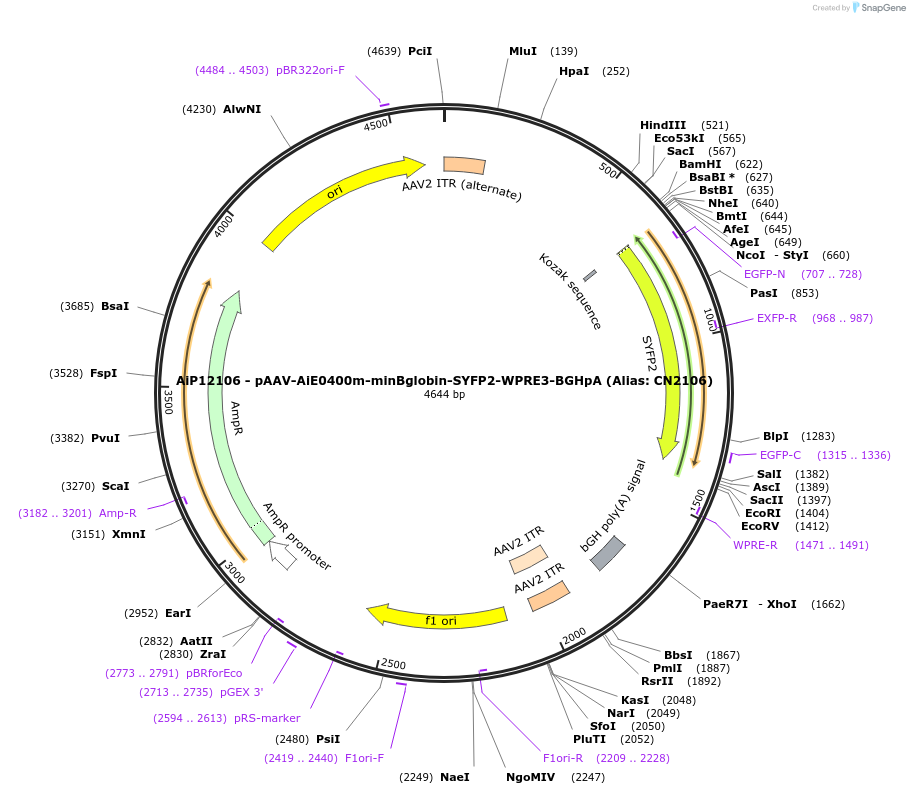
Plasmid#208152PurposeDirect-expressing SYFP2 in oligodendrocytes AAV VirusDepositorInsertSYFP2
UseAAVMutationNAPromoterBeta Globin minimal promoterAvailable SinceOct. 23, 2023AvailabilityAcademic Institutions and Nonprofits only -
AiP12162 - pAAV-AiE0400h-minBglobin-SYFP2-WPRE3-BGHpA (Alias: CN2162)
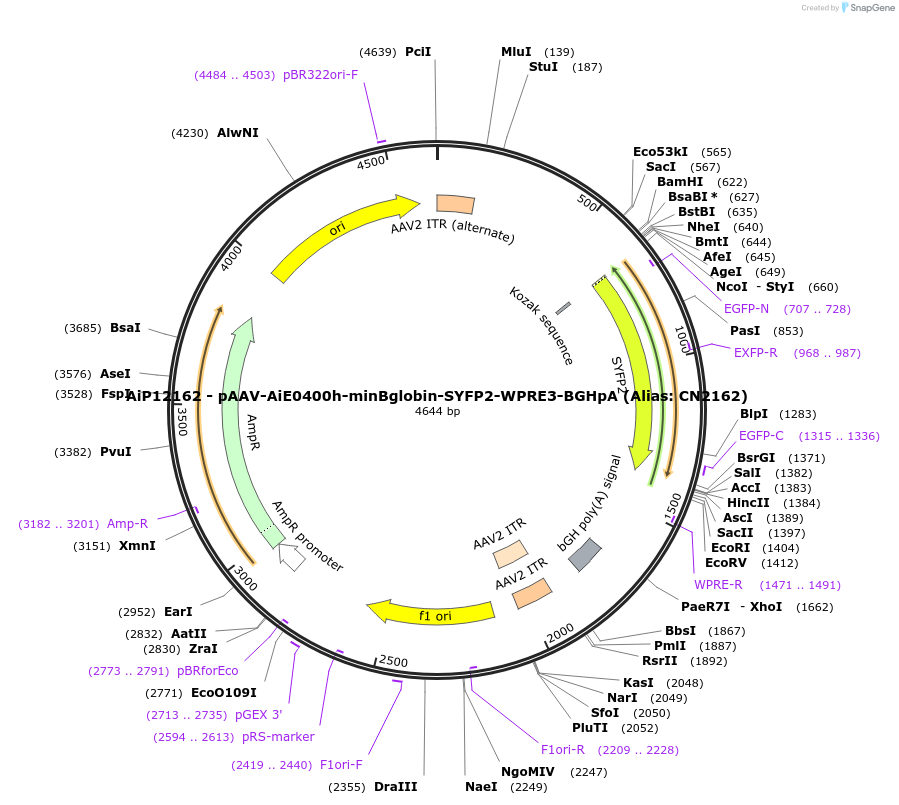
Plasmid#208151PurposeDirect-expressing SYFP2 in oligodendrocytes AAV VirusDepositorInsertSYFP2
UseAAVMutationNAPromoterBeta Globin minimal promoterAvailable SinceOct. 23, 2023AvailabilityAcademic Institutions and Nonprofits only -
AiP12099 - pAAV-AiE0396m-minBglobin-SYFP2-WPRE3-BGHpA (Alias: CN2099)
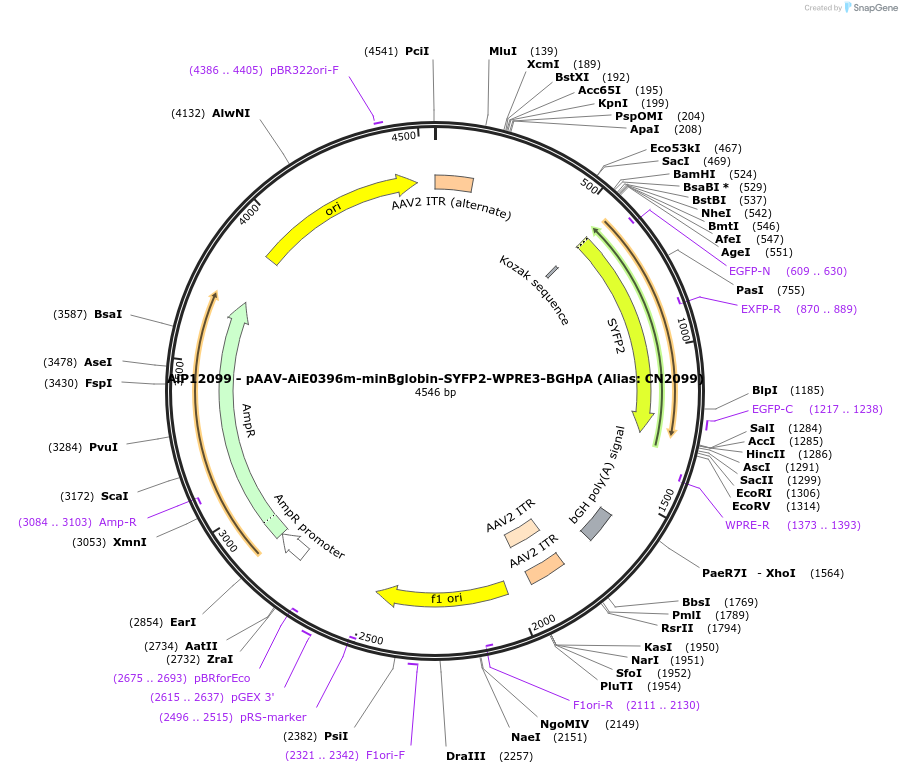
Plasmid#208149PurposeDirect-expressing SYFP2 in oligodendrocytes AAV VirusDepositorInsertSYFP2
