We narrowed to 9,261 results for: Arch
-
Plasmid#125407PurposeCRISPR-mediated repression of human islet enhancer containing T2D-associated variant rs7163757 (C2CD4A/B GWAS locus)DepositorInsertKRAB-dCas9-2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceJuly 24, 2019AvailabilityAcademic Institutions and Nonprofits only -
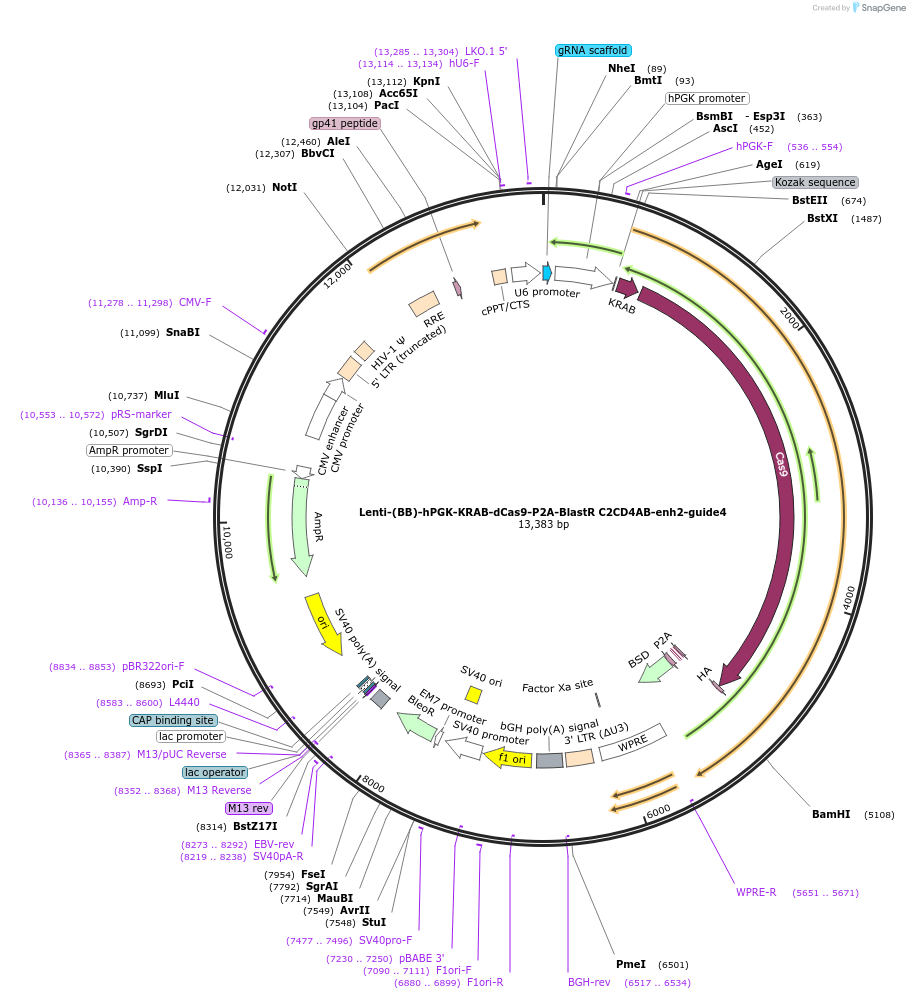
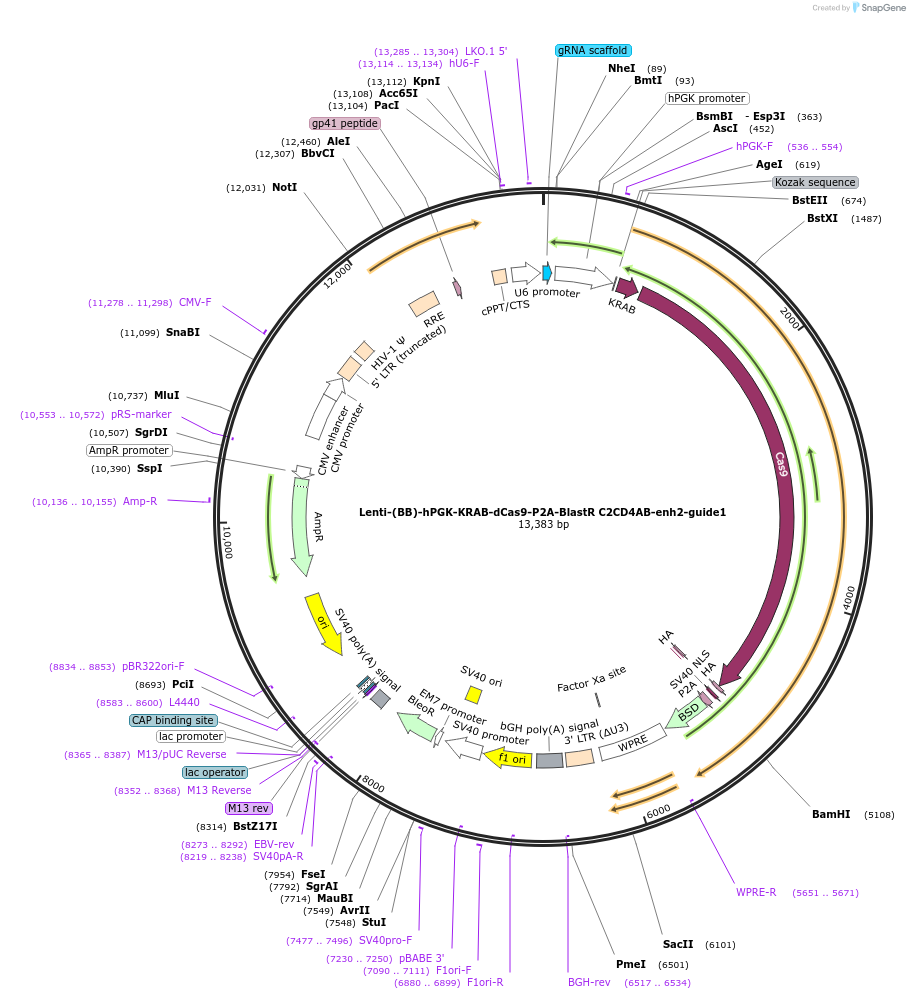
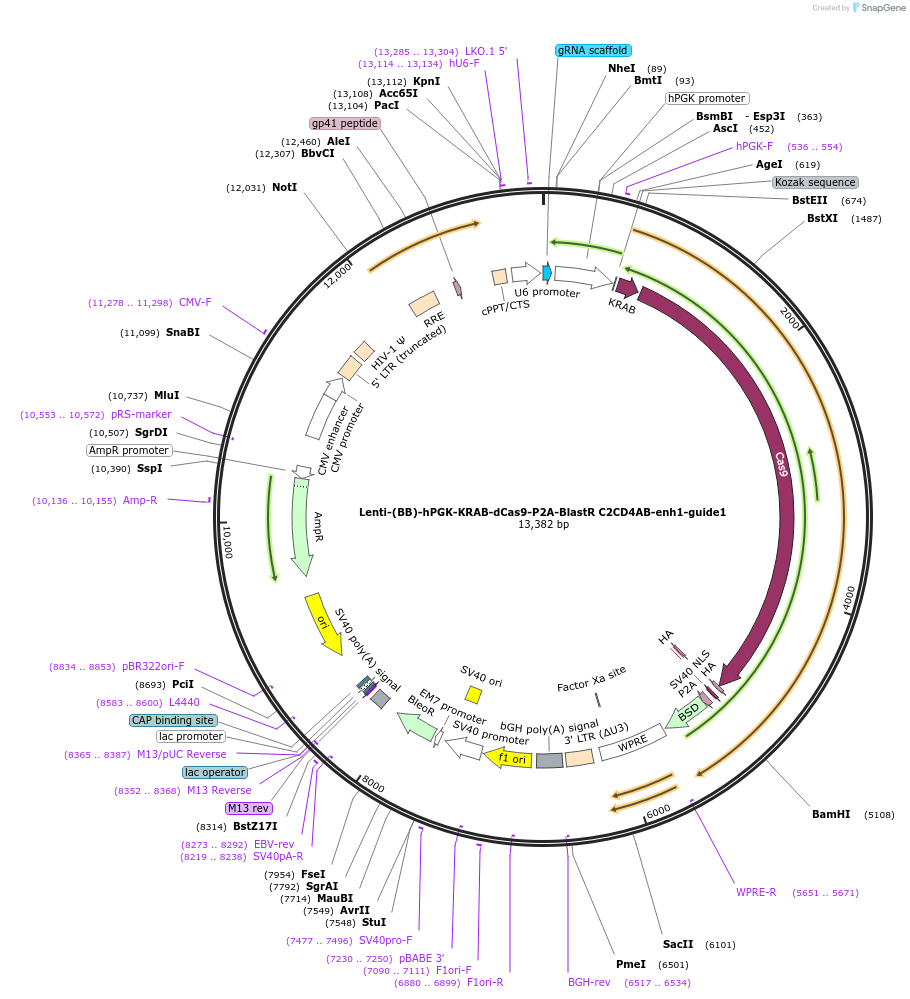
Lenti-(BB)-hPGK-KRAB-dCas9-P2A-BlastR C2CD4AB-enh2-guide1
Plasmid#125404PurposeCRISPR-mediated repression of human islet enhancer containing T2D-associated variant rs7163757 (C2CD4A/B GWAS locus)DepositorInsertKRAB-dCas9-2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceJuly 23, 2019AvailabilityAcademic Institutions and Nonprofits only -
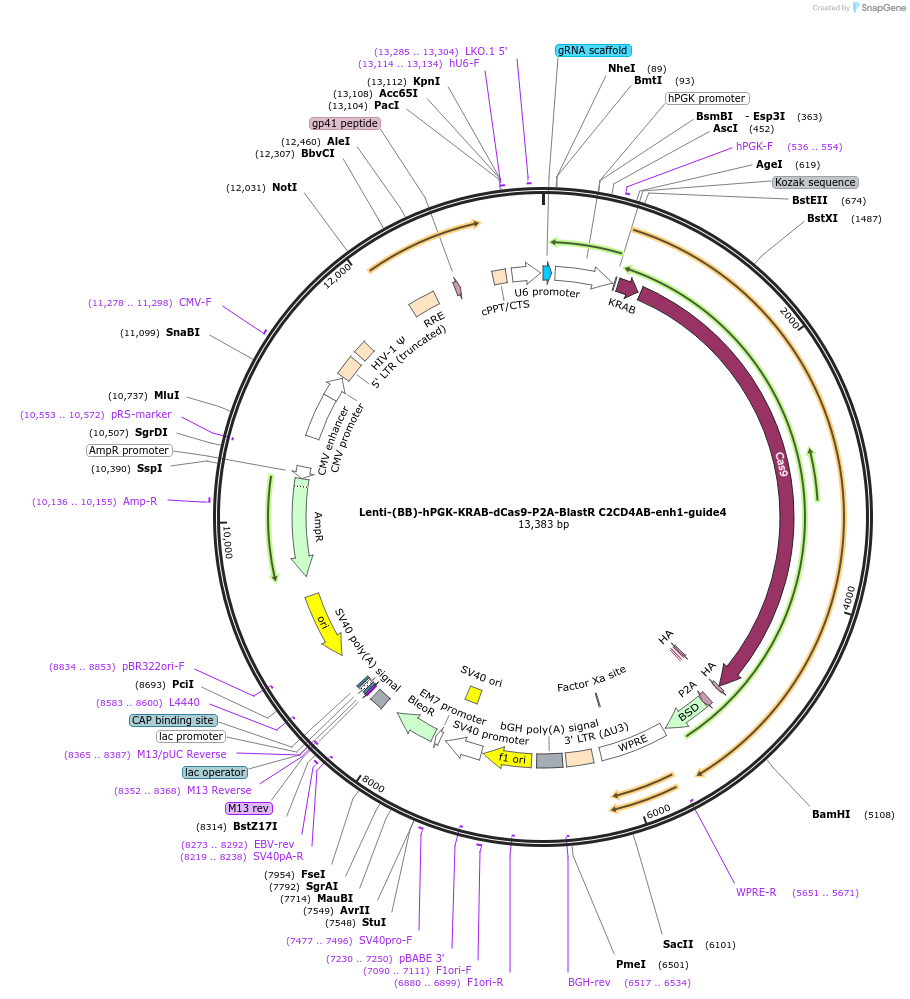
Lenti-(BB)-hPGK-KRAB-dCas9-P2A-BlastR C2CD4AB-enh1-guide4
Plasmid#125403PurposeCRISPR-mediated repression of human islet enhancer containing T2D-associated variant rs182222907 (C2CD4A/B GWAS locus)DepositorInsertKRAB-dCas9-2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceJuly 23, 2019AvailabilityAcademic Institutions and Nonprofits only -
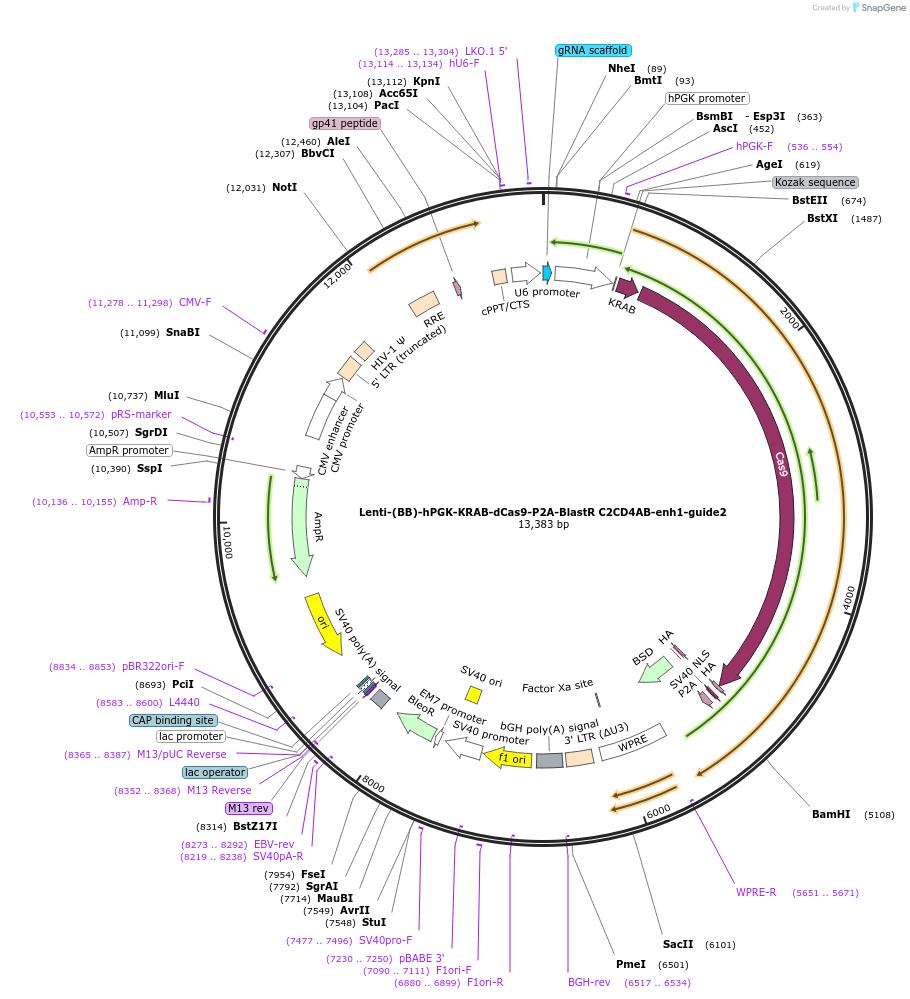
Lenti-(BB)-hPGK-KRAB-dCas9-P2A-BlastR C2CD4AB-enh1-guide2
Plasmid#125401PurposeCRISPR-mediated repression of human islet enhancer containing T2D-associated variant rs182222907 (C2CD4A/B GWAS locus)DepositorInsertKRAB-dCas9-2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceJuly 23, 2019AvailabilityAcademic Institutions and Nonprofits only -
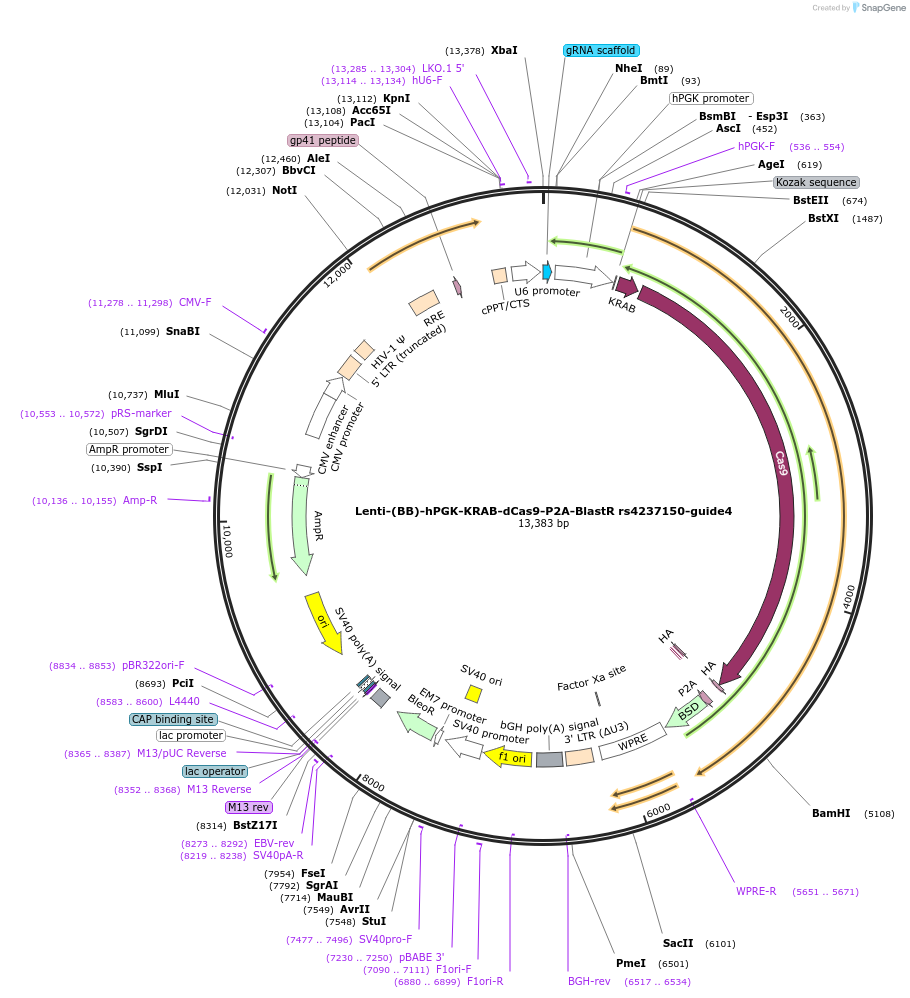
Lenti-(BB)-hPGK-KRAB-dCas9-P2A-BlastR rs4237150-guide4
Plasmid#125399PurposeCRISPR-mediated repression of human islet enhancer containing T1D/T2D-associated variant rs4237150 (GLIS3 GWAS locus)DepositorInsertKRAB-dCas9-2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceJuly 23, 2019AvailabilityAcademic Institutions and Nonprofits only -
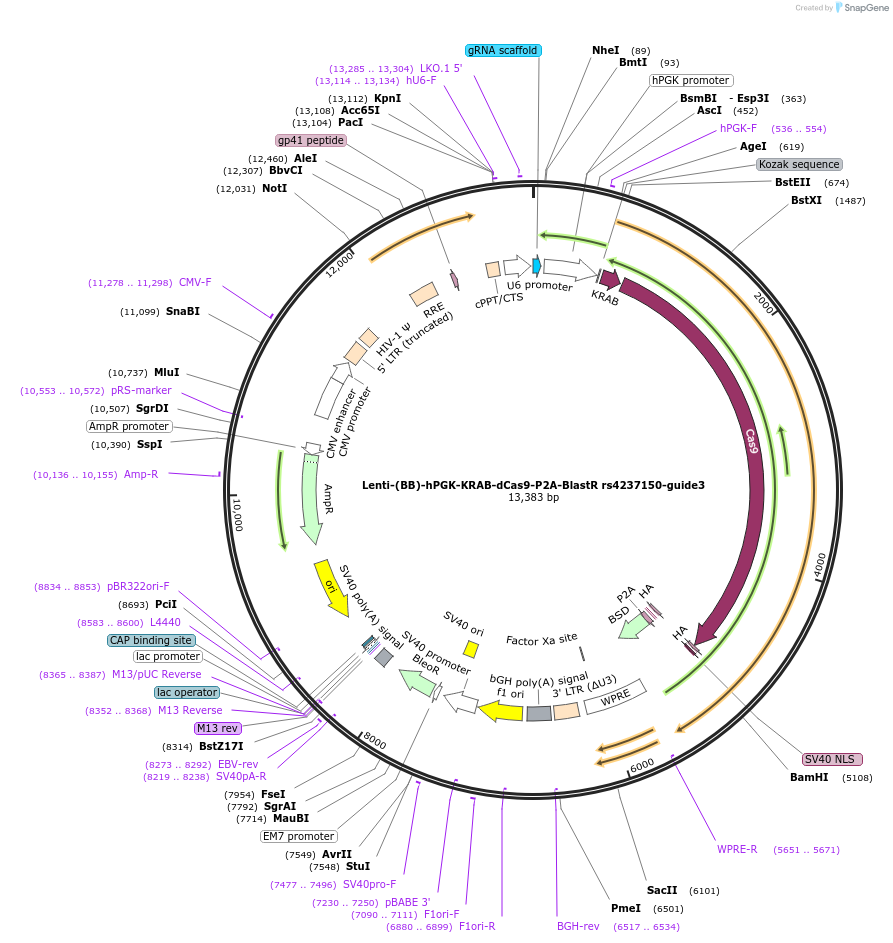
Lenti-(BB)-hPGK-KRAB-dCas9-P2A-BlastR rs4237150-guide3
Plasmid#125398PurposeCRISPR-mediated repression of human islet enhancer containing T1D/T2D-associated variant rs4237150 (GLIS3 GWAS locus)DepositorInsertKRAB-dCas9-2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceJuly 23, 2019AvailabilityAcademic Institutions and Nonprofits only -
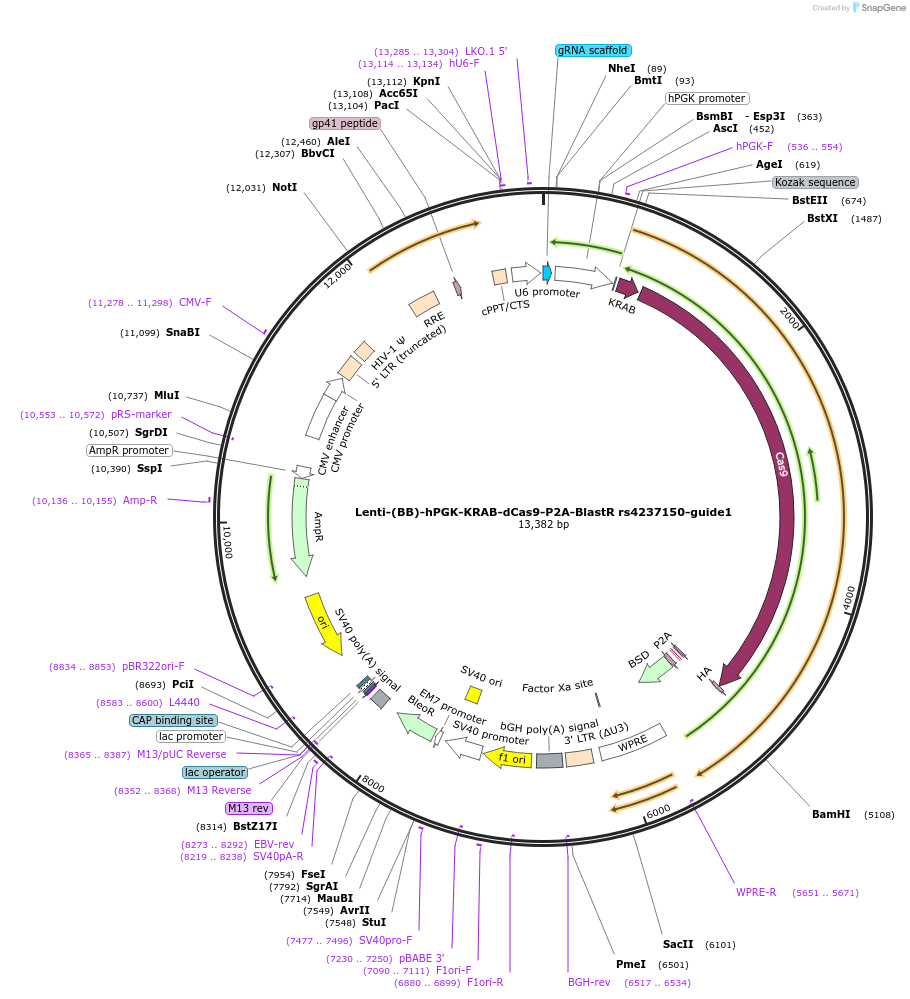
Lenti-(BB)-hPGK-KRAB-dCas9-P2A-BlastR rs4237150-guide1
Plasmid#125397PurposeCRISPR-mediated repression of human islet enhancer containing T1D/T2D-associated variant rs4237150 (GLIS3 GWAS locus)DepositorInsertKRAB-dCas9-2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceJuly 23, 2019AvailabilityAcademic Institutions and Nonprofits only -
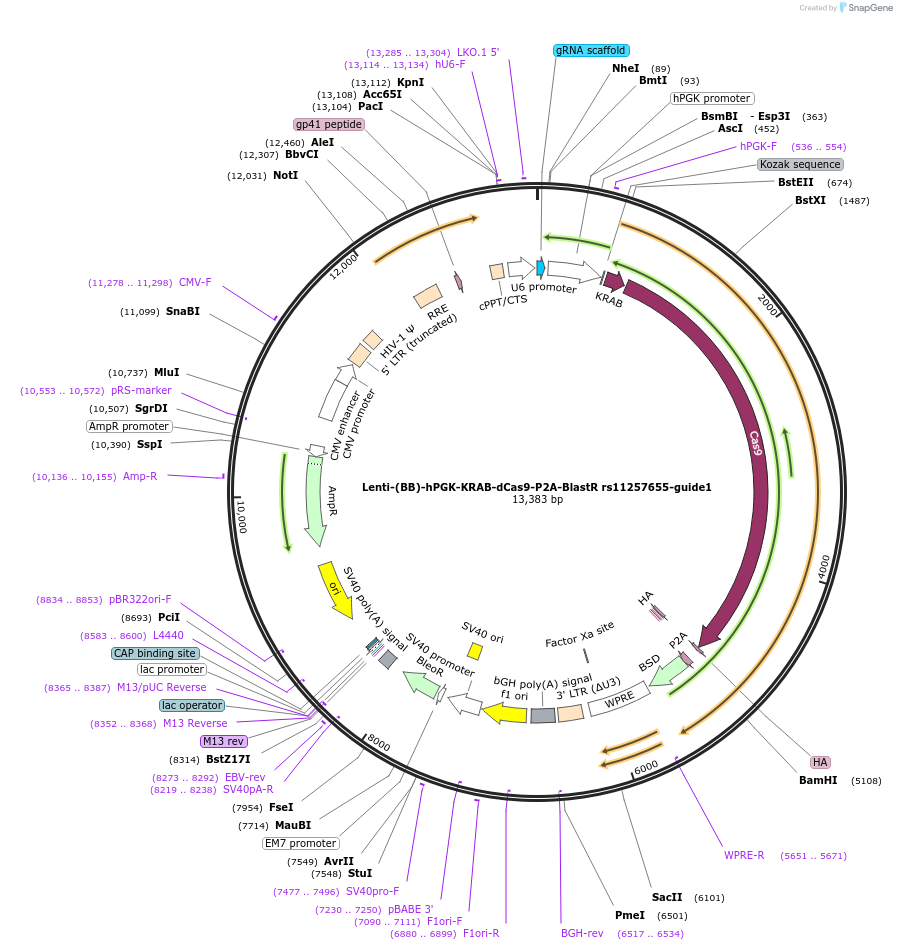
Lenti-(BB)-hPGK-KRAB-dCas9-P2A-BlastR rs11257655-guide1
Plasmid#125393PurposeCRISPR-mediated repression of human islet enhancer containing T2D-associated variant rs11257655 (CDC123/CAMK1D GWAS locus)DepositorInsertKRAB-dCas9-2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceJuly 23, 2019AvailabilityAcademic Institutions and Nonprofits only -
Lenti-(BB)-hPGK-KRAB-dCas9-P2A-BlastR C2CD4AB-enh1-guide1
Plasmid#125400PurposeCRISPR-mediated repression of human islet enhancer containing T2D-associated variant rs182222907 (C2CD4A/B GWAS locus)DepositorInsertKRAB-dCas9-2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceJuly 9, 2019AvailabilityAcademic Institutions and Nonprofits only -
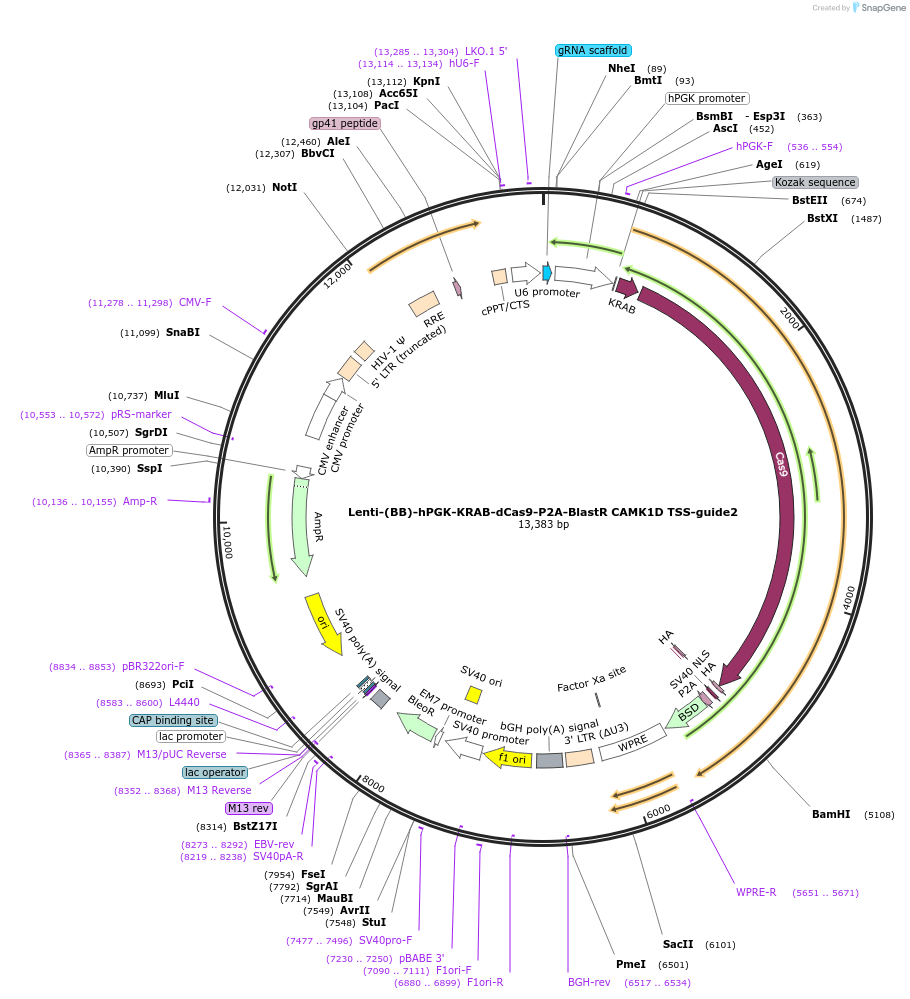
Lenti-(BB)-hPGK-KRAB-dCas9-P2A-BlastR CAMK1D TSS-guide2
Plasmid#118178PurposeCRISPR-mediated repression of CAMK1D. KRAB domain and catalytically inactive Cas9 from S. pyogenes with 2A-BlastR under the hPGK promoter.DepositorInsertKRAB-dCas9-2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceJune 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
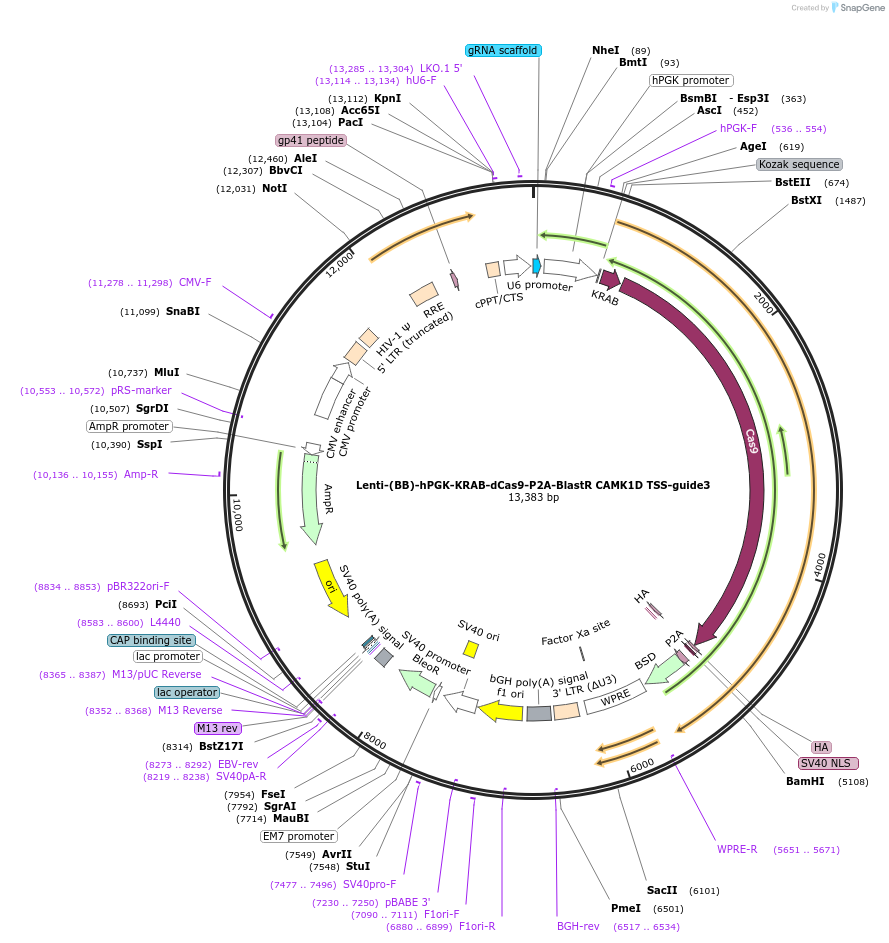
Lenti-(BB)-hPGK-KRAB-dCas9-P2A-BlastR CAMK1D TSS-guide3
Plasmid#118179PurposeCRISPR-mediated repression of CAMK1D. KRAB domain and catalytically inactive Cas9 from S. pyogenes with 2A-BlastR under the hPGK promoter.DepositorInsertKRAB-dCas9-2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceApril 3, 2019AvailabilityAcademic Institutions and Nonprofits only -
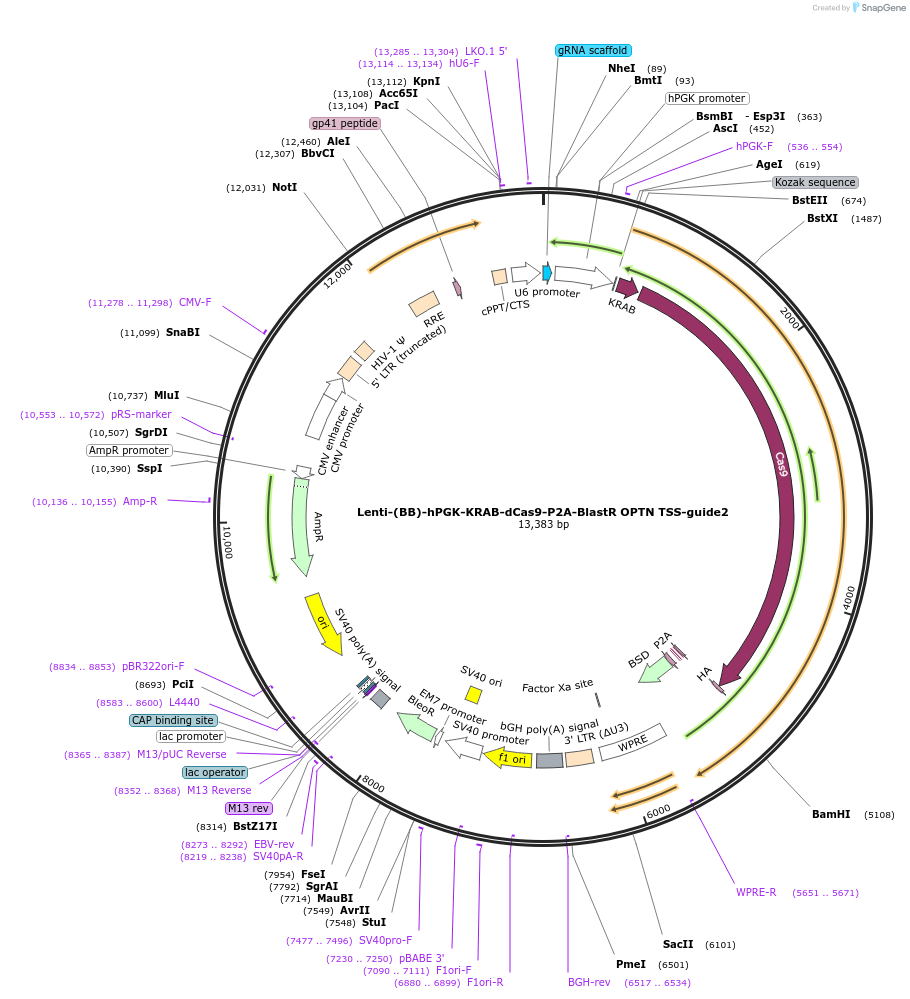
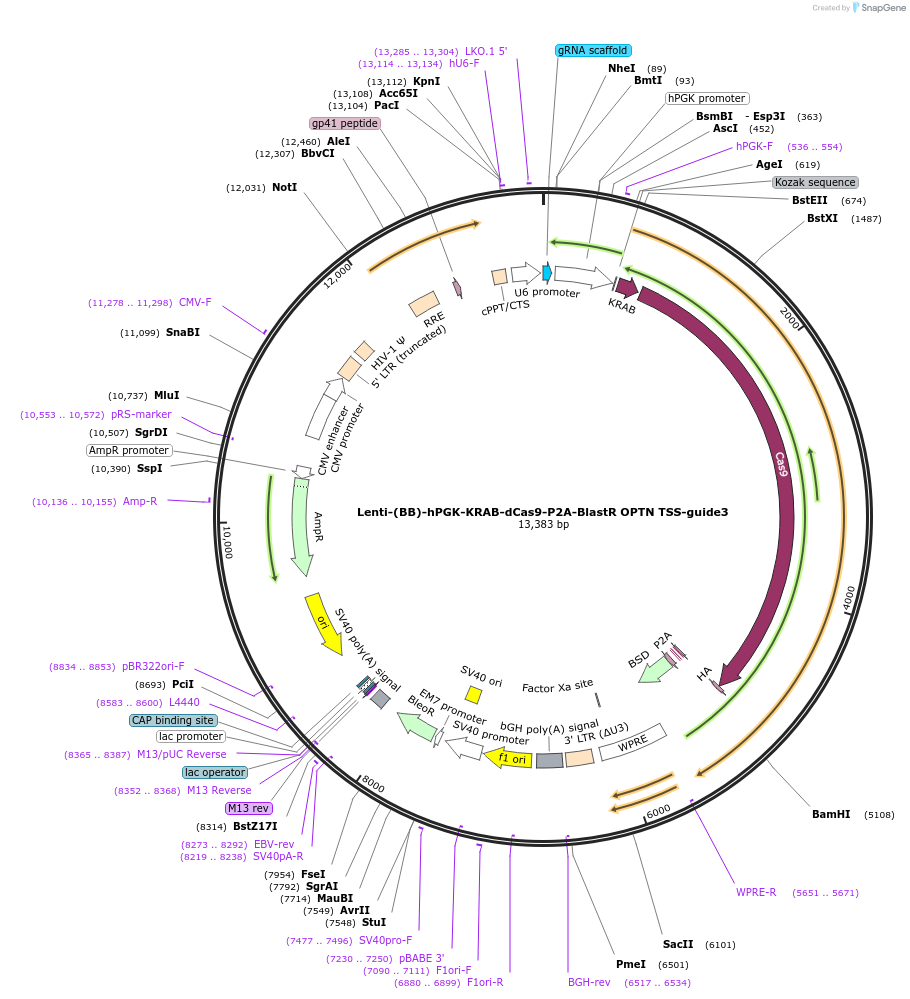
Lenti-(BB)-hPGK-KRAB-dCas9-P2A-BlastR OPTN TSS-guide2
Plasmid#118182PurposeCRISPR-mediated repression of OPTN. KRAB domain and catalytically inactive Cas9 from S. pyogenes with 2A-BlastR under the hPGK promoter.DepositorInsertKRAB-dCas9-2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceApril 3, 2019AvailabilityAcademic Institutions and Nonprofits only -
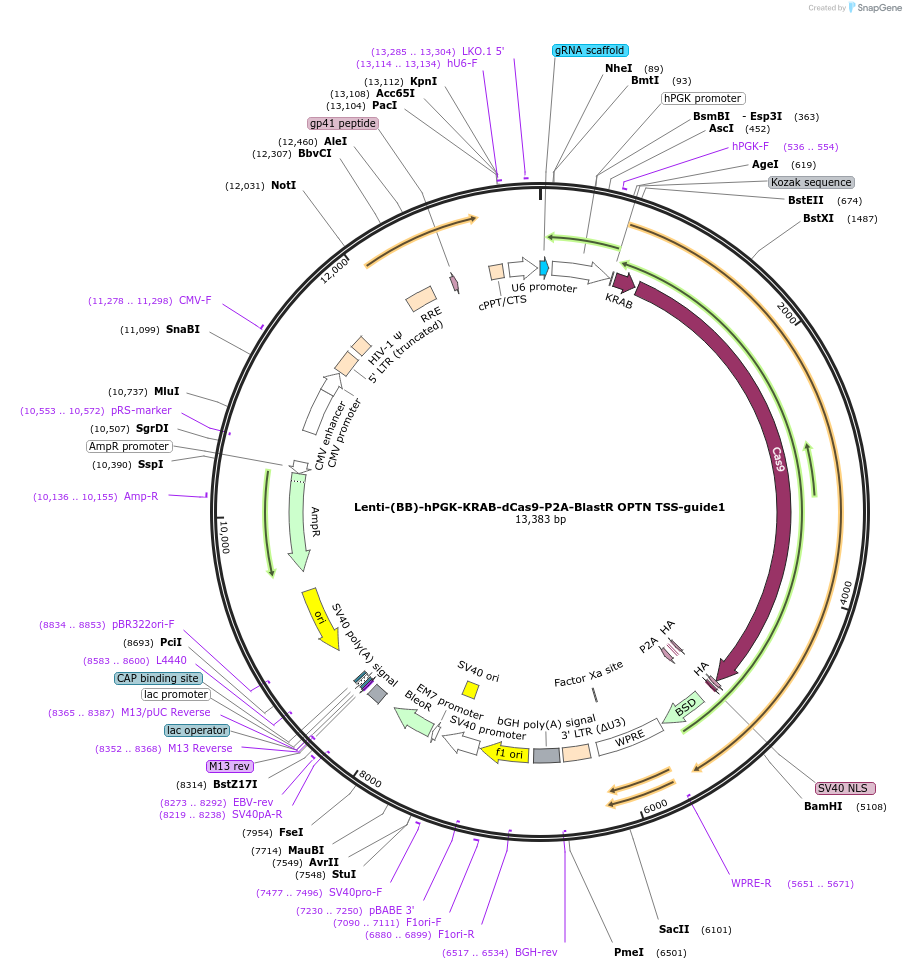
Lenti-(BB)-hPGK-KRAB-dCas9-P2A-BlastR OPTN TSS-guide1
Plasmid#118181PurposeCRISPR-mediated repression of OPTN. KRAB domain and catalytically inactive Cas9 from S. pyogenes with 2A-BlastR under the hPGK promoter.DepositorInsertKRAB-dCas9-2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceApril 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
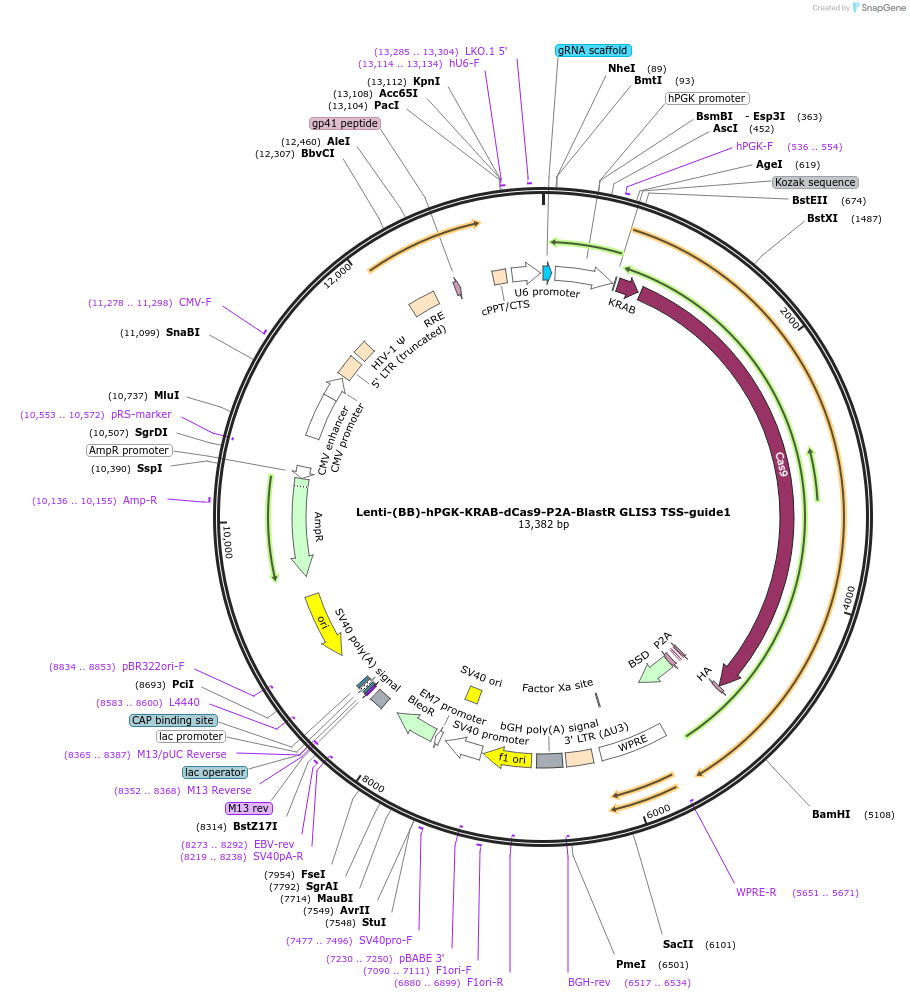
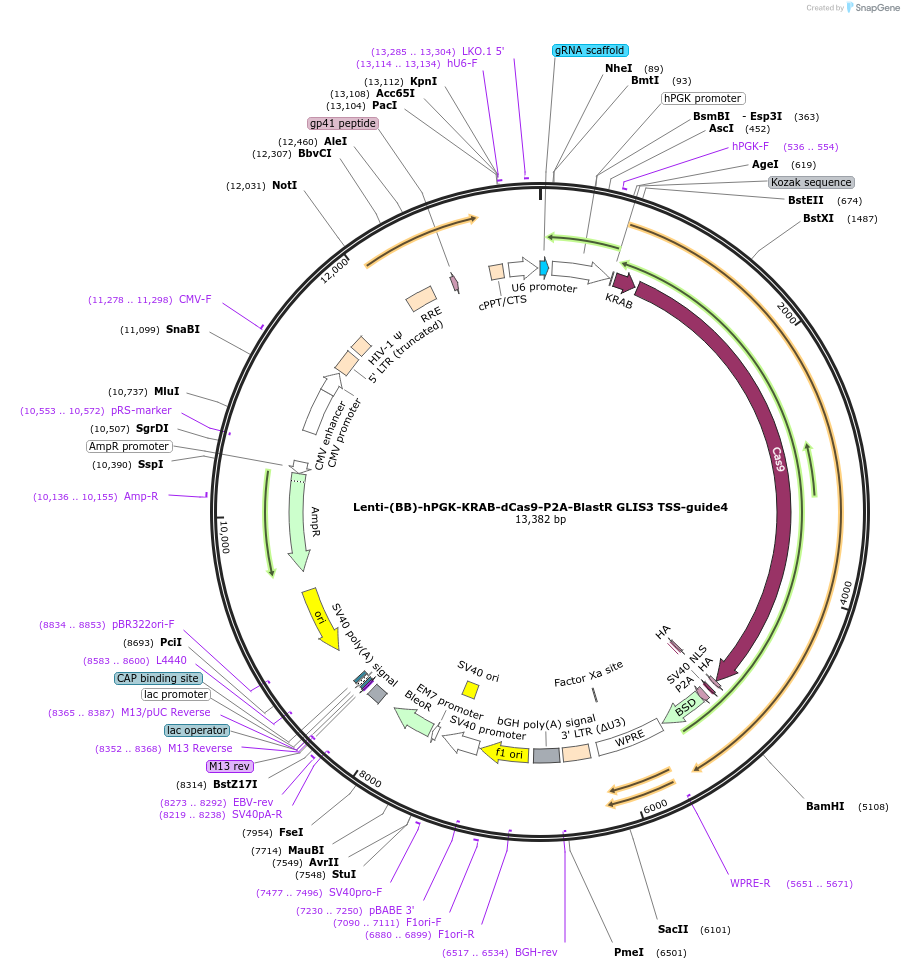
Lenti-(BB)-hPGK-KRAB-dCas9-P2A-BlastR GLIS3 TSS-guide1
Plasmid#118173PurposeCRISPR-mediated repression of GLIS3. KRAB domain and catalytically inactive Cas9 from S. pyogenes with 2A-BlastR under the hPGK promoter.DepositorInsertKRAB-dCas9-2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceApril 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
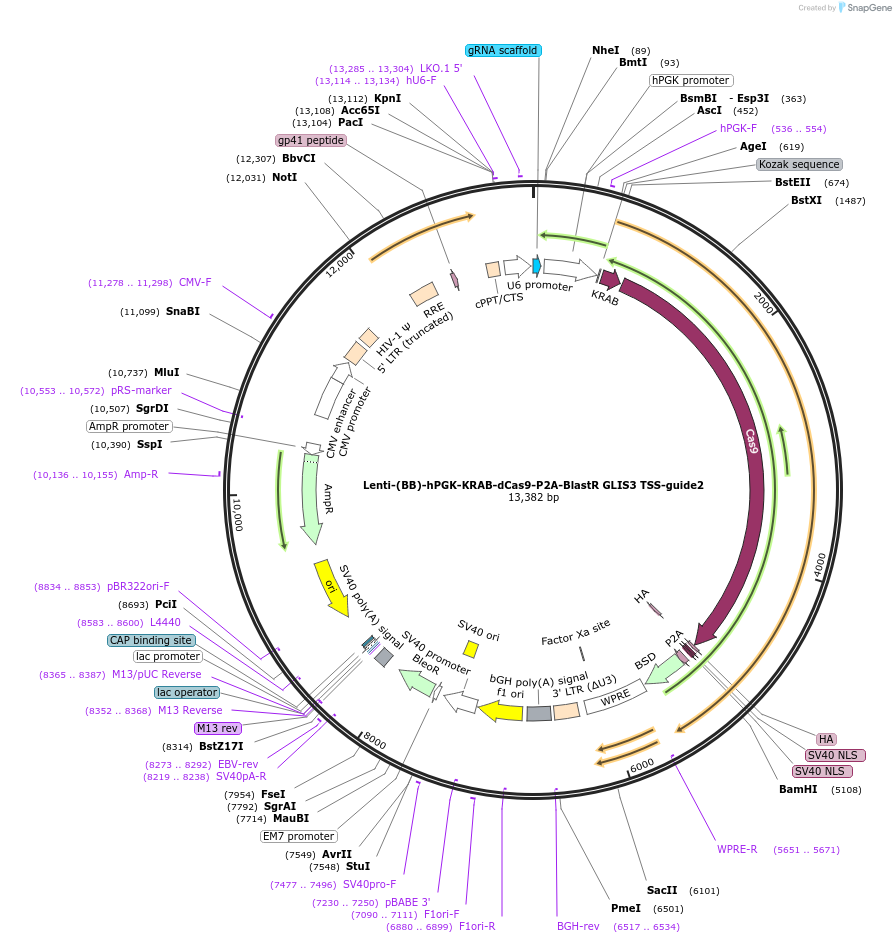
Lenti-(BB)-hPGK-KRAB-dCas9-P2A-BlastR GLIS3 TSS-guide2
Plasmid#118174PurposeCRISPR-mediated repression of GLIS3. KRAB domain and catalytically inactive Cas9 from S. pyogenes with 2A-BlastR under the hPGK promoter.DepositorInsertKRAB-dCas9-2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceApril 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
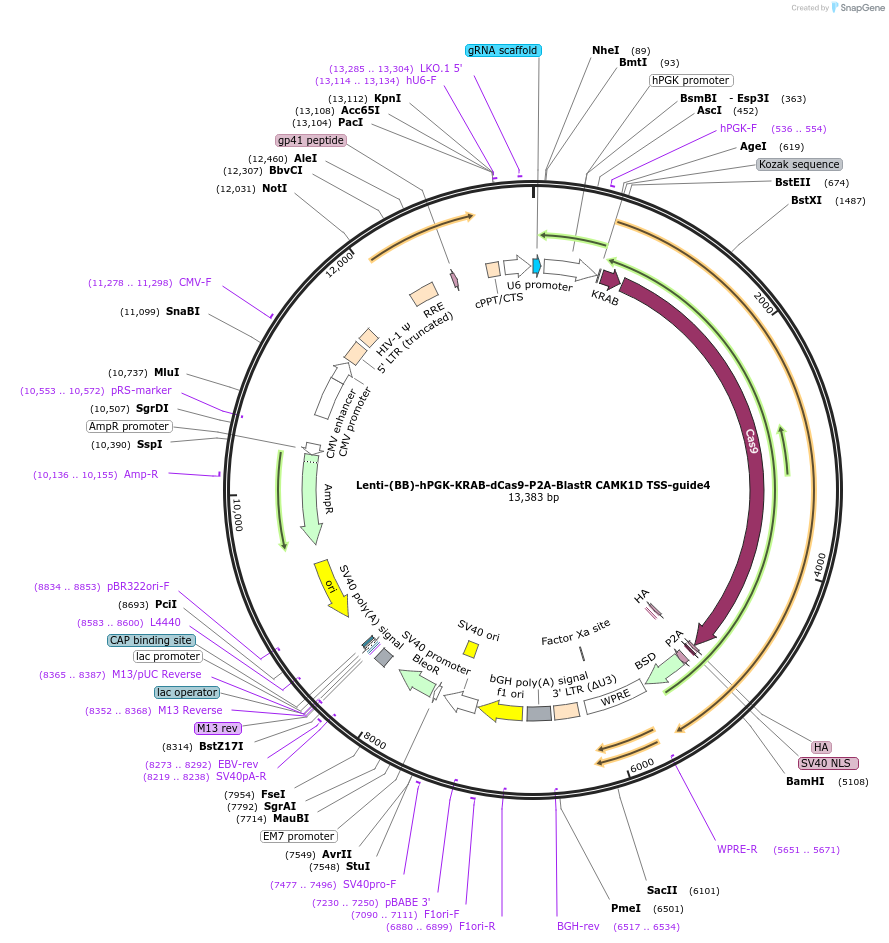
Lenti-(BB)-hPGK-KRAB-dCas9-P2A-BlastR CAMK1D TSS-guide4
Plasmid#118180PurposeCRISPR-mediated repression of CAMK1D. KRAB domain and catalytically inactive Cas9 from S. pyogenes with 2A-BlastR under the hPGK promoter.DepositorInsertKRAB-dCas9-2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceApril 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
Lenti-(BB)-hPGK-KRAB-dCas9-P2A-BlastR OPTN TSS-guide3
Plasmid#118183PurposeCRISPR-mediated repression of OPTN. KRAB domain and catalytically inactive Cas9 from S. pyogenes with 2A-BlastR under the hPGK promoter.DepositorInsertKRAB-dCas9-2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceApril 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
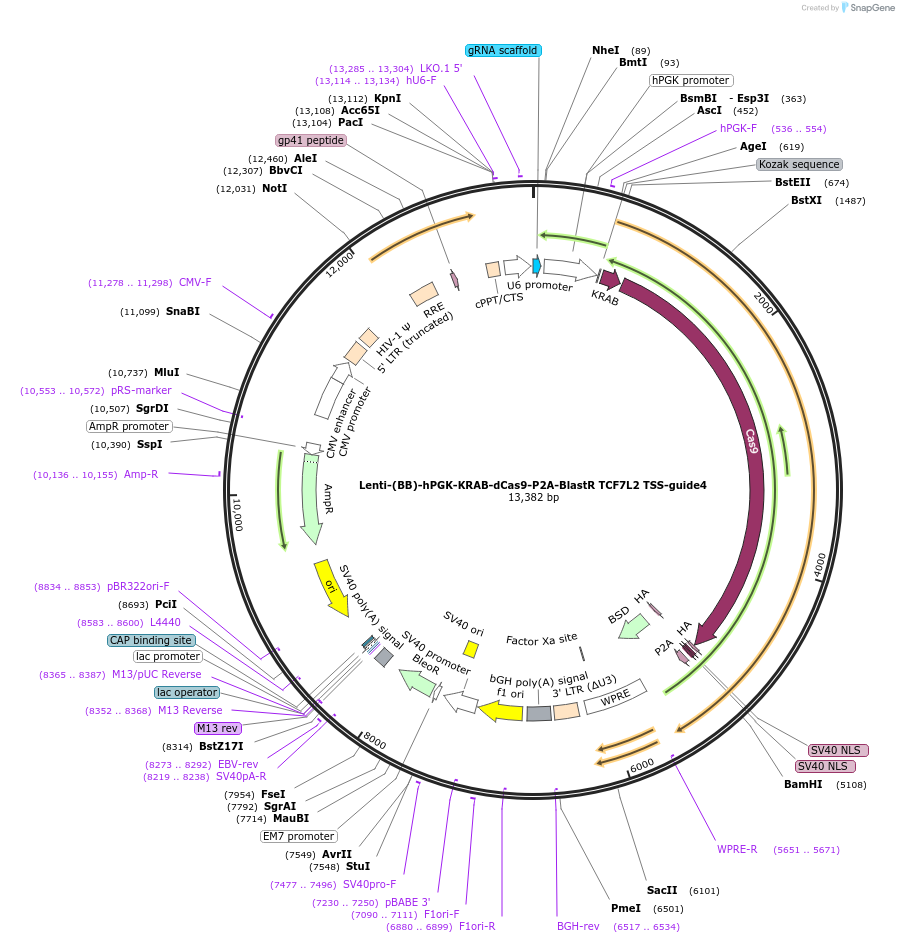
Lenti-(BB)-hPGK-KRAB-dCas9-P2A-BlastR GLIS3 TSS-guide4
Plasmid#118176PurposeCRISPR-mediated repression of GLIS3. KRAB domain and catalytically inactive Cas9 from S. pyogenes with 2A-BlastR under the hPGK promoter.DepositorInsertKRAB-dCas9-2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceApril 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
Lenti-(BB)-hPGK-KRAB-dCas9-P2A-BlastR TCF7L2 TSS-guide4
Plasmid#118172PurposeCRISPR-mediated repression of TCF7L2. KRAB domain and catalytically inactive Cas9 from S. pyogenes with 2A-BlastR under the hPGK promoter.DepositorInsertKRAB-dCas9-2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceApril 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
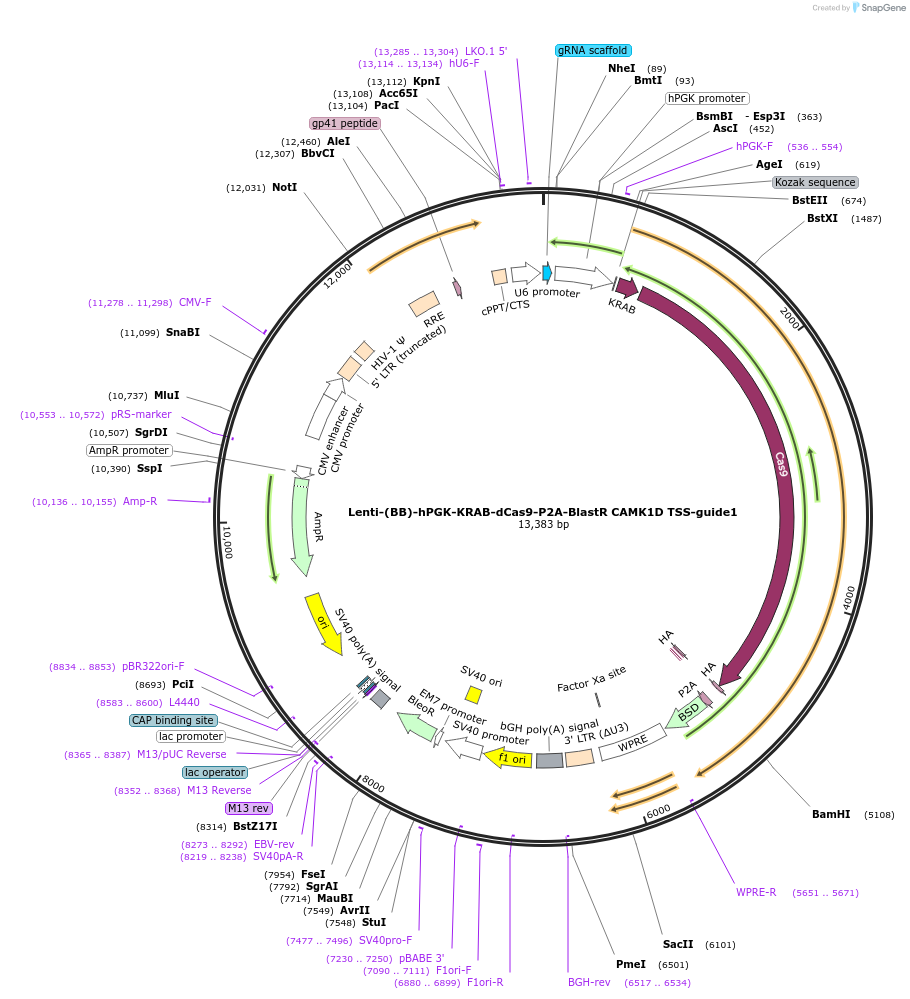
Lenti-(BB)-hPGK-KRAB-dCas9-P2A-BlastR CAMK1D TSS-guide1
Plasmid#118177PurposeCRISPR-mediated repression of CAMK1D. KRAB domain and catalytically inactive Cas9 from S. pyogenes with 2A-BlastR under the hPGK promoter.DepositorInsertKRAB-dCas9-2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceApril 1, 2019AvailabilityAcademic Institutions and Nonprofits only