We narrowed to 20,614 results for: ACE;
-
Plasmid#65172PurposeEncodes tENO2 as a Type 4b part to be used in the Dueber YTK systemDepositorInserttENO2
ExpressionBacterialAvailable SinceJune 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
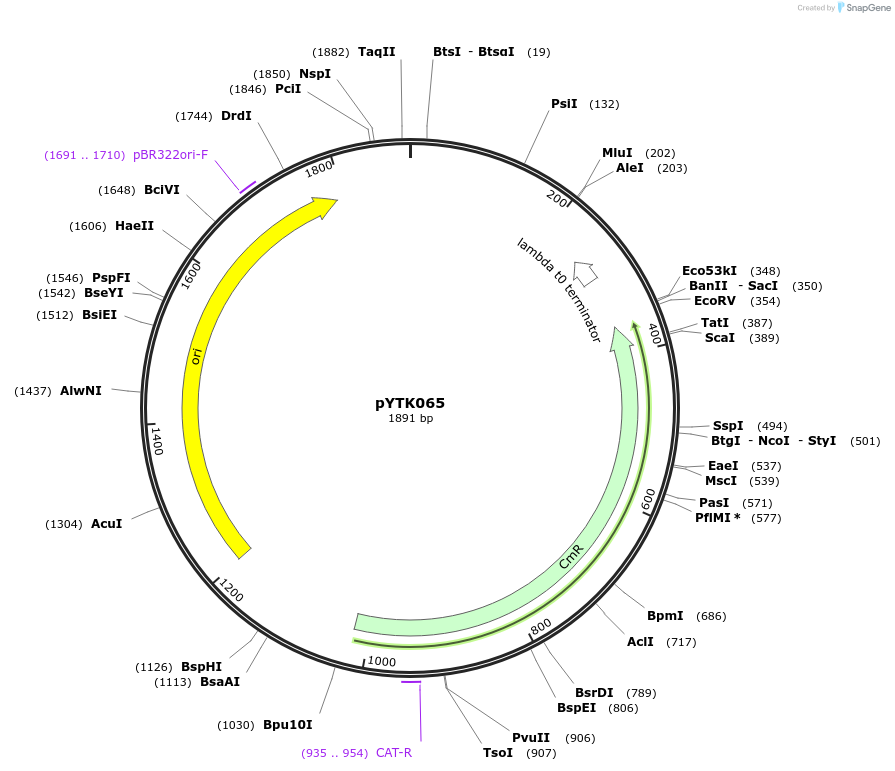
pYTK015
Plasmid#65122PurposeEncodes pHHF1 as a Type 2 part to be used in the Dueber YTK systemDepositorInsertpHHF1
ExpressionBacterialAvailable SinceJune 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
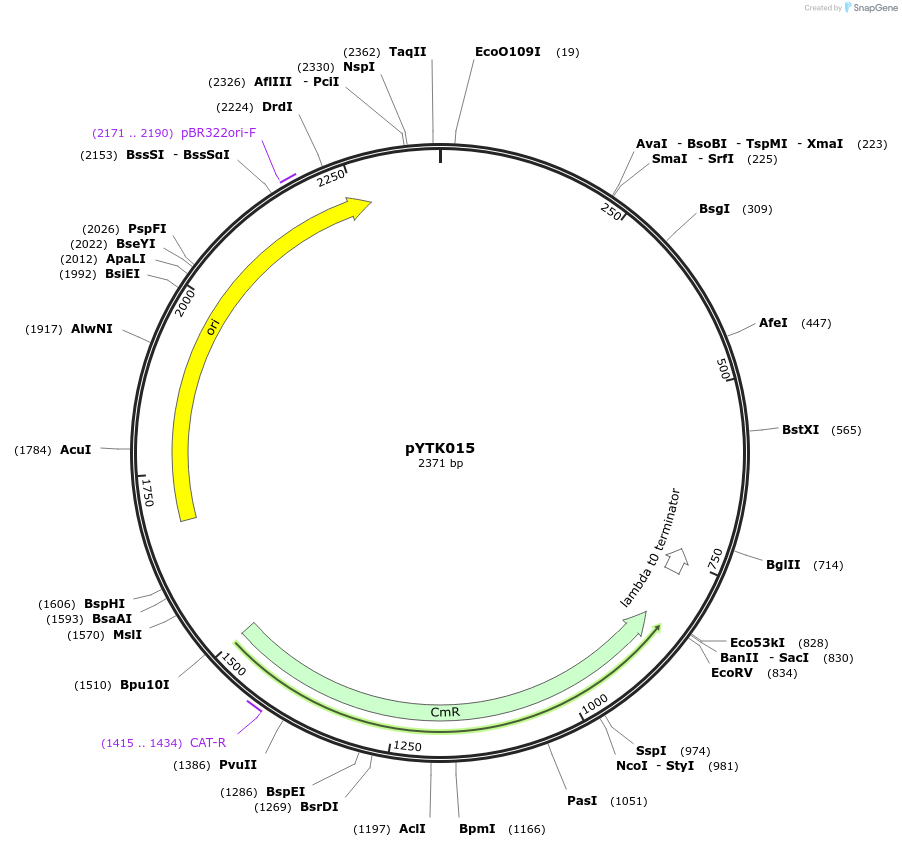
pYTK016
Plasmid#65123PurposeEncodes pHTB2 as a Type 2 part to be used in the Dueber YTK systemDepositorInsertpHTB2
ExpressionBacterialAvailable SinceJune 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
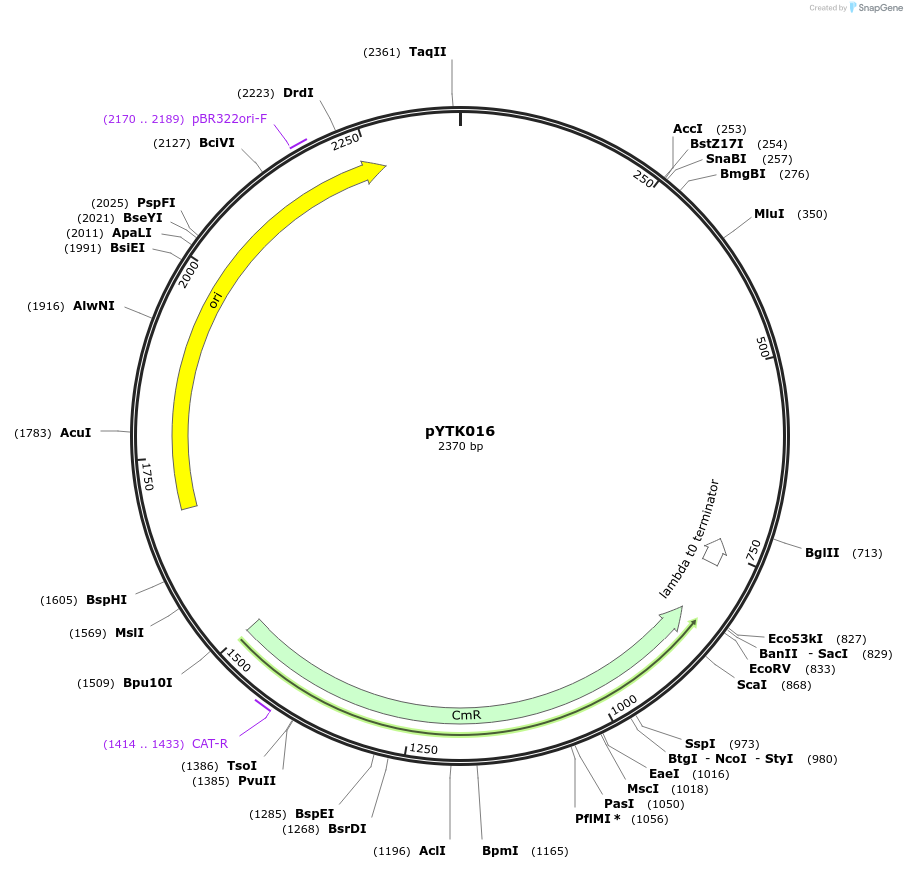
pYTK018
Plasmid#65125PurposeEncodes pALD6 as a Type 2 part to be used in the Dueber YTK systemDepositorInsertpALD6
ExpressionBacterialAvailable SinceJune 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
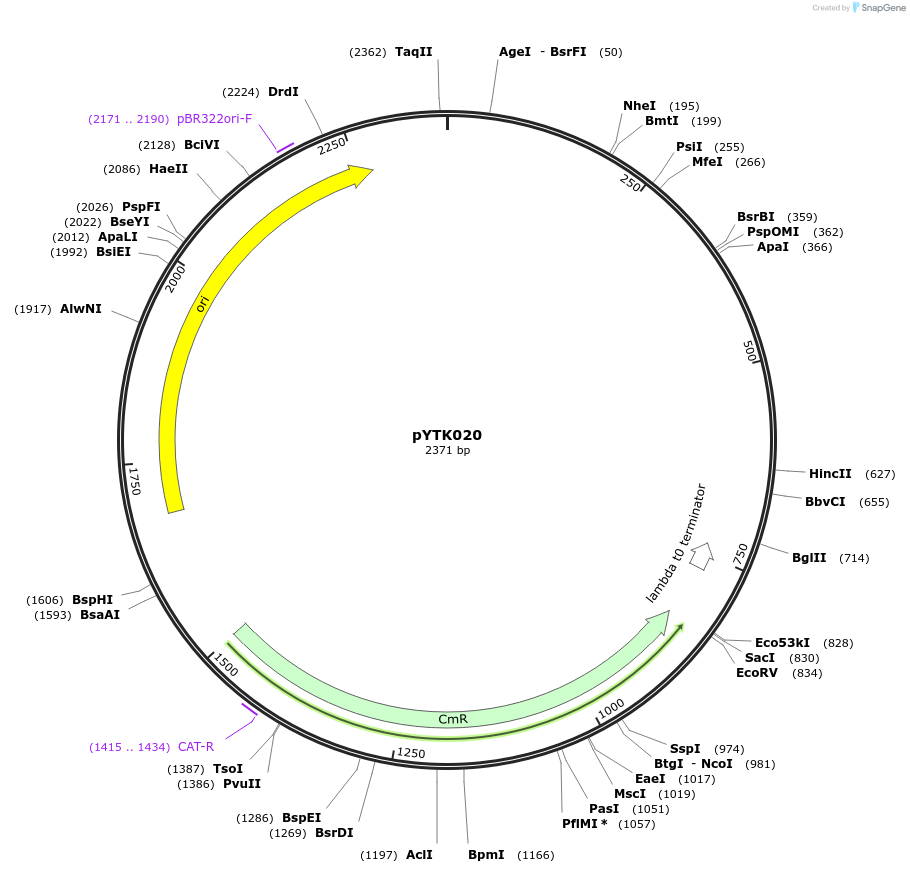
pYTK020
Plasmid#65127PurposeEncodes pRET2 as a Type 2 part to be used in the Dueber YTK systemDepositorInsertpRET2
ExpressionBacterialAvailable SinceJune 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
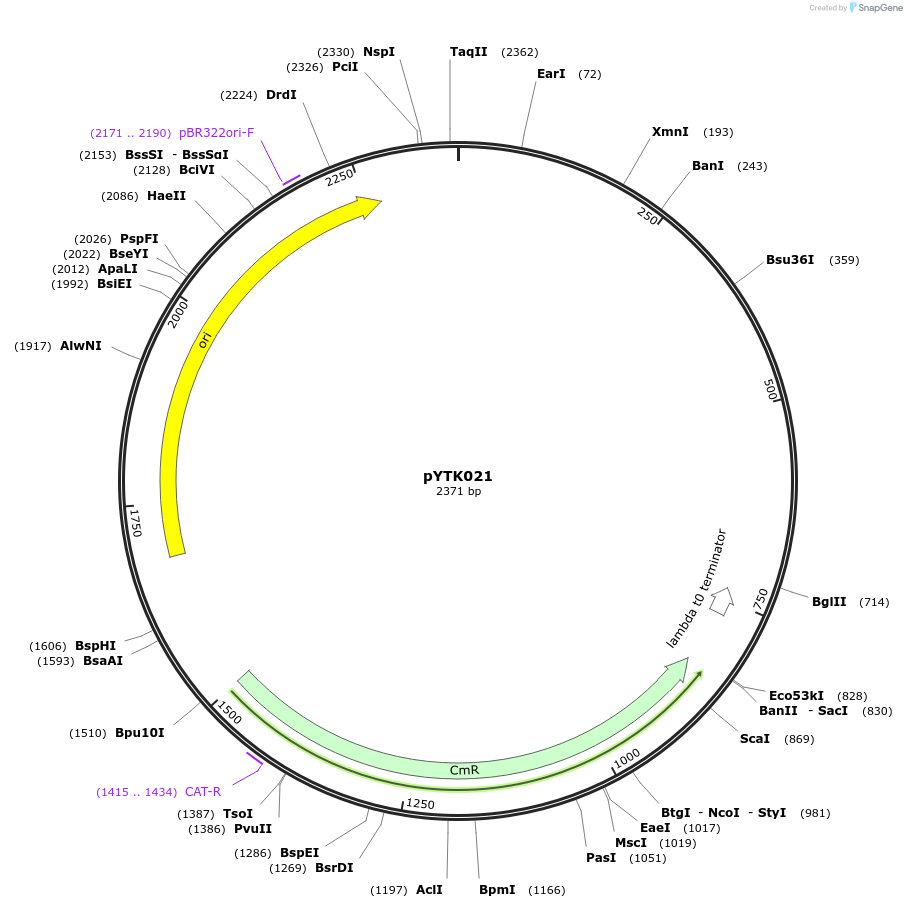
pYTK021
Plasmid#65128PurposeEncodes pRNR1 as a Type 2 part to be used in the Dueber YTK systemDepositorInsertpRNR1
ExpressionBacterialAvailable SinceJune 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
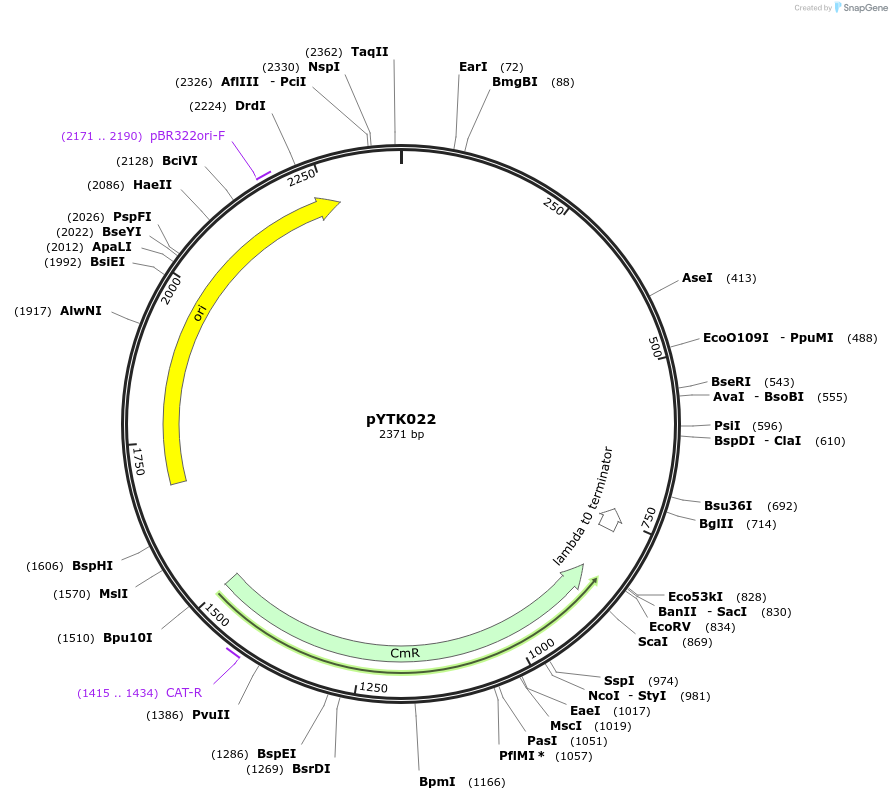
pYTK022
Plasmid#65129PurposeEncodes pSAC6 as a Type 2 part to be used in the Dueber YTK systemDepositorInsertpSAC6
ExpressionBacterialAvailable SinceJune 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
pRPR1_c2gRNA_RPR1t
Plasmid#64380Purposeencodes c2 gRNADepositorInsertc2 gRNA
ExpressionYeastAvailable SinceMay 5, 2015AvailabilityAcademic Institutions and Nonprofits only -
pRPR1_g1gRNA_RPR1t
Plasmid#64387Purposeencodes g1 gRNADepositorInsertg1 gRNA
ExpressionYeastAvailable SinceMay 5, 2015AvailabilityAcademic Institutions and Nonprofits only -
pRPR1_c7gRNA_RPR1t
Plasmid#64385Purposeencodes c7 gRNADepositorInsertc7 gRNA
ExpressionYeastAvailable SinceMay 5, 2015AvailabilityAcademic Institutions and Nonprofits only -
pRPR1_c1gRNA_RPR1t
Plasmid#64379Purposeencodes c1 gRNADepositorInsertc1 gRNA
ExpressionYeastAvailable SinceMay 5, 2015AvailabilityAcademic Institutions and Nonprofits only -
pRPR1_c5gRNA_RPR1t
Plasmid#64383Purposeencodes c5 gRNADepositorInsertc5 gRNA
ExpressionYeastAvailable SinceMay 5, 2015AvailabilityAcademic Institutions and Nonprofits only -
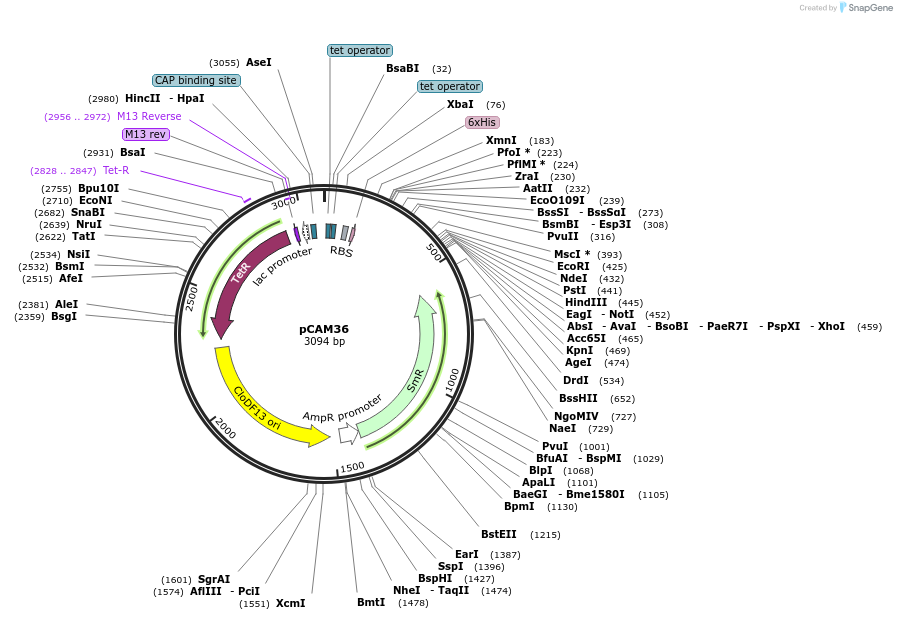
pCAM36
Plasmid#86476PurposeExpresses 6His-p9DepositorInsert6His-p9
UseSynthetic BiologyTags6HisExpressionBacterialPromoterpTetAvailabilityAcademic Institutions and Nonprofits only -
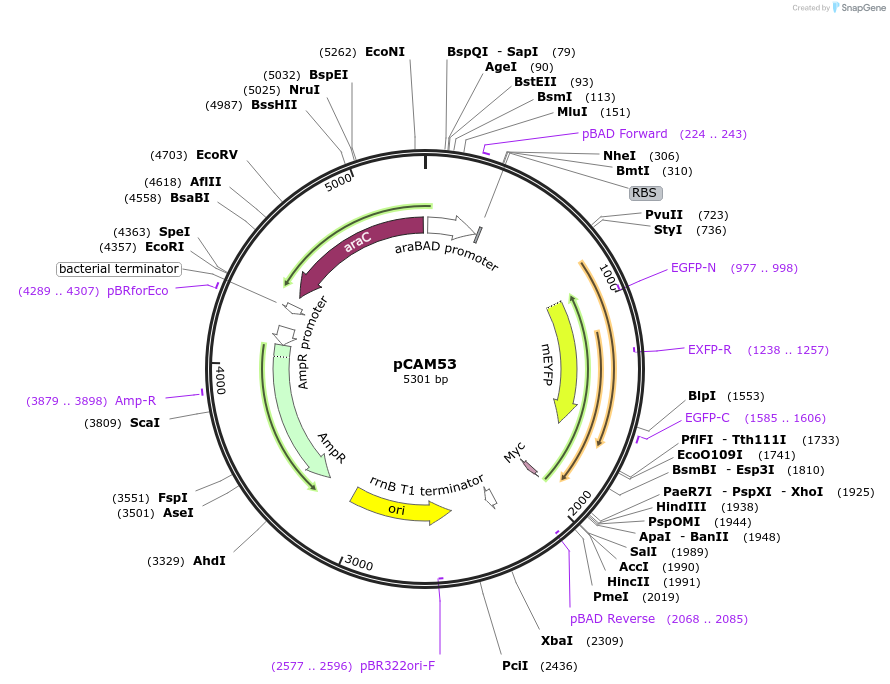
pCAM53
Plasmid#86477PurposeExpresses p12 and mCitrine-p9DepositorInsertp12, mCitrine-p9
UseSynthetic BiologyTagsmCitrineExpressionBacterialPromoterpBADAvailabilityAcademic Institutions and Nonprofits only -
E. coli BW25113 ΔtnaA ΔtrpR
Bacterial Strain#205015PurposeUsed for the characterization of the TrpR1-PtrpO1 biosensor system with the deletion of tnaA gene and trpR gene (no antibiotics marker)DepositorBacterial ResistanceNoneSpeciesEscherichia coliAvailable SinceAug. 3, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
ATMY-C321
Bacterial Strain#218774PurposeE. coli strain, genotype: C321.ΔA pUltraBR-MjY ΔtyrS tyrTV::tolC tyrU::GentR lambda::ZeoRDepositorBacterial ResistanceSpectinomycin (50 ug/mL), Gentamycin (10 ug/mL)SpeciesEscherichia coliAvailable SinceJune 28, 2024AvailabilityAcademic Institutions and Nonprofits only -
JEC027
Bacterial Strain#180310PurposeE. coli MG1655 strain with knockout of endogenous Type I-E CRISPR Cas systemDepositorBacterial ResistanceNoneAvailable SinceMay 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
rEc.13.delA
Bacterial Strain#69495PurposeRecoded strain from MG1655, prfA deletedDepositorBacterial ResistanceAmpicillinAvailable SinceOct. 1, 2015AvailabilityAcademic Institutions and Nonprofits only -
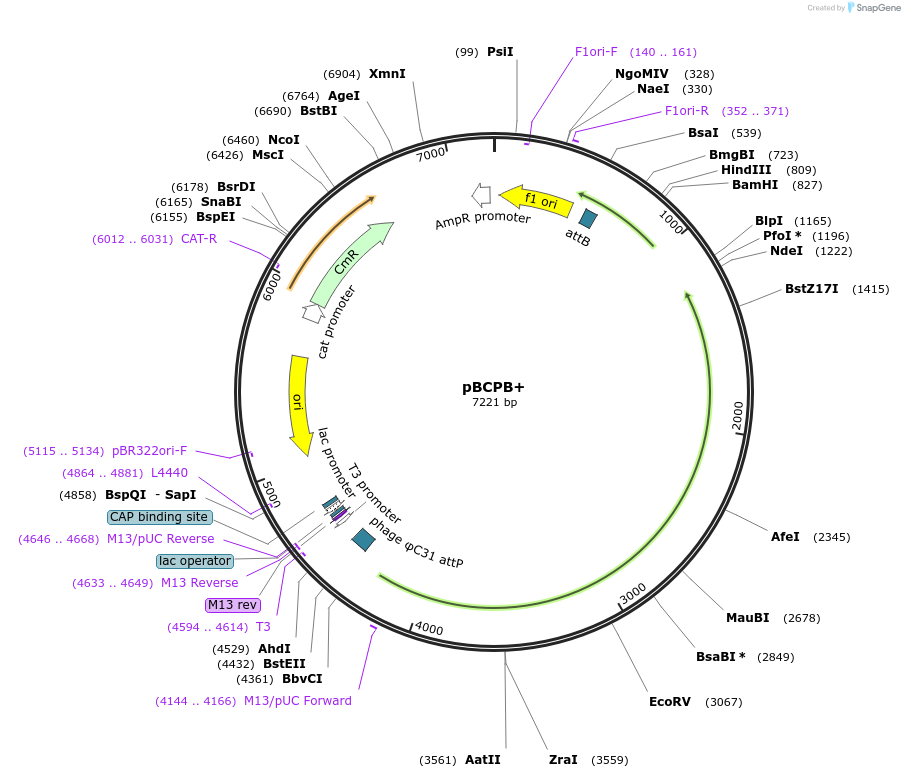
pBCPB+
Plasmid#18940DepositorInsertattP, lacZ, attB
ExpressionBacterialAvailable SinceAug. 13, 2008AvailabilityAcademic Institutions and Nonprofits only -
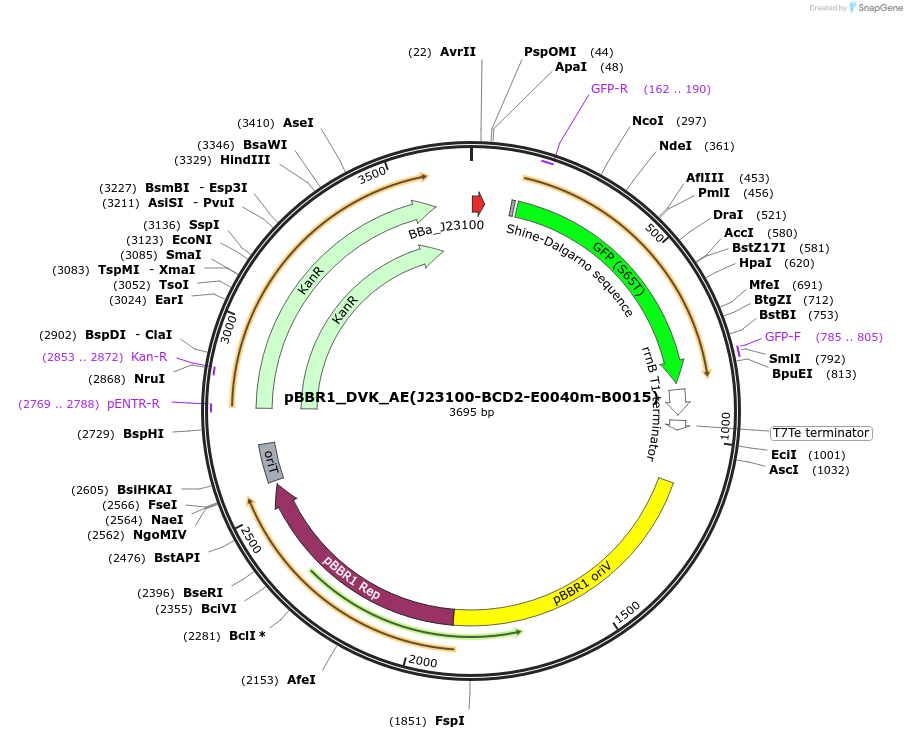
pBBR1_DVK_AE(J23100-BCD2-E0040m-B0015)
Plasmid#197660PurposeExpresses GFP under the control of J23100 and BCD2DepositorInsertGFP
UseSynthetic BiologyExpressionBacterialAvailable SinceJune 12, 2023AvailabilityAcademic Institutions and Nonprofits only