We narrowed to 10,079 results for: SUB
-
Plasmid#10986DepositorInsertrpb1-D261N (RPO21 Budding Yeast)
ExpressionBacterial and YeastMutationMutation 781 G to A in the RPB1 ORF resulting i…Available SinceDec. 16, 2005AvailabilityAcademic Institutions and Nonprofits only -
pL-rpb1-R320C
Plasmid#10987DepositorInsertrpb1-R320C (RPO21 Budding Yeast)
ExpressionBacterial and YeastMutationMutation 958 C to T in the RPB1 ORF resulting i…Available SinceDec. 16, 2005AvailabilityAcademic Institutions and Nonprofits only -
pL-rpb1-E1230K
Plasmid#10990DepositorInsertrpb1-E1230K (RPO21 Budding Yeast)
ExpressionBacterial and YeastMutationMutation 3688 G to A in the RPB1 ORF resulting in…Available SinceDec. 16, 2005AvailabilityAcademic Institutions and Nonprofits only -
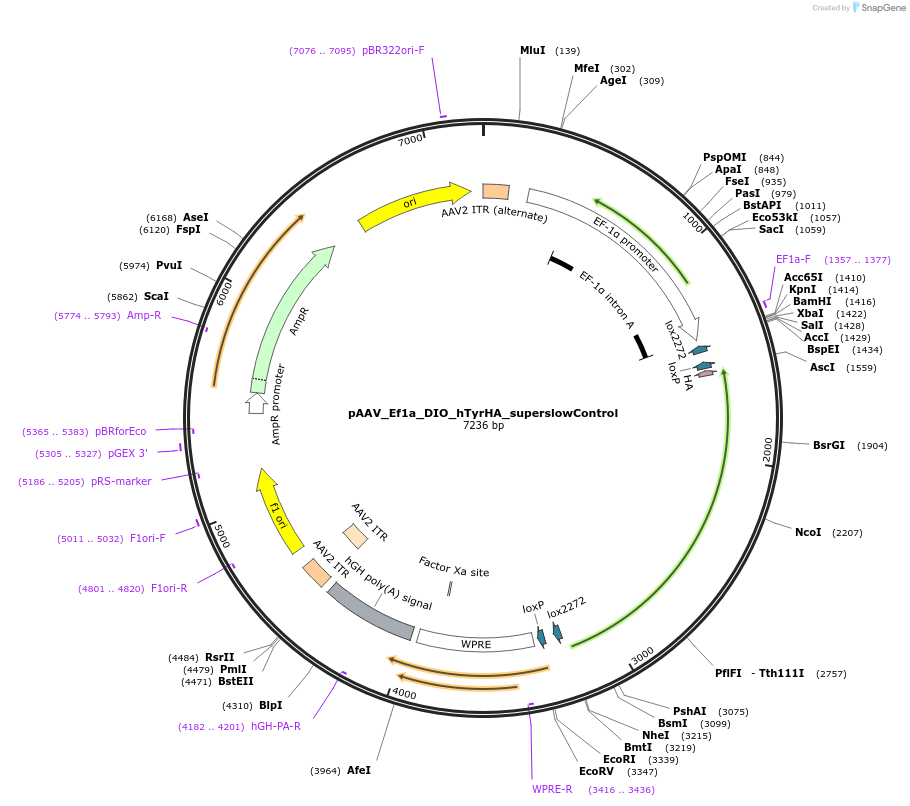
pAAV_Ef1a_DIO_hTyrHA_superslowControl
Plasmid#249346PurposeControl hTyr (D383N/A206T) — human tyrosinase carrying D383N and A206T substitutions; catalytic activity super slow; intended as an enzymatically-impaired control.DepositorInsertmutated super slow human Tyrosinase (TYR Human)
UseAAVTagsHA tagMutationD383N/A206TPromoterEF1aAvailable SinceJan. 12, 2026AvailabilityAcademic Institutions and Nonprofits only -
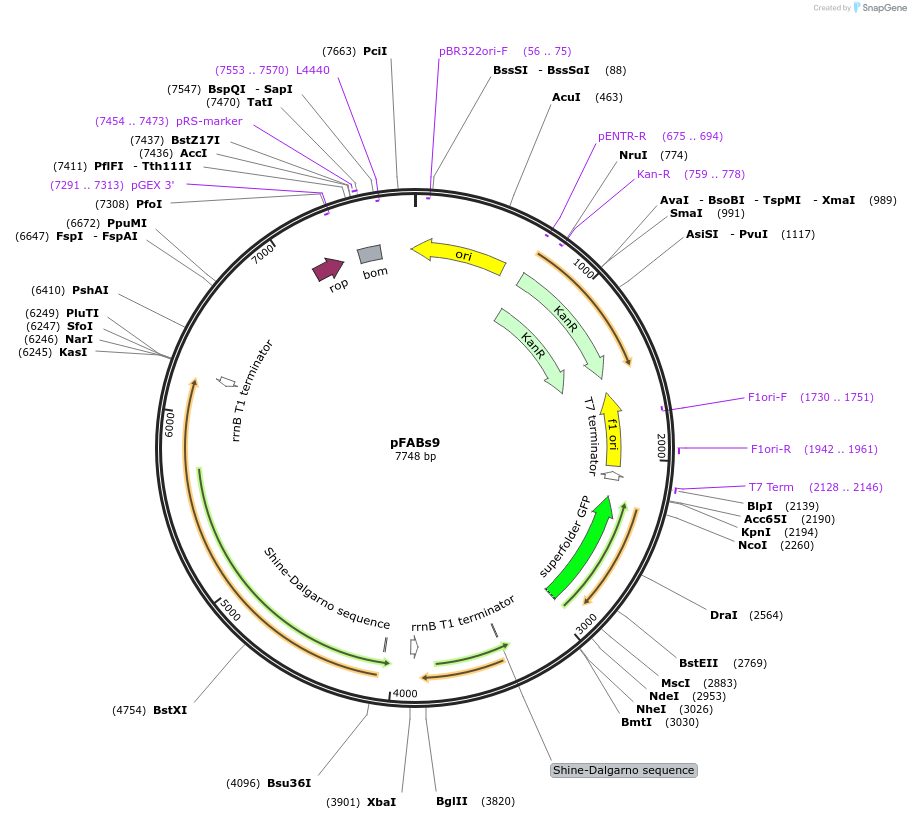
pFABs9
Plasmid#241905PurposeFerrulic acid biosensor - expresses sfGFP in response to substituted cinnamic acidDepositorInsertsfGFP, ferC, ferA
UseSynthetic BiologyMutationnoneAvailable SinceSept. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
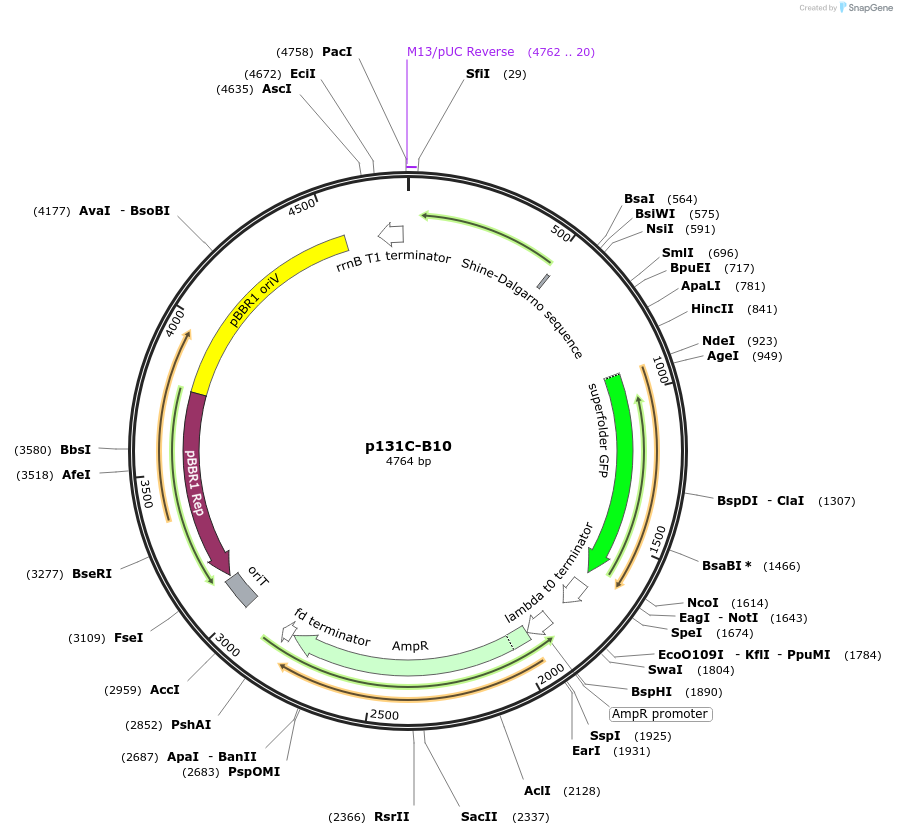
p131C-B10
Plasmid#241908PurposeOptimised PCA biosensor produced sfGFP in response to hydroxyl-substituted benzoic acids, including protocatechuic acid (PCA)DepositorInsertpcaV, sfGFP
UseSynthetic BiologyMutationnoneAvailable SinceAug. 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
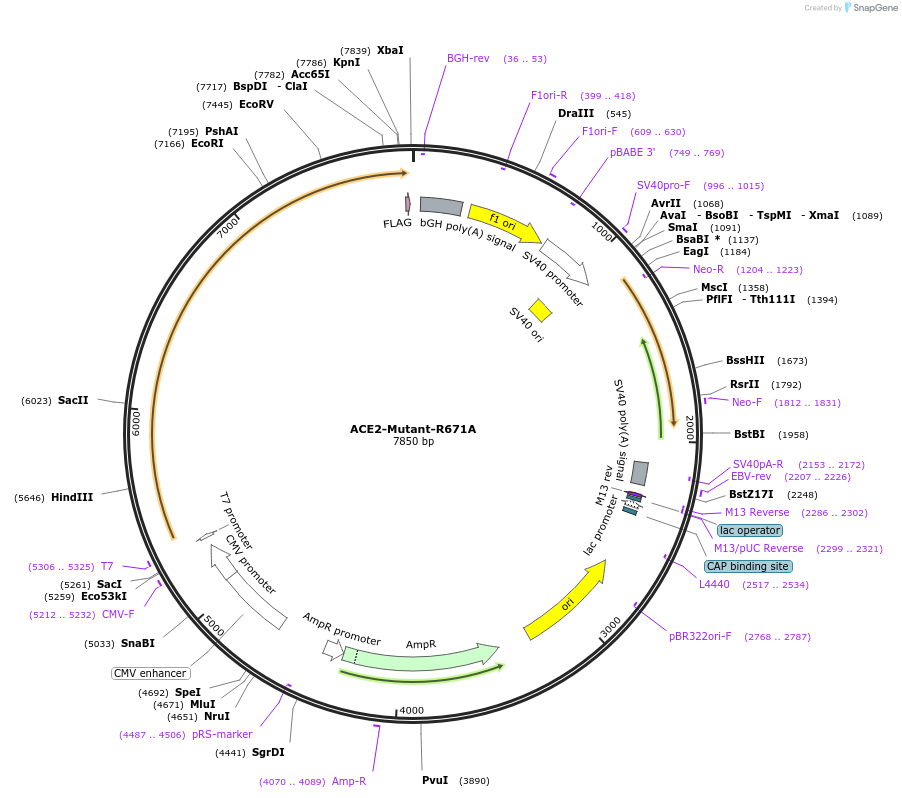
ACE2-Mutant-R671A
Plasmid#240606PurposeTo test ACE2 cleavage by TMPRSS2. This protease targets R and K amino acids of ACE2; R was substituted by A at Residue 671 of ACE2 in the collectrin-like siteDepositorInsertAngiotensin-converting enzyme 2 (ACE2 Human)
TagsFLAG tagExpressionMammalianMutationArginine (R) at position 671 on ACE2 is replaced …PromoterCMVAvailable SinceAug. 11, 2025AvailabilityAcademic Institutions and Nonprofits only -
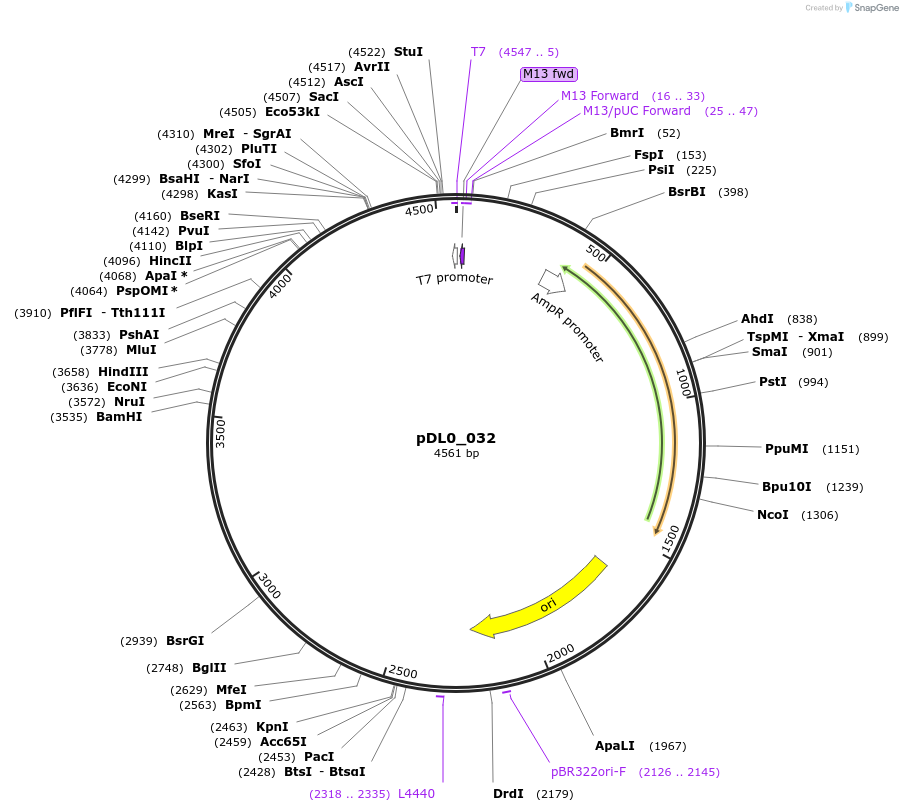
pDL0_032
Plasmid#210756PurposePU_Oryza sativa Ubiquitin3 promoterDepositorInsertOsUbi3 promoter
UseSynthetic BiologyAvailable SinceJune 25, 2025AvailabilityAcademic Institutions and Nonprofits only -
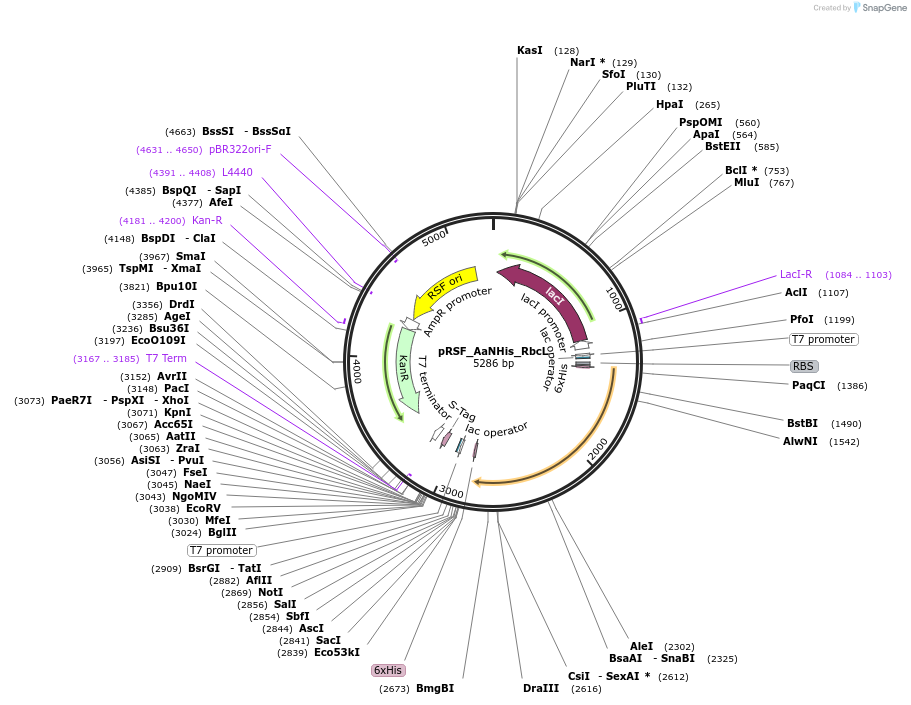
pRSF_AaNHis_RbcL
Plasmid#229517PurposeExpresses Anthoceros agrestis Rubisco large (N terminus hexahistidine)DepositorInsertRubisco large subunit
TagsHexahistidine tagExpressionBacterialPromoterT7Available SinceJune 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
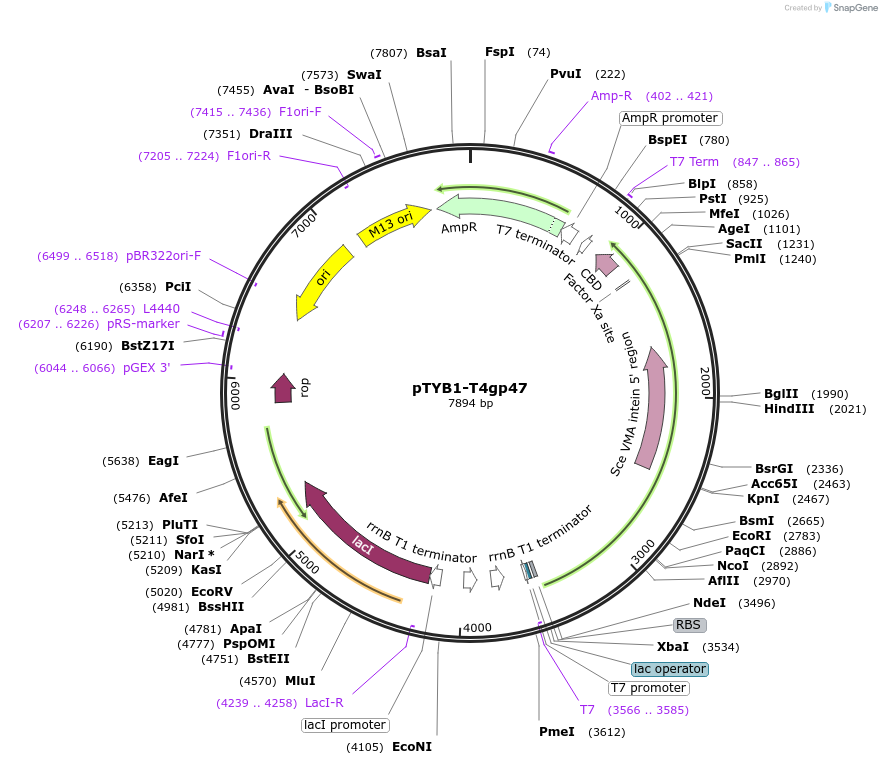
pTYB1-T4gp47
Plasmid#228212PurposeExpression in BL21(DE3) of T4 SbcD-like subunit of palindrome specific endonucleaseDepositorInsert47 (47 Escherichia phage T4)
TagsSce VMA intein-chitin binding domainExpressionBacterialPromoterT7Available SinceDec. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
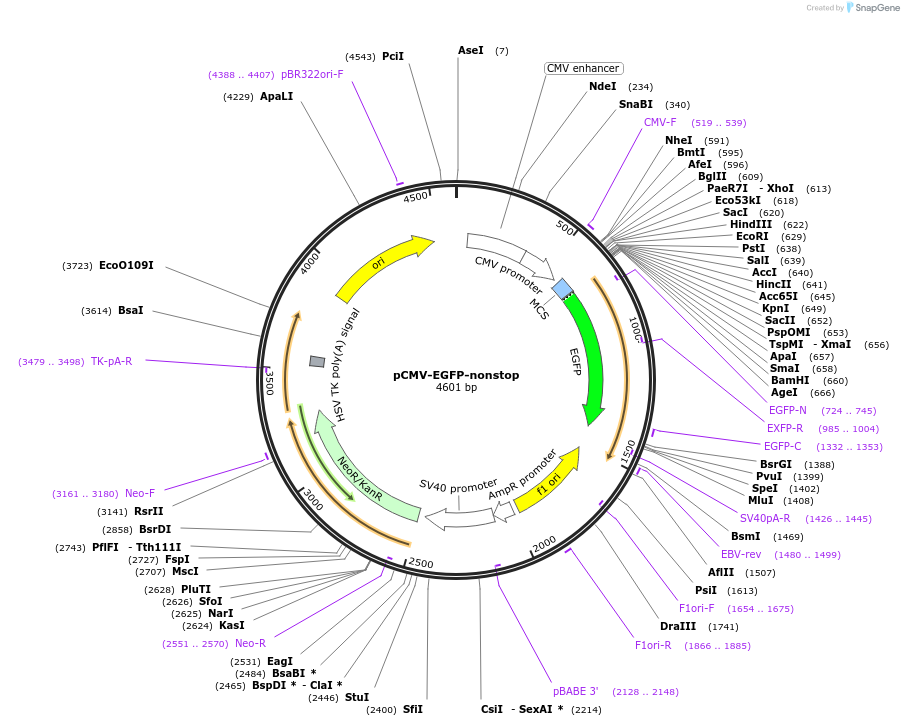
pCMV-EGFP-nonstop
Plasmid#226406PurposeExpresses EGFP without a stop codon that produces a GFP-tagged readthrough RQC substrate.DepositorInsertEGFP without stop codon
ExpressionMammalianPromoterCMVAvailable SinceNov. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
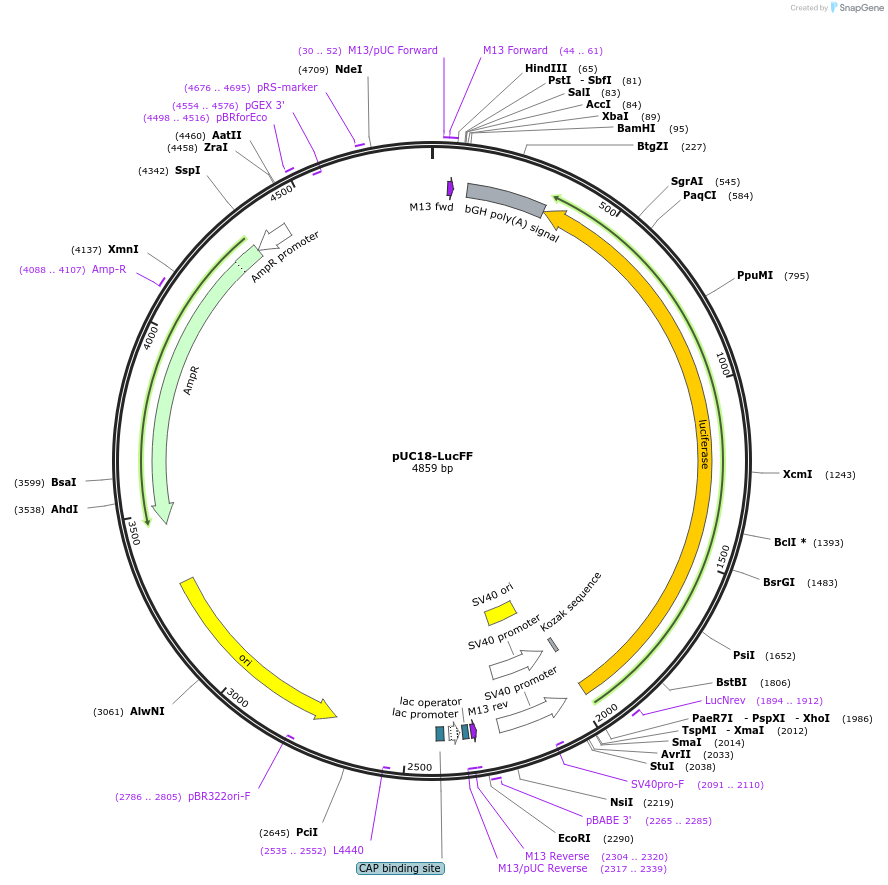
pUC18-LucFF
Plasmid#220496PurposeTo generate substrates to monitor Ku overloading and its impact on transcription at the DNA ends vicinity.DepositorInsertFirefly Luciferase
ExpressionMammalianPromoterSV40Available SinceJuly 3, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
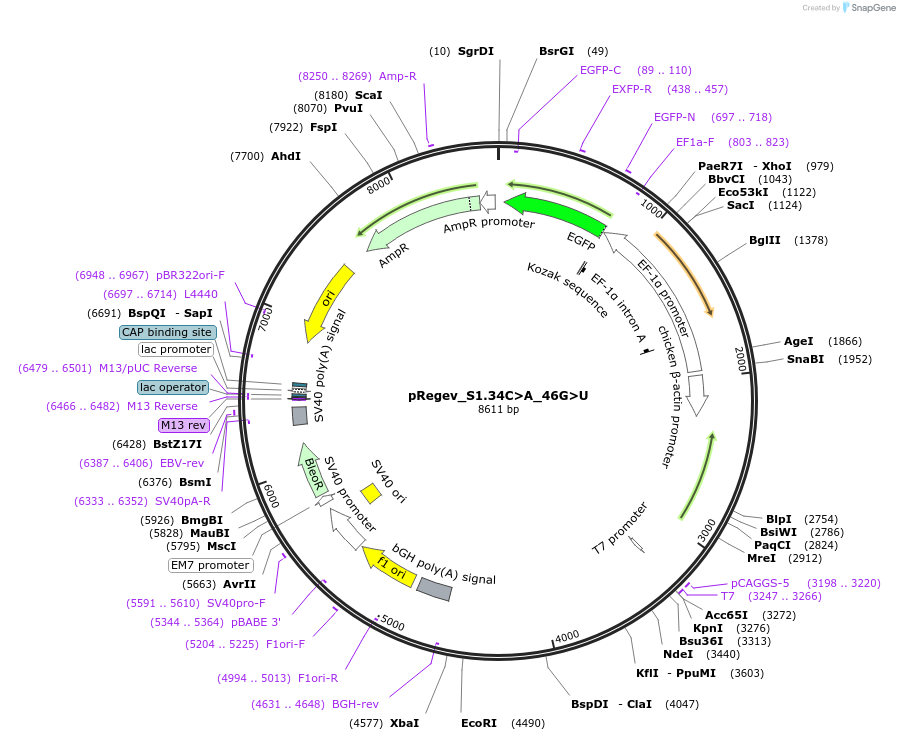
pRegev_S1.34C>A_46G>U
Plasmid#204161PurposepRegev_S1 with double mutations that restore secondary structure.DepositorInsertS1 exon with 34C>A and 46G>U substitutions in beta globin splicing minigene
UseSynthetic BiologyExpressionMammalianMutation34C>A and 46G>U mutationsPromoterTruncated CAGAvailable SinceAug. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
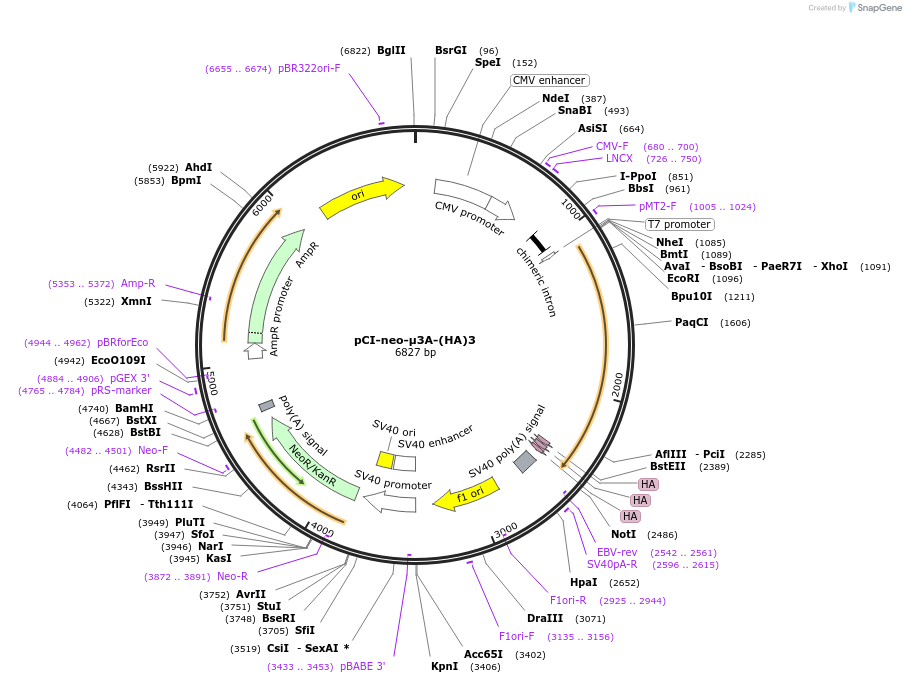
pCI-neo-μ3A-(HA)3
Plasmid#198179PurposeExpression of HA-tagged AP-3 μ3A in mammalian cellsDepositorInsertAP-3 μ3A
Tagstriple HA tagExpressionMammalianMutationsilent substitution in codon 225PromoterCMVAvailable SinceApril 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
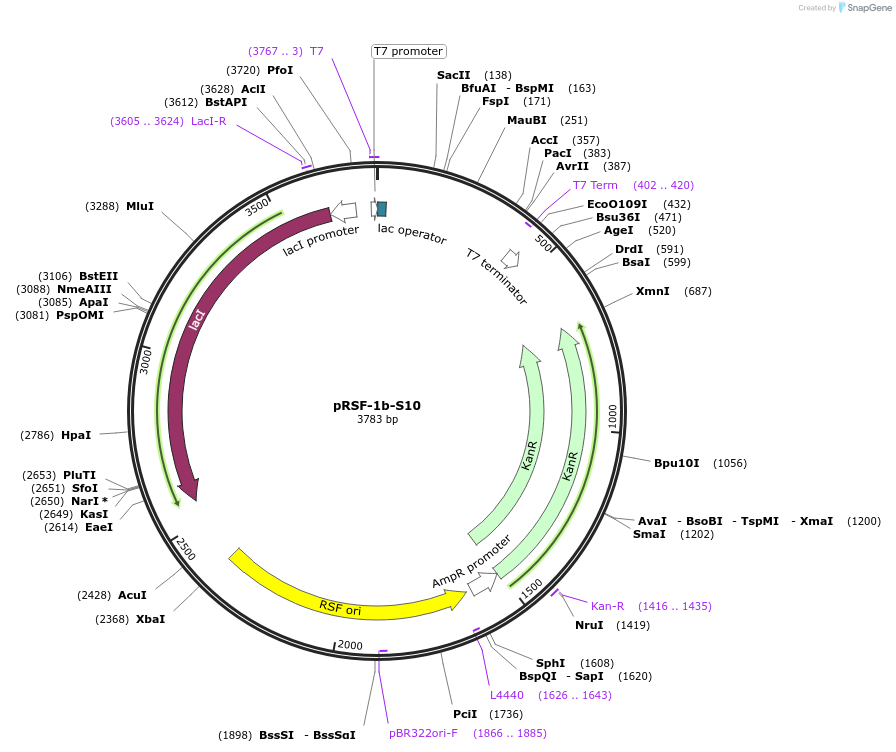
pRSF-1b-S10
Plasmid#128594PurposeExpresses tagless S10 ribosomal protein in E.coliDepositorAvailable SinceMay 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
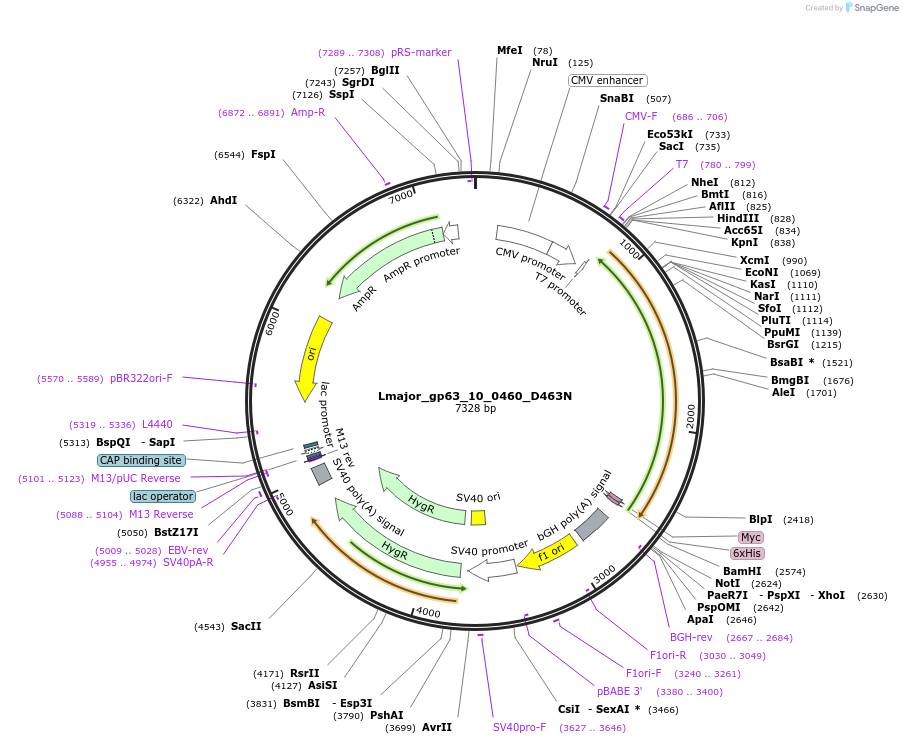
Lmajor_gp63_10_0460_D463N
Plasmid#171644PurposeExpression of L. major glcyoprotein-63 (D463N) with substrate binding site mutation from chromosome 10 in mammalian cellsDepositorInsertLmjF.10.0460 D463N
TagsMyc-HisExpressionMammalianMutationD463NAvailable SinceDec. 20, 2021AvailabilityAcademic Institutions and Nonprofits only -
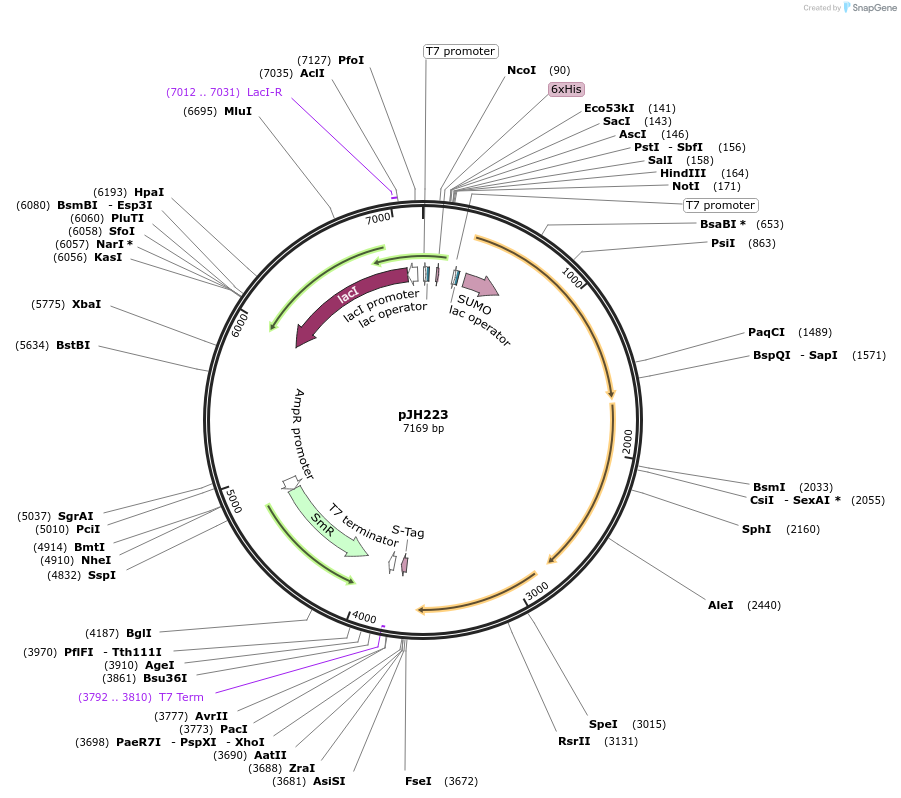
pJH223
Plasmid#172151PurposepylBCD variant in pCDF backboneDepositorInsertPT7/lac (pylBCD 34B_sub-pop5); LacI
ExpressionBacterialMutationPylB(N61S, E178K, E251D) PylD(A252V)Available SinceSept. 24, 2021AvailabilityAcademic Institutions and Nonprofits only -
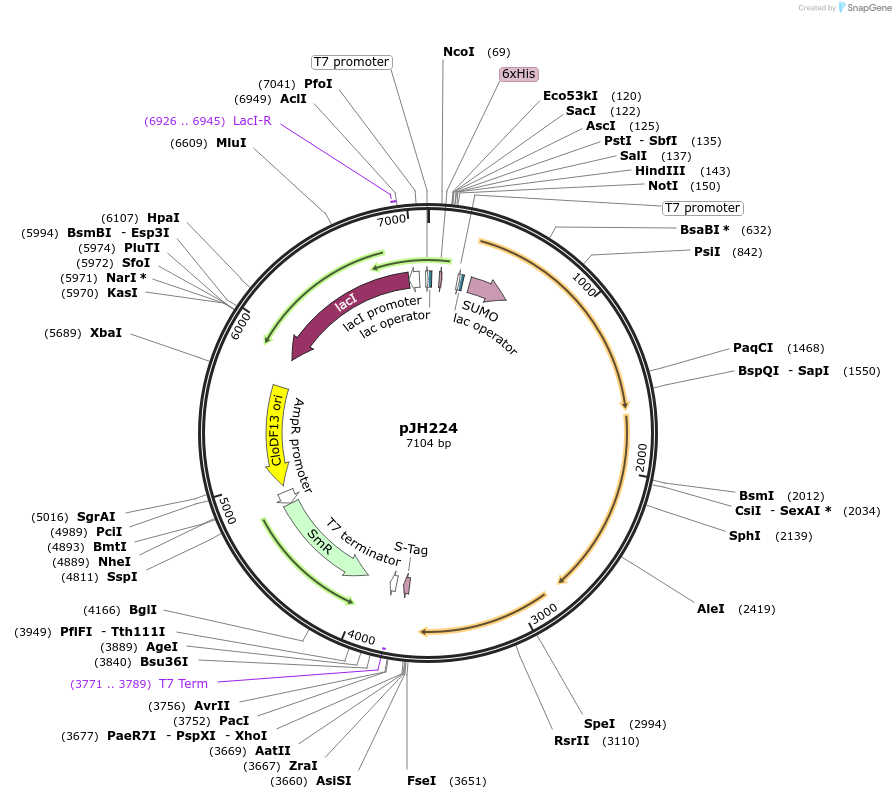
pJH224
Plasmid#172152PurposepylBCD variant in pCDF backboneDepositorInsertPT7/lac (pylBCD 40C_sub-pop6); LacI
ExpressionBacterialMutationPylB(N61S, E175K, E178K)Available SinceSept. 24, 2021AvailabilityAcademic Institutions and Nonprofits only -
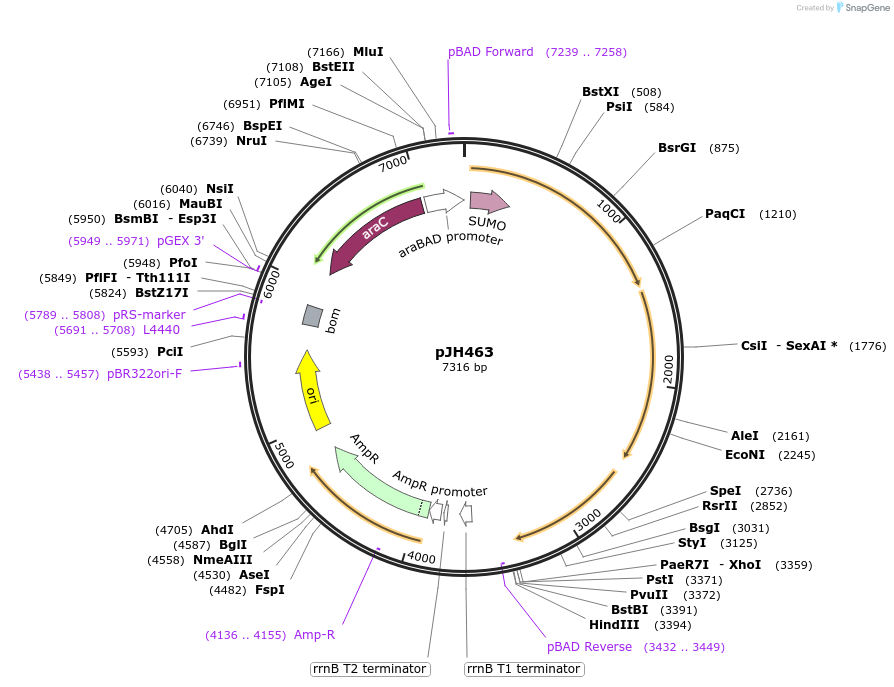
pJH463
Plasmid#172155PurposepylBCD variant in pBAD backboneDepositorInsertPara (pylBCD 36A_sub-pop1); AraC
ExpressionBacterialMutationPylB(N61S, E122K, E178K, G188R)Available SinceSept. 24, 2021AvailabilityAcademic Institutions and Nonprofits only -
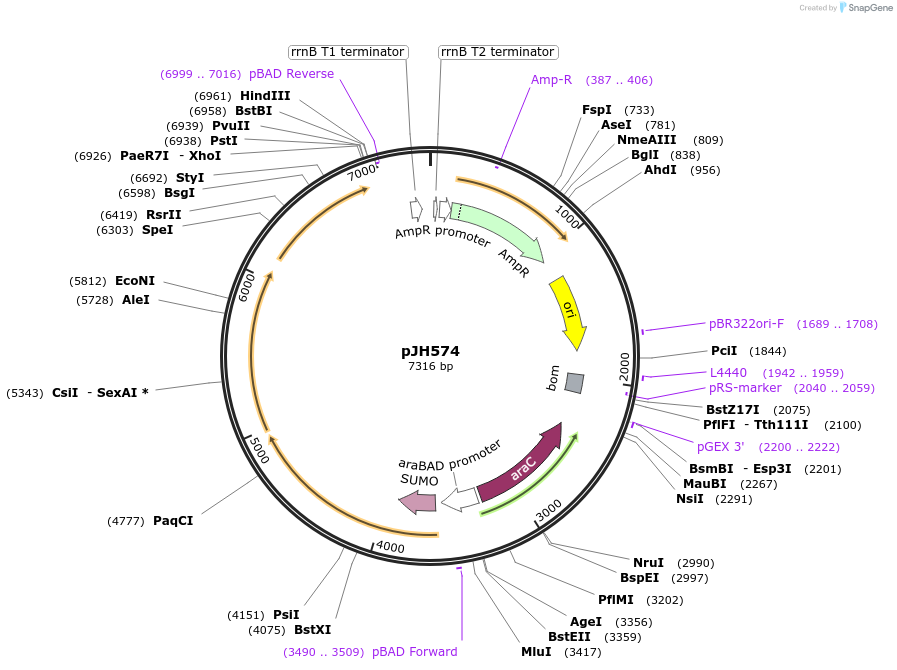
pJH574
Plasmid#172158PurposepylBCD variant in pBAD backboneDepositorInsertPara (pylBCD 36A_sub-pop2); AraC
ExpressionBacterialMutationPylB(N61S, E84K, E122K) PylC(E316D)Available SinceSept. 24, 2021AvailabilityAcademic Institutions and Nonprofits only