We narrowed to 2,840 results for: fas
-
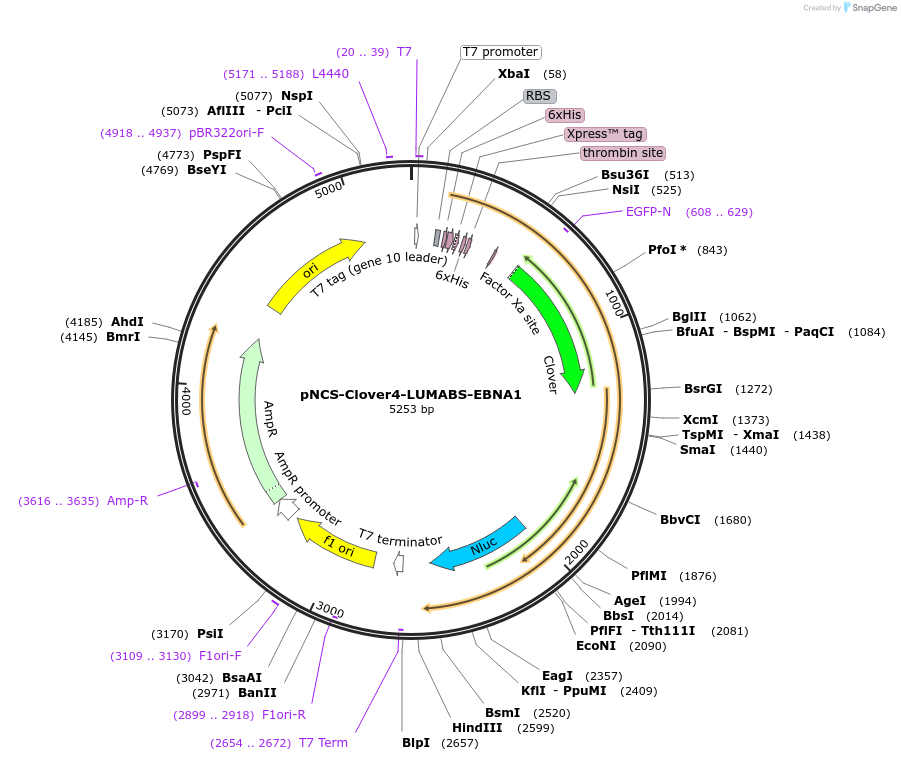
Plasmid#183488PurposeTo express the Clover4-LUMABS-EBNA1 sensor for Epstein-barr Virus antibody dection.DepositorInsertClover4-LUMABS-EBNA1
ExpressionBacterialAvailable SinceAug. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
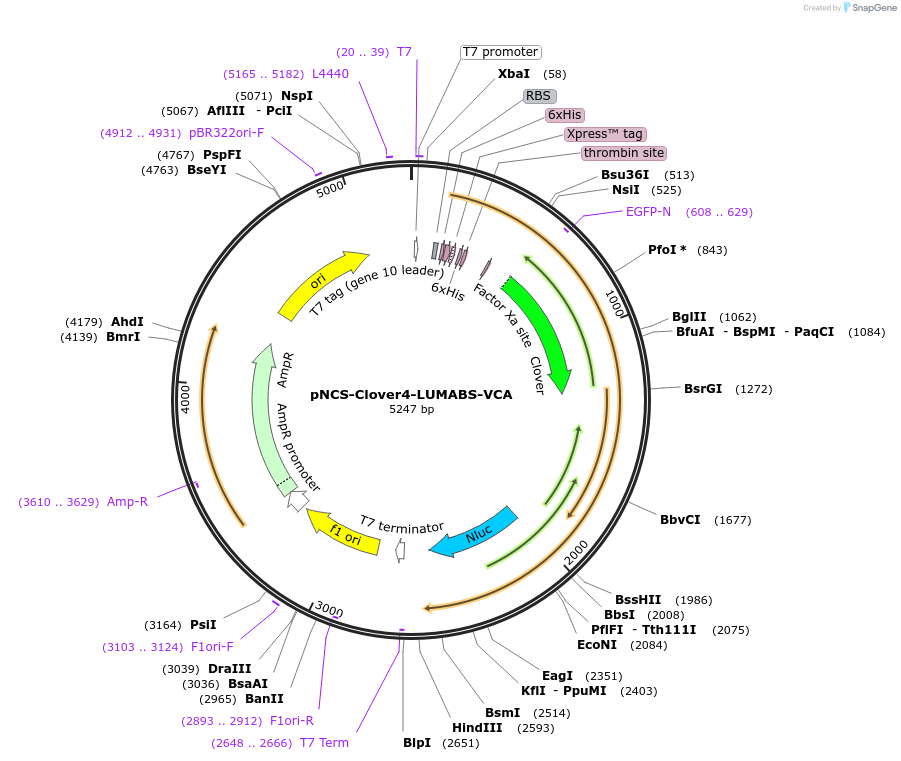
pNCS-Clover4-LUMABS-VCA
Plasmid#183489PurposeTo express the Clover4-LUMABS-VCA sensor for Epstein-barr Virus antibody dection.DepositorInsertClover4-LUMABS-VCA
ExpressionBacterialAvailable SinceAug. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
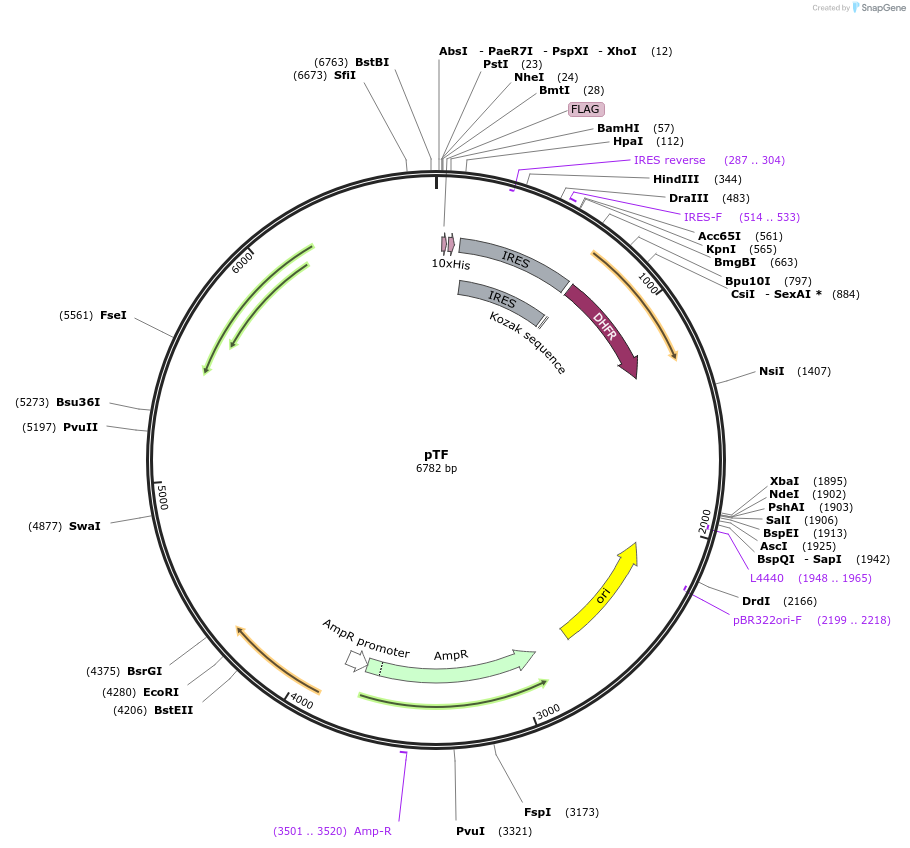
pTF
Plasmid#162784PurposeProtein expression in CHO cells - C-terminal FLAG and 10xhis tags are codedDepositorTypeEmpty backboneTags10xHis and FLAGExpressionMammalianPromoterCHO Elongation factor 1AAvailable SinceJan. 13, 2022AvailabilityAcademic Institutions and Nonprofits only -
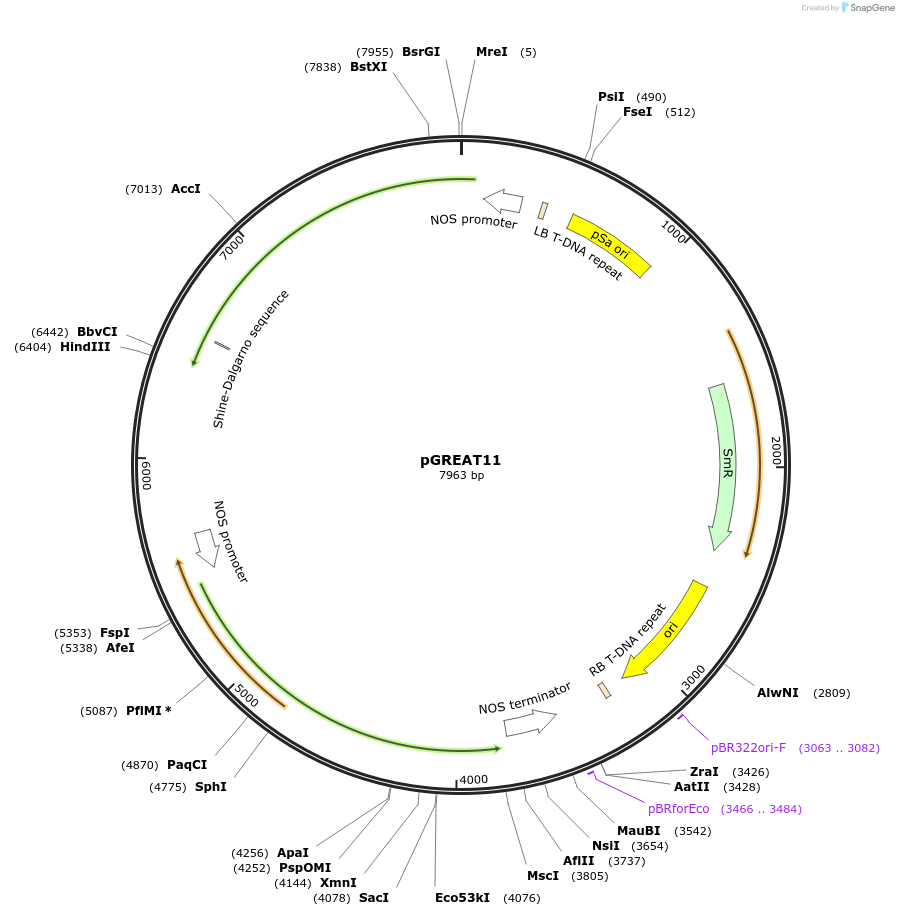
pGREAT11
Plasmid#170899PurposeGoldenBraid Double Transcriptional Unit - pNOS_E-Luc_AtACT2t+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…PromoterpNOSAvailable SinceAug. 25, 2021AvailabilityAcademic Institutions and Nonprofits only -
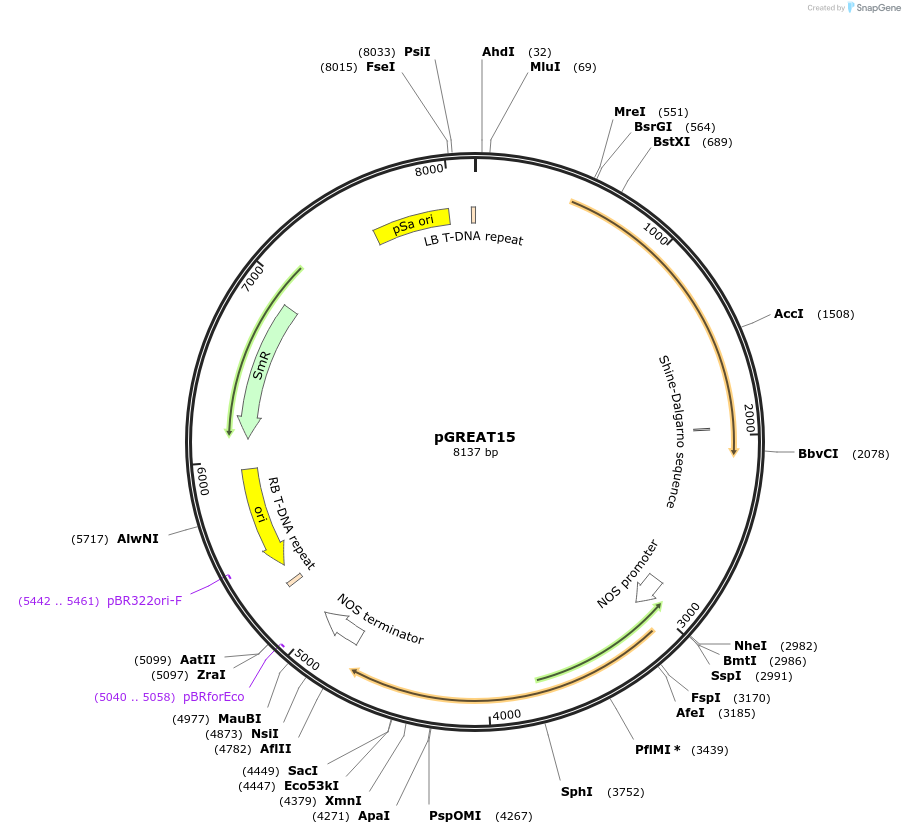
pGREAT15
Plasmid#170903PurposeGoldenBraid Double Transcriptional Unit - pCmYLCV911_E-Luc_AtACT2t+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…PromoterpCmYLCV911Available SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
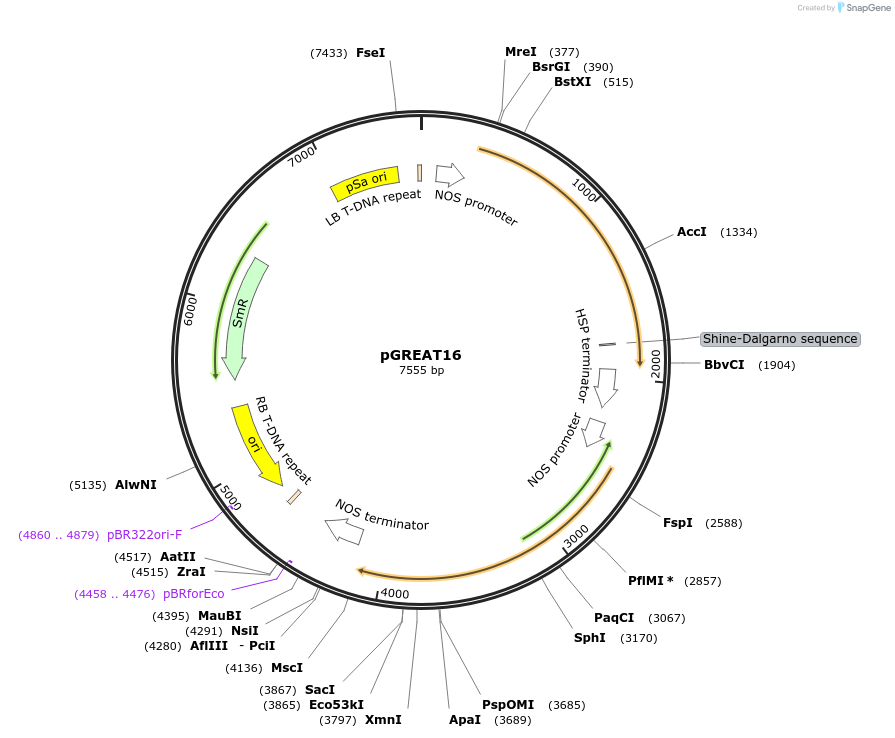
pGREAT16
Plasmid#170904PurposeGoldenBraid Double Transcriptional Unit - pNOS_E-Luc_AtHSP18.2t+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…PromoterpNOSAvailable SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
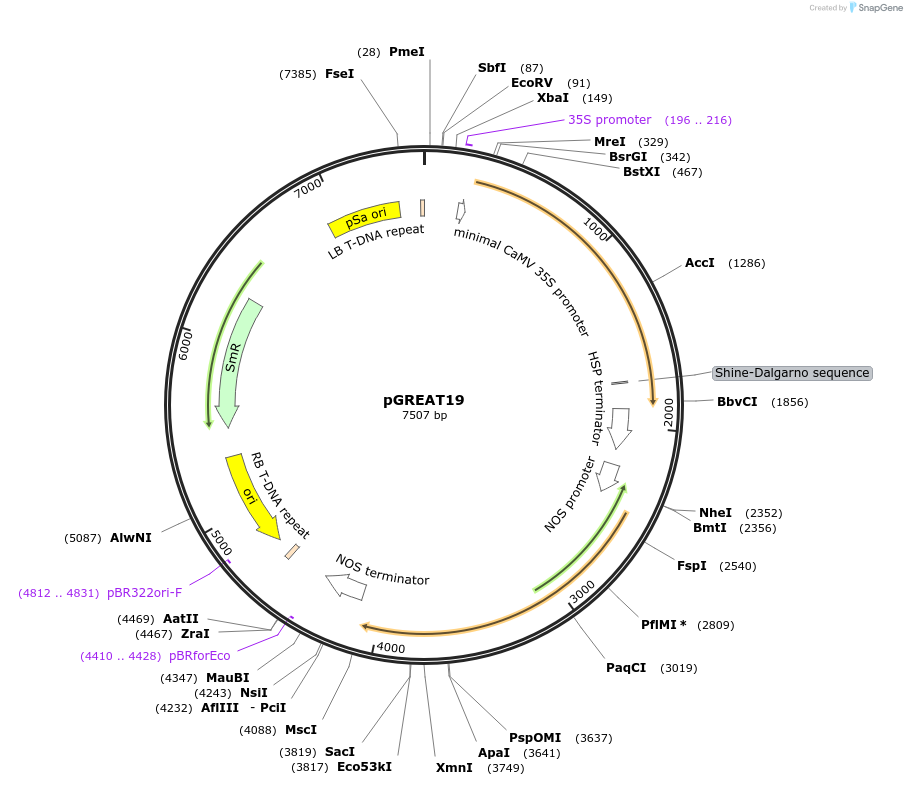
pGREAT19
Plasmid#170907PurposeGoldenBraid Double Transcriptional Unit - pG10-90_E-Luc_AtHSP18.2t+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…PromoterpG10-90Available SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
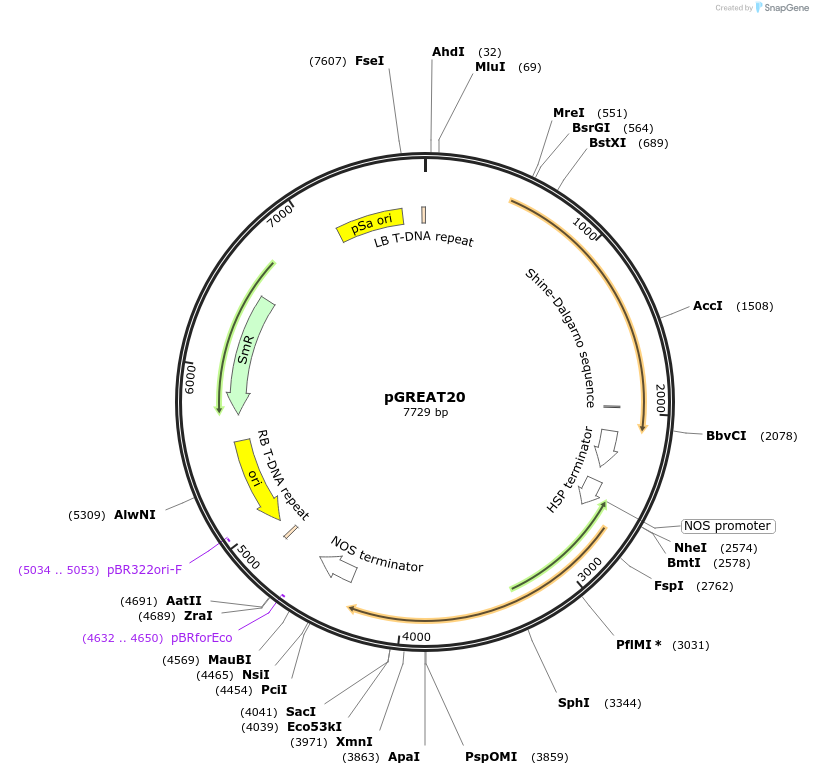
pGREAT20
Plasmid#170908PurposeGoldenBraid Double Transcriptional Unit - pCmYLCV911_E-Luc_AtHSP18.2t+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…PromoterpCmYLCV911Available SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
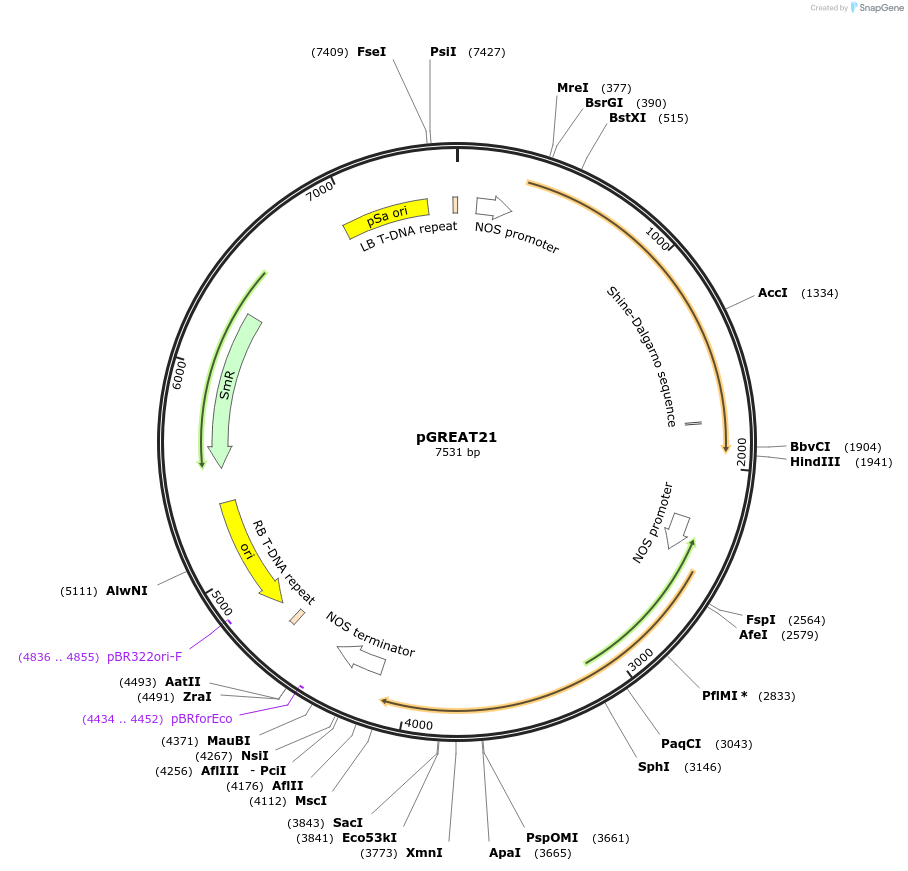
pGREAT21
Plasmid#170909PurposeGoldenBraid Double Transcriptional Unit - pNOS_E-Luc_35St+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…PromoterpNOSAvailable SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
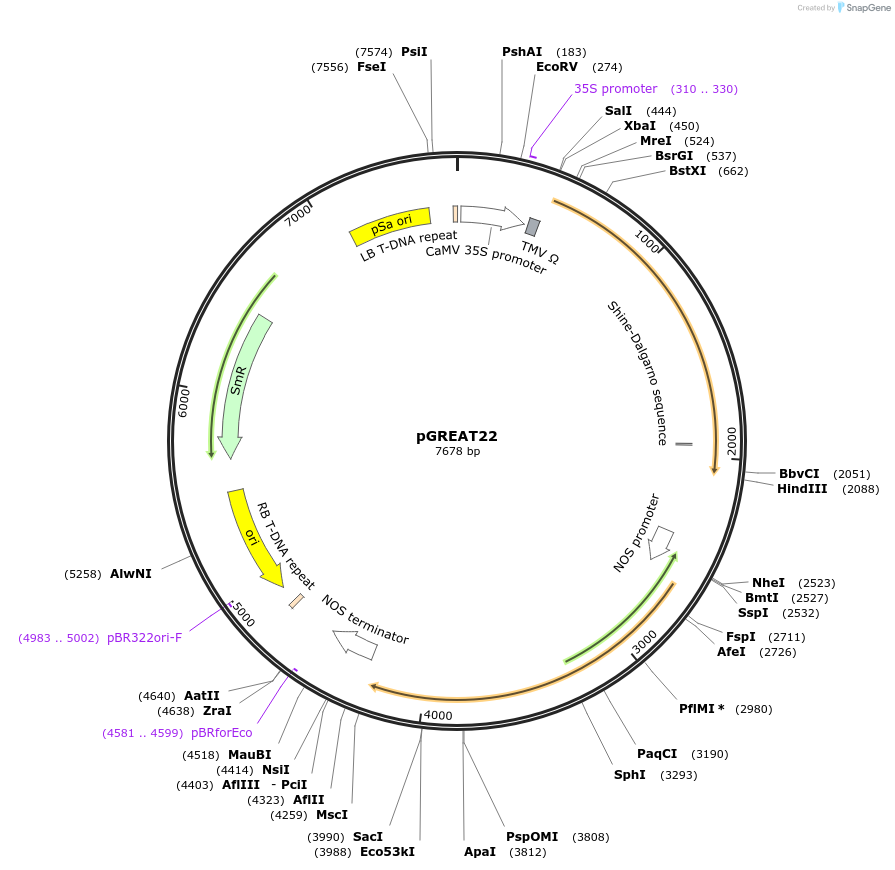
pGREAT22
Plasmid#170910PurposeGoldenBraid Double Transcriptional Unit - p35SoTMV_E-Luc_35St+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…Promoterp35SoTMVAvailable SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
