We narrowed to 3,022 results for: Rho;
-
Plasmid#195006Purposeterminator from Agrobacterium tumefaciens nopaline synthase (tNOS, MF116010) for T2 position (H/I)DepositorInsert(H) tNOS (I)
UseTagsExpressionBacterialMutationPromoterAvailable SinceFeb. 28, 2023AvailabilityAcademic Institutions and Nonprofits only -
(E) tNOS (F) _T1 (SBE118)
Plasmid#194997Purposeterminator from Agrobacterium tumefaciens nopaline synthase (tNOS, MF116010) for T1 position (E/F)DepositorInsert(E) tNOS (F)
UseTagsExpressionBacterialMutationPromoterAvailable SinceFeb. 28, 2023AvailabilityAcademic Institutions and Nonprofits only -
(I) pXYL (J) P3 (SBE108)
Plasmid#194991Purposepromoter of the gene xylose reductase from R. toruloides for a position P3DepositorInsert(I) pXYL (J)
UseTagsExpressionBacterialMutationPromoterAvailable SinceFeb. 28, 2023AvailabilityAcademic Institutions and Nonprofits only -
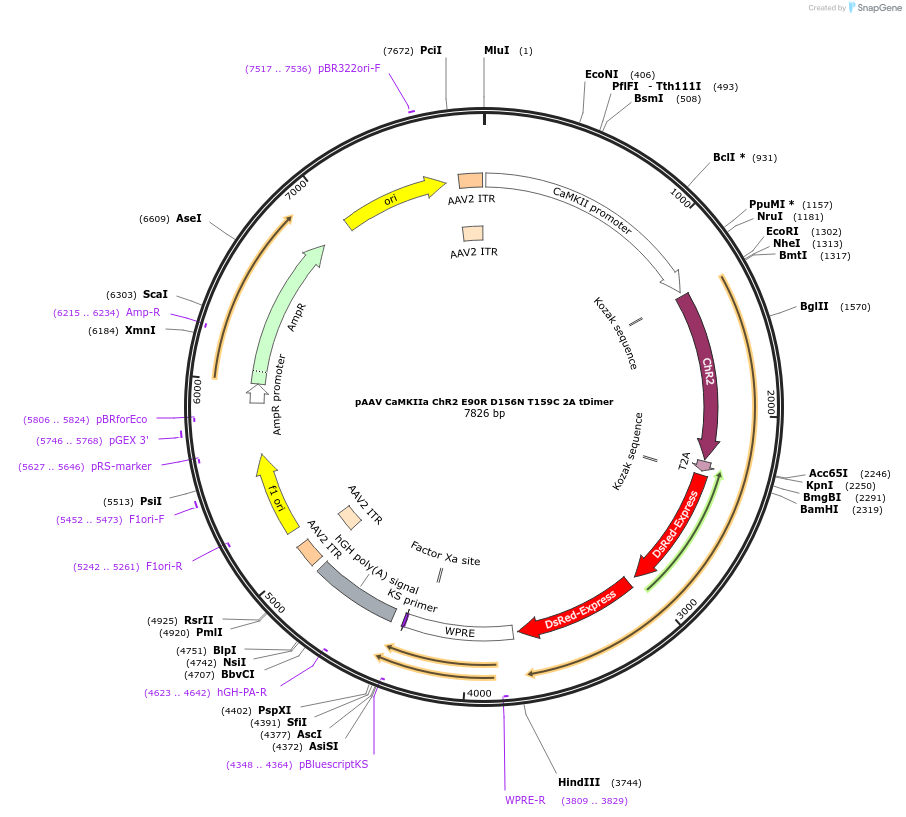
pAAV CaMKIIa ChR2 E90R D156N T159C 2A tDimer
Plasmid#52878PurposeChloride-conducting Channelrhodopsin with slow kinetics, neuron-specific promoter, plus red fluorescent protein, excitatory neuronsDepositorInsertsChannelrhodopsin-2
red fluorescent protein
UseAAVTagsExpressionMammalianMutationE90R, D156N, T159CPromoterCaMKIIaAvailable SinceJuly 16, 2014AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3.1-GcACR_457-EYFP
Plasmid#103137PurposeAnion channelrhodopsin from Geminigera cryophila (CCMP2564) for expression in mammalian cellsDepositorInsertAnion channelrhodopsin GcACR_457
UseTagsEYFPExpressionMammalianMutationhuman codon optimizedPromoterCMVAvailable SinceNov. 29, 2017AvailabilityAcademic Institutions and Nonprofits only -
pcDNA5_NLS-Halo-eGFP-BASU
Plasmid#218449Purposeexpressed the NLS-Halo-eGFP-BASU fusion protein via stable Flp-In integrationDepositorInsertNLS-Halo-eGFP-BASU
UseTagsExpressionMammalianMutationPromoterCMVAvailable SinceJune 4, 2024AvailabilityAcademic Institutions and Nonprofits only -
(R)pGPD1(D)_P1(SBE109)
Plasmid#194993Purposepromoter of the gene glyceraldehyde-3-phosphate dehydrogenase from R. toruloides for a P1 position (overhangs R/D)DepositorInsert(R) pGPD (D)
UseTagsExpressionBacterialMutationPromoterAvailable SinceFeb. 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
(F)pGPD(G)_P2 (SBE110)
Plasmid#194992Purposepromoter of the gene glyceraldehyde-3-phosphate dehydrogenase from R. toruloides for a P2 position (overhangs F/G)DepositorInsert(F) pGPD (G)
UseTagsExpressionBacterialMutationPromoterAvailable SinceDec. 7, 2023AvailabilityAcademic Institutions and Nonprofits only -
(I) pADH2 (J) P3 (SBE114)
Plasmid#187629Purposepromoter of the gene alcohol dehydrogenase 2 from R. toruloides for the P3 position (I/J)DepositorInsert(I) pADH2 (J)
UseTagsExpressionBacterialMutationPromoterAvailable SinceDec. 7, 2023AvailabilityAcademic Institutions and Nonprofits only -
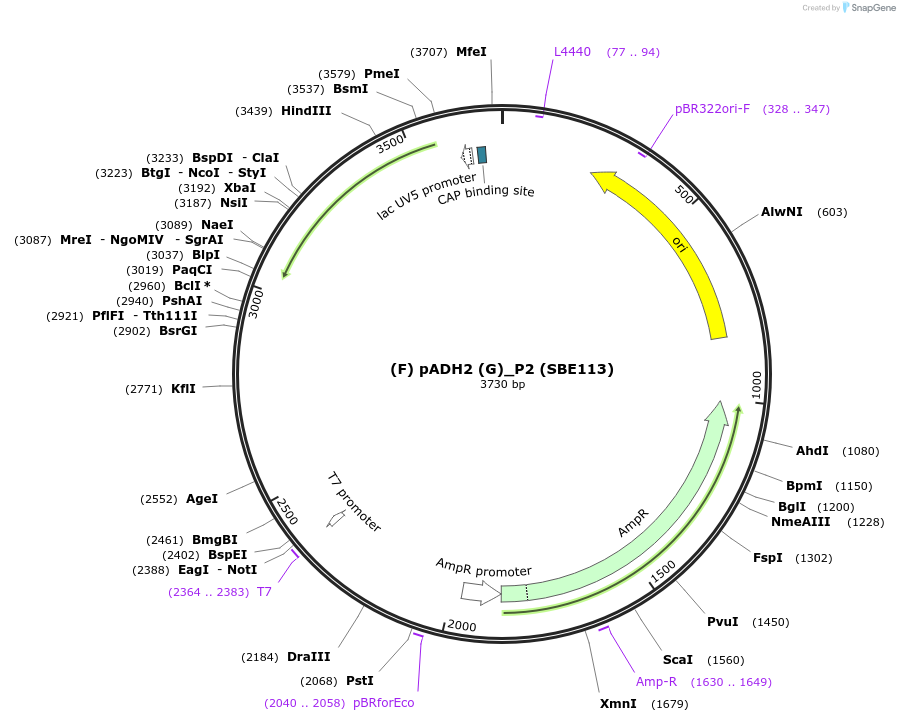
(F) pADH2 (G)_P2 (SBE113)
Plasmid#187628Purposepromoter of the gene alcohol dehydrogenase 2 from R. toruloides fro the P2 position (F/G)DepositorInsert(F) pADH2 (G)
UseTagsExpressionBacterialMutationPromoterAvailable SinceDec. 7, 2023AvailabilityAcademic Institutions and Nonprofits only -
(F) pXYL (G) P1 (SBE216)
Plasmid#195017Purposepromoter of the gene xylose reductase from R. toruloides for the position P2 (F/G)DepositorInsert(F) pXYL (G)
UseTagsExpressionBacterialMutationPromoterAvailable SinceNov. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
(R) pXYL (D) P1 (SBE215)
Plasmid#195016Purposepromoter of the gene xylose reductase from R. toruloides for the position P1 (R/D)DepositorInsert(R) pXYL (D)
UseTagsExpressionBacterialMutationPromoterAvailable SinceNov. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
(R)pADH2(D)_P1(SBE214)
Plasmid#195015Purposepromoter of the gene alcohol dehydrogenase 2 from R. toruloides for the position P1DepositorInsert(B) pADH2 (R)
UseTagsExpressionBacterialMutationPromoterAvailable SinceNov. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
(I) pGPD (J)_P3 (SBE111)
Plasmid#194990Purposepromoter of the gene glyceraldehyde-3-phosphate dehydrogenase from R. toruloides for a P3 position (I/J)DepositorInsert(I) pGPD (J)
UseTagsExpressionBacterialMutationPromoterAvailable SinceNov. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
(S) insD_ku70 (Q) (SBE139)
Plasmid#187627Purposethe insertional region downstream for the deletion of the KU70 gene in R. toruloides. Overhangs S/QDepositorInsert(S) insD KU70 (Q)
UseTagsExpressionBacterialMutationPromoterAvailable SinceNov. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
(P) insUP_ku70 (SBE138) (B)
Plasmid#187626Purposethe insertional region upstream for the deletion of the KU70 gene in R. toruloides. Overhangs P/BDepositorInsert(P) insUP KU70 (B)
UseTagsExpressionBacterialMutationPromoterAvailable SinceNov. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3.1-PsuACR_353-EYFP
Plasmid#111076PurposeAnion channelrhodopsin from Proteomonas sulcata (CCMP704) for expression in mammalian cellsDepositorInsertAnion channelrhodopsin PsuACR_353
UseTagsEYFPExpressionMammalianMutationPromoterAvailable SinceJune 4, 2018AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3.1-C1ACR_023-EYFP
Plasmid#103966PurposeAnion channelrhodopsin from unclassified cryptophyte (CCMP2293) for expression in mammalian cellsDepositorInsertAnion channelrhodopsin C1ACR_023
UseTagsEYFPExpressionMammalianMutationhuman codon optimizedPromoterCMVAvailable SinceNov. 30, 2017AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3.1-C1ACR_887-EYFP
Plasmid#103965PurposeAnion channelrhodopsin from unclassified cryptophyte (CCMP2293) for expression in mammalian cellsDepositorInsertAnion channelrhodopsin C1ACR_887
UseTagsEYFPExpressionMammalianMutationhuman codon optimizedPromoterCMVAvailable SinceNov. 30, 2017AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3.1-G1ACR_203-EYFP
Plasmid#103143PurposeAnion channelrhodopsin from Geminigera sp. (Caron Lab Isolate) for expression in mammalian cellsDepositorInsertAnion channelrhodopsin G1ACR_203
UseTagsEYFPExpressionMammalianMutationhuman codon optimizedPromoterCMVAvailable SinceNov. 29, 2017AvailabilityAcademic Institutions and Nonprofits only