UseAAVMutationNAPromoterBeta Globin minimal promoterAvailable SinceOct. 23, 2023AvailabilityAcademic Institutions and Nonprofits only -
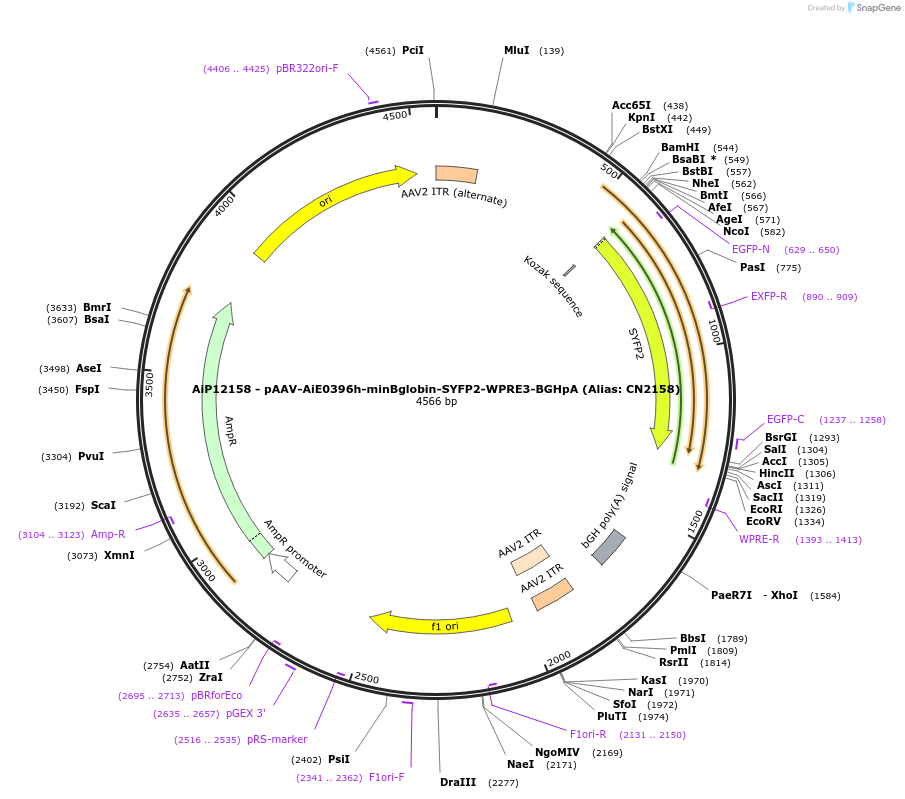
AiP12158 - pAAV-AiE0396h-minBglobin-SYFP2-WPRE3-BGHpA (Alias: CN2158)
Plasmid#208148PurposeDirect-expressing SYFP2 in oligodendrocytes AAV VirusDepositorInsertSYFP2
UseAAVMutationNAPromoterBeta Globin minimal promoterAvailable SinceOct. 20, 2023AvailabilityAcademic Institutions and Nonprofits only -
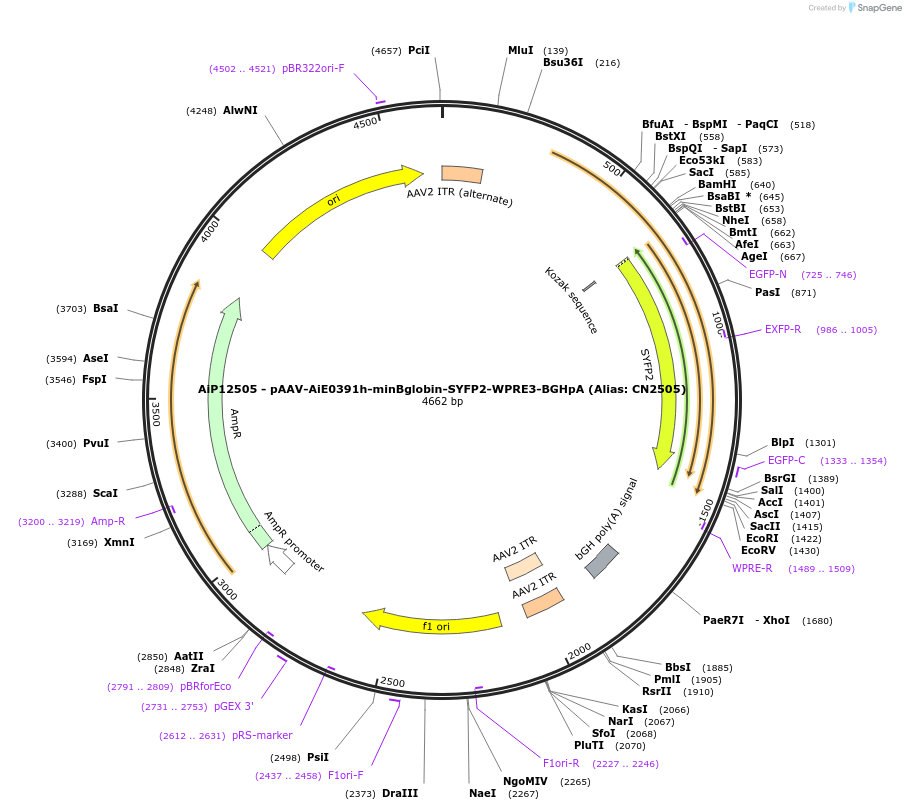
AiP12505 - pAAV-AiE0391h-minBglobin-SYFP2-WPRE3-BGHpA (Alias: CN2505)
Plasmid#208145PurposeDirect-expressing SYFP2 in oligodendrocytes AAV VirusDepositorInsertSYFP2
UseAAVMutationNAPromoterBeta Globin minimal promoterAvailable SinceOct. 20, 2023AvailabilityAcademic Institutions and Nonprofits only -
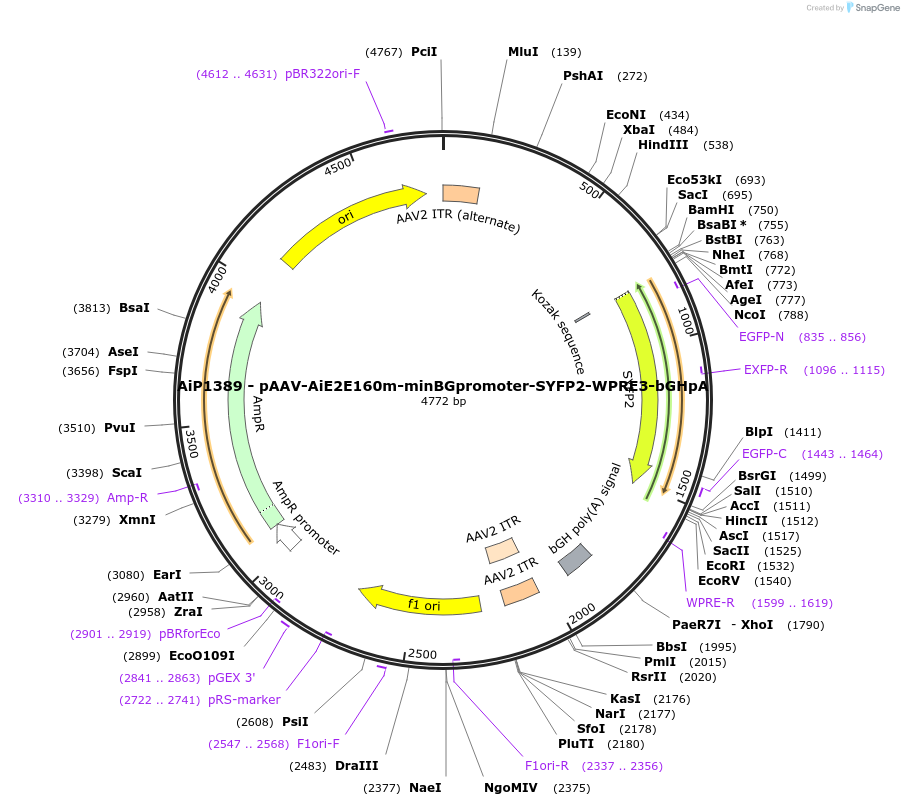
AiP1389 - pAAV-AiE2E160m-minBGpromoter-SYFP2-WPRE3-bGHpA
Plasmid#208140PurposeDirect-expressing SYFP2 in astrocytes AAV Virus. Alias: AiP1389 - pAAV-AiE2160m-minBG-SYFP2-WPRE-BGHpADepositorInsertSYFP2
UseAAVMutationNAPromoterBeta Globin minimal promoterAvailable SinceOct. 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
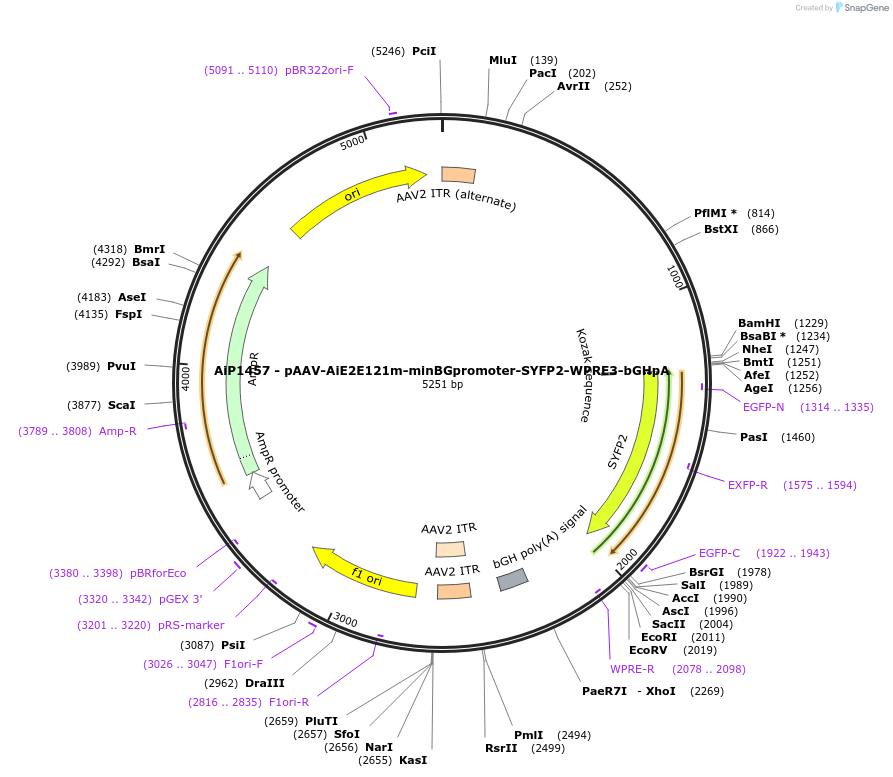
AiP1457 - pAAV-AiE2E121m-minBGpromoter-SYFP2-WPRE3-bGHpA
Plasmid#208138PurposeDirect-expressing SYFP2 in astrocytes AAV Virus. Alias: AiP1457 - pAAV-AiE2121m-minBG-SYFP2-WPRE-BGHpADepositorInsertSYFP2
UseAAVMutationNAPromoterBeta Globin minimal promoterAvailable SinceOct. 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
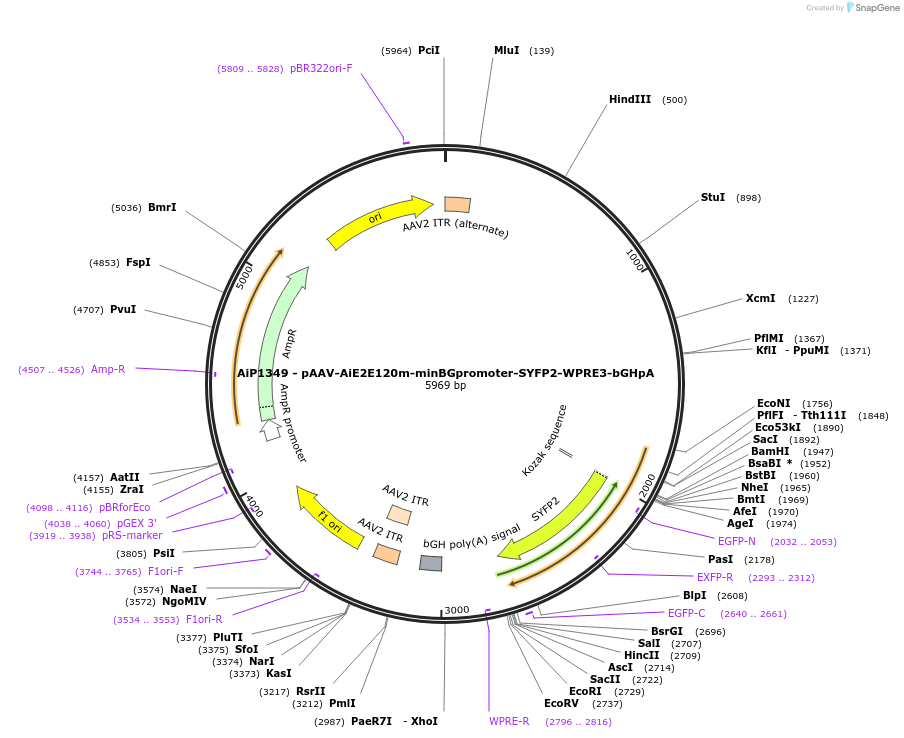
AiP1349 - pAAV-AiE2E120m-minBGpromoter-SYFP2-WPRE3-bGHpA
Plasmid#208137PurposeDirect-expressing SYFP2 in astrocytes AAV Virus. Alias: AiP1349 - pAAV-AiE2120m-minBG-SYFP2-WPRE-BGHpADepositorInsertSYFP2
UseAAVMutationNAPromoterBeta Globin minimal promoterAvailable SinceOct. 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
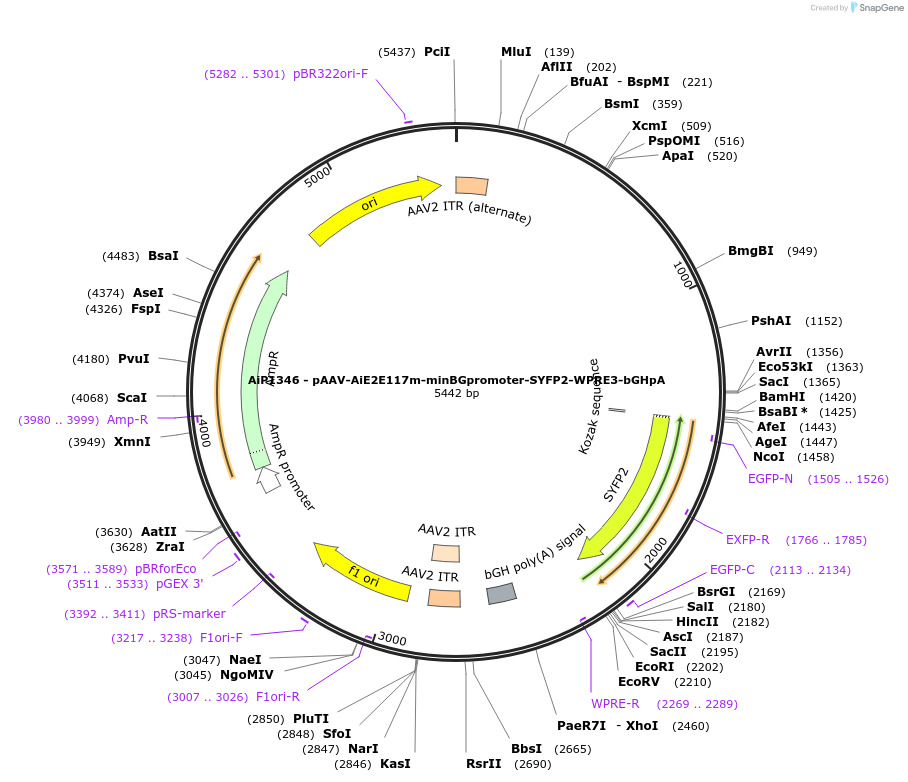
AiP1346 - pAAV-AiE2E117m-minBGpromoter-SYFP2-WPRE3-bGHpA
Plasmid#208135PurposeDirect-expressing SYFP2 in astrocytes AAV Virus. Alias: AiP1346 - pAAV-AiE2117m-minBG-SYFP2-WPRE-BGHpADepositorInsertSYFP2
UseAAVMutationNAPromoterBeta Globin minimal promoterAvailable SinceOct. 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
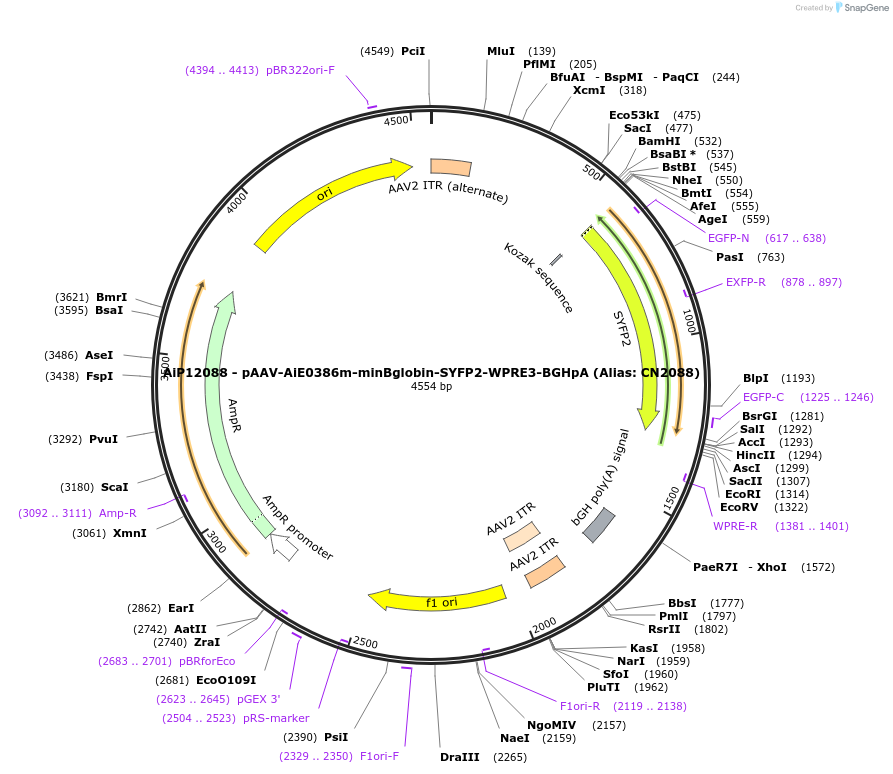
AiP12088 - pAAV-AiE0386m-minBglobin-SYFP2-WPRE3-BGHpA (Alias: CN2088)
Plasmid#208132PurposeDirect-expressing SYFP2 in astrocytes AAV VirusDepositorInsertSYFP2
UseAAVMutationNAPromoterBeta Globin minimal promoterAvailable SinceOct. 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
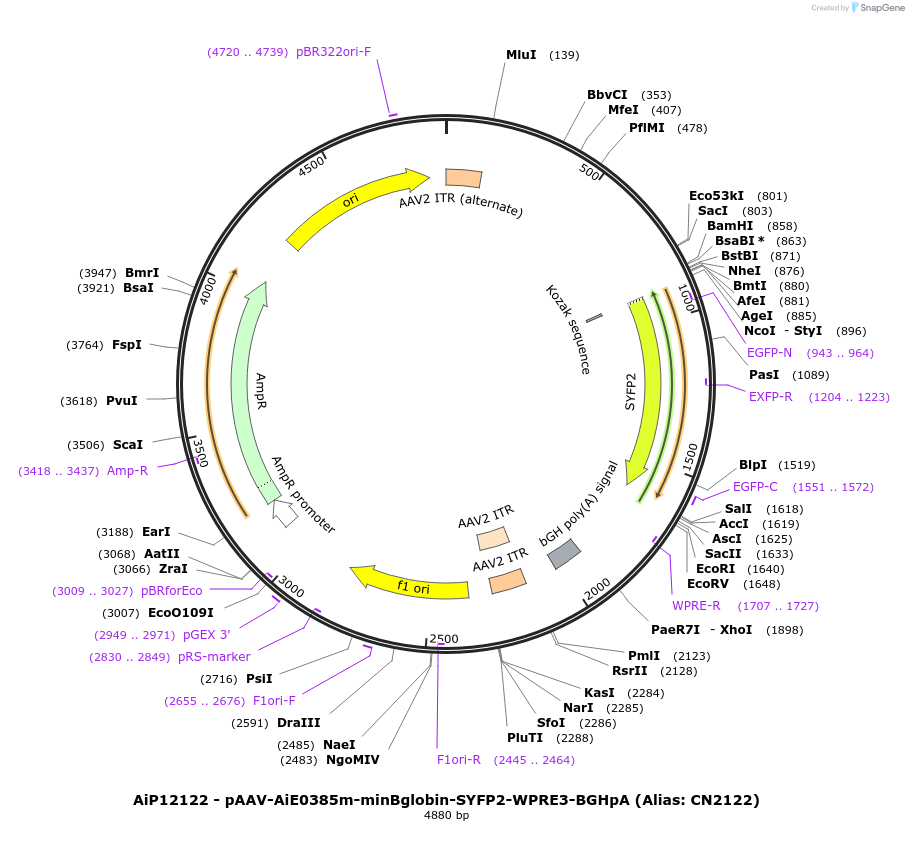
AiP12122 - pAAV-AiE0385m-minBglobin-SYFP2-WPRE3-BGHpA (Alias: CN2122)
Plasmid#208131PurposeDirect-expressing SYFP2 in astrocytes AAV VirusDepositorInsertSYFP2
UseAAVMutationNAPromoterBeta Globin minimal promoterAvailable SinceOct. 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
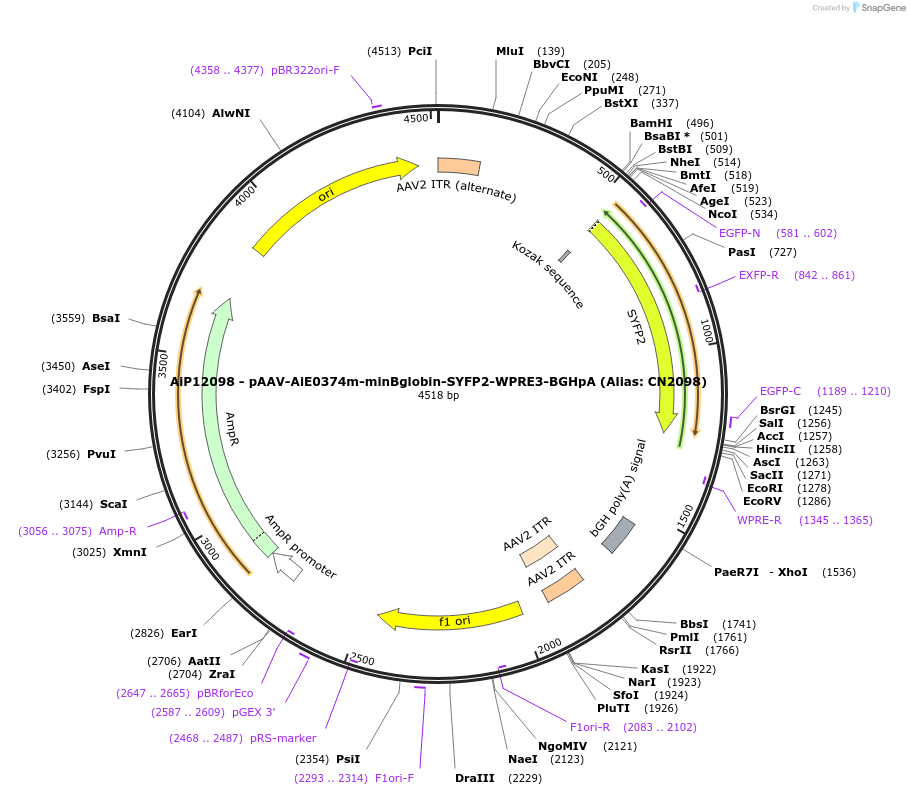
AiP12098 - pAAV-AiE0374m-minBglobin-SYFP2-WPRE3-BGHpA (Alias: CN2098)
Plasmid#208129PurposeDirect-expressing SYFP2 in astrocytes AAV VirusDepositorInsertSYFP2
UseAAVMutationNAPromoterBeta Globin minimal promoterAvailable SinceOct. 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
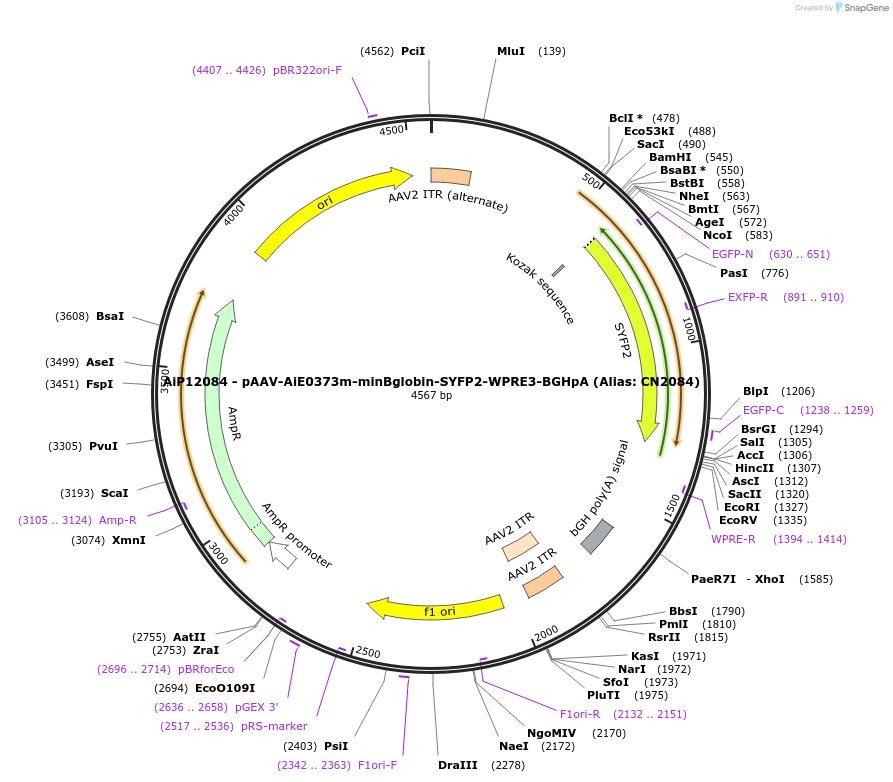
AiP12084 - pAAV-AiE0373m-minBglobin-SYFP2-WPRE3-BGHpA (Alias: CN2084)
Plasmid#208128PurposeDirect-expressing SYFP2 in astrocytes AAV VirusDepositorInsertSYFP2
UseAAVMutationNAPromoterBeta Globin minimal promoterAvailable SinceOct. 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
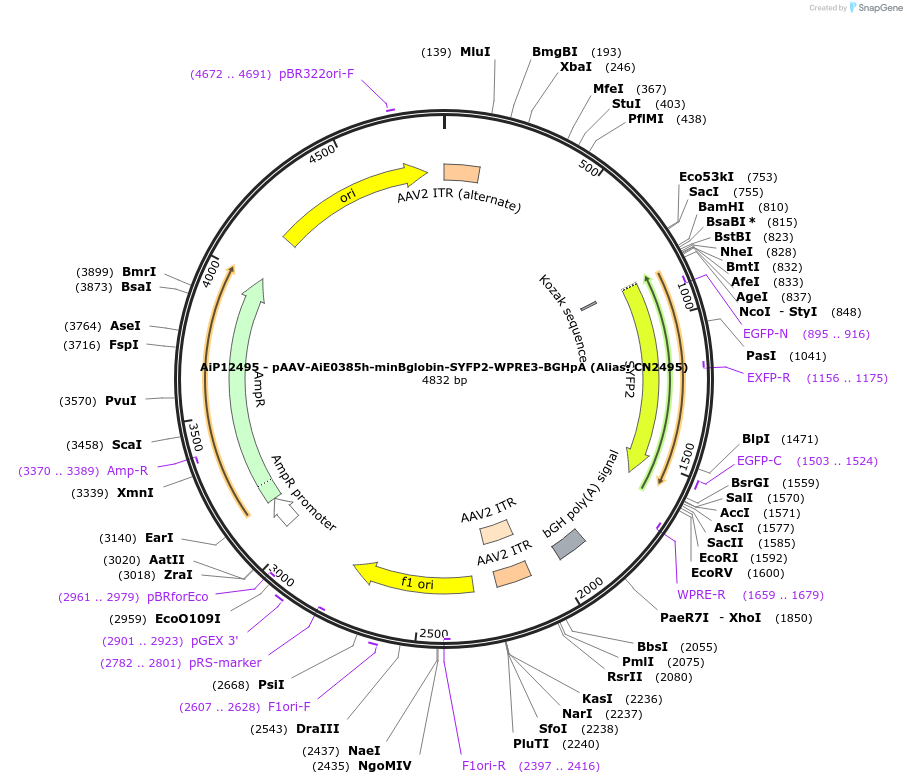
AiP12495 - pAAV-AiE0385h-minBglobin-SYFP2-WPRE3-BGHpA (Alias: CN2495)
Plasmid#208125PurposeDirect-expressing SYFP2 in astrocytes AAV VirusDepositorInsertSYFP2
UseAAVMutationNAPromoterBeta Globin minimal promoterAvailable SinceOct. 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
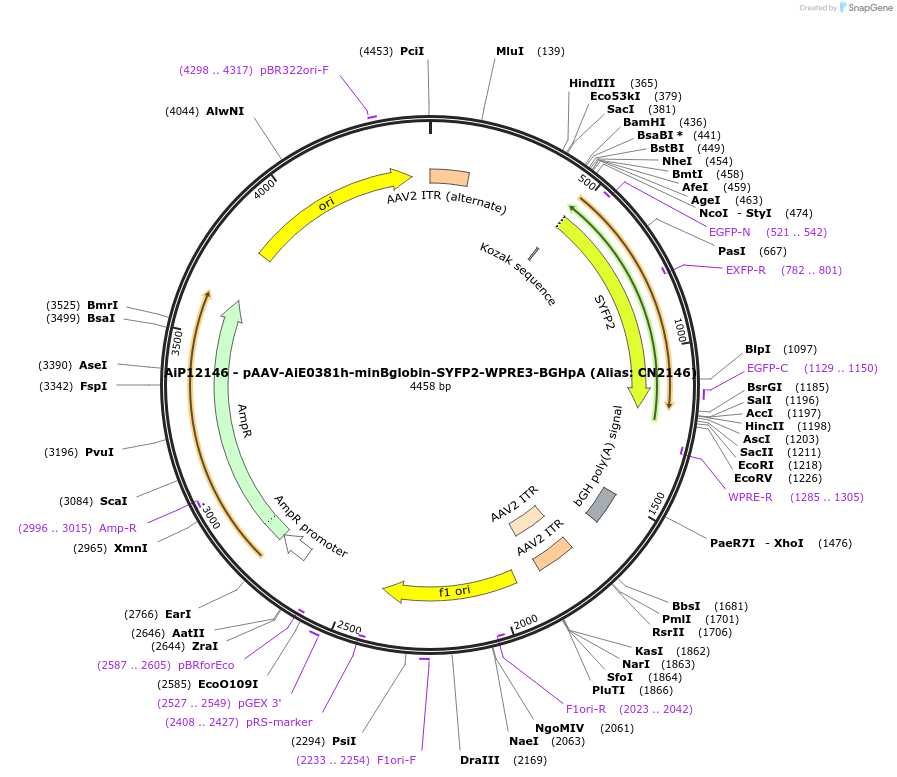
AiP12146 - pAAV-AiE0381h-minBglobin-SYFP2-WPRE3-BGHpA (Alias: CN2146)
Plasmid#208124PurposeDirect-expressing SYFP2 in astrocytes AAV VirusDepositorInsertSYFP2
UseAAVMutationNAPromoterBeta Globin minimal promoterAvailable SinceOct. 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
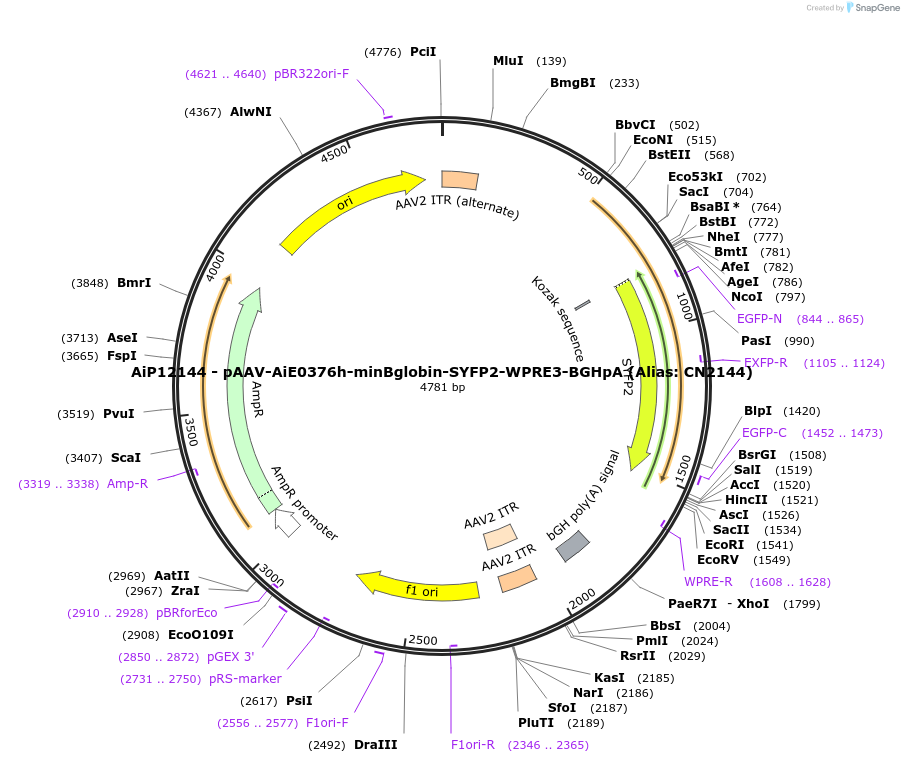
AiP12144 - pAAV-AiE0376h-minBglobin-SYFP2-WPRE3-BGHpA (Alias: CN2144)
Plasmid#208122PurposeDirect-expressing SYFP2 in astrocytes AAV VirusDepositorInsertSYFP2
UseAAVMutationNAPromoterBeta Globin minimal promoterAvailable SinceOct. 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
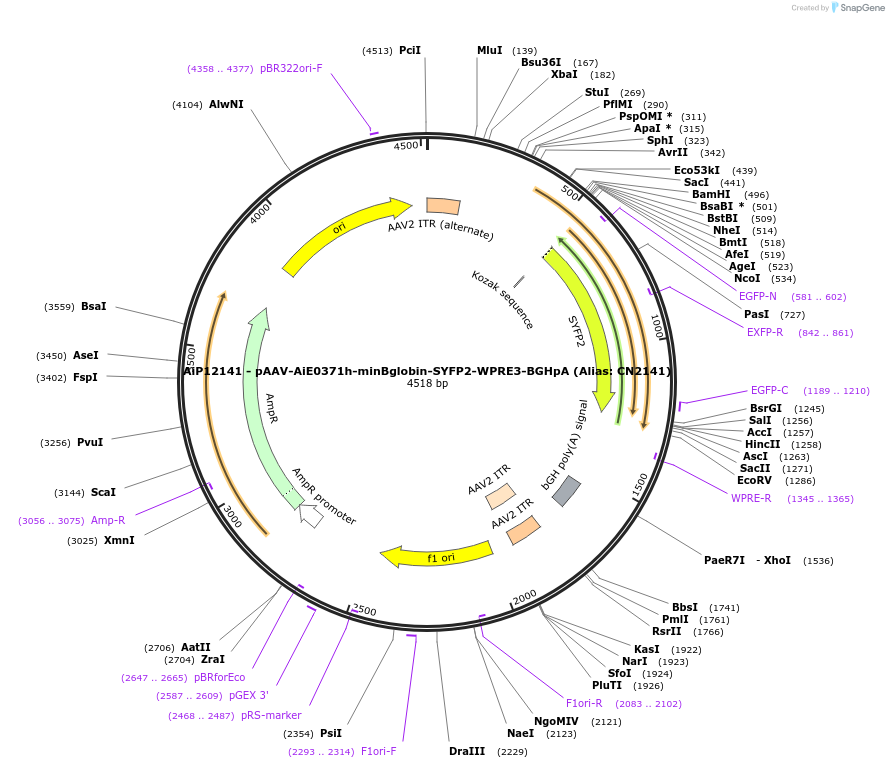
AiP12141 - pAAV-AiE0371h-minBglobin-SYFP2-WPRE3-BGHpA (Alias: CN2141)
Plasmid#208121PurposeDirect-expressing SYFP2 in astrocytes AAV VirusDepositorInsertSYFP2
UseAAVMutationNAPromoterBeta Globin minimal promoterAvailable SinceOct. 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
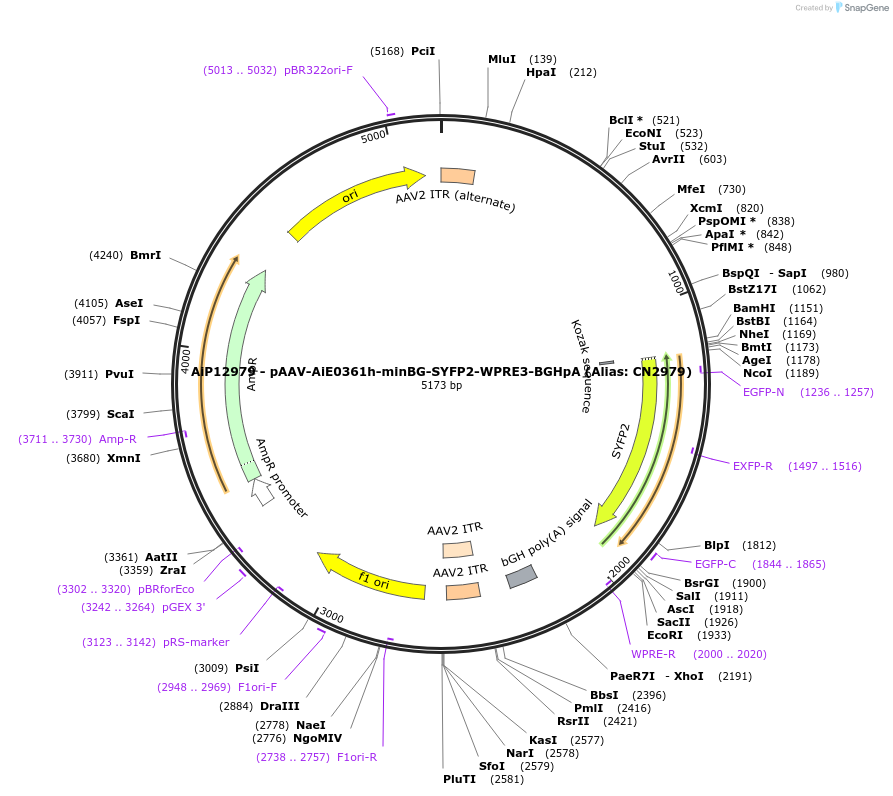
AiP12979 - pAAV-AiE0361h-minBG-SYFP2-WPRE3-BGHpA (Alias: CN2979)
Plasmid#208120PurposeAiE0361h is an enhancer sequence designed to drive AAV-mediated transgene expression in brain astrocytesDepositorInsertSYFP2
UseAAVMutationNAPromoterBeta Globin minimal promoterAvailable SinceOct. 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
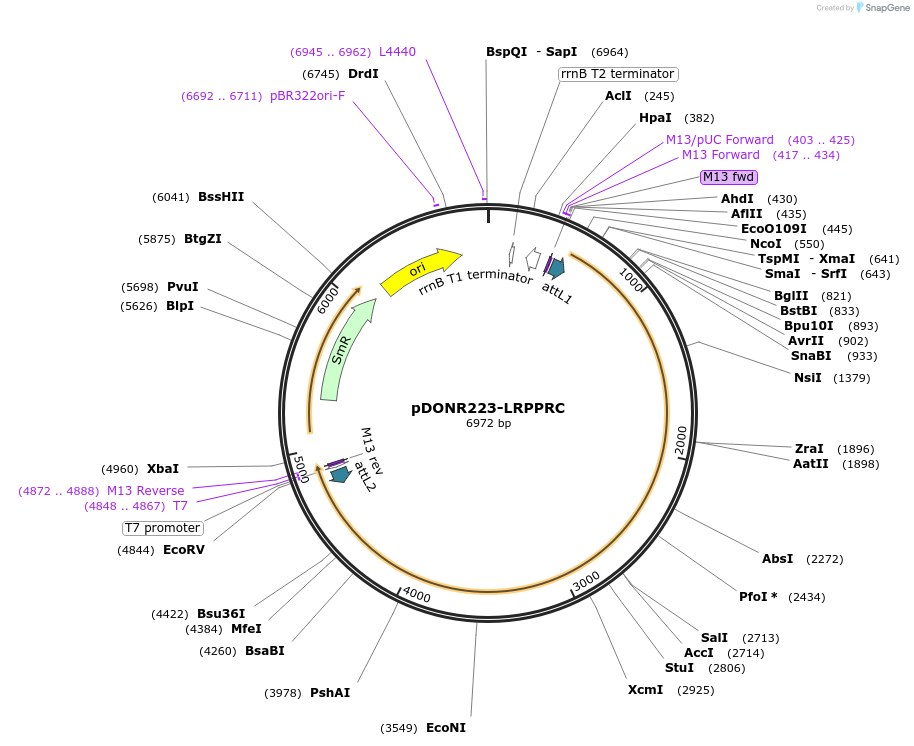
pDONR223-LRPPRC
Plasmid#23521DepositorAvailable SinceJuly 30, 2010AvailabilityAcademic Institutions and Nonprofits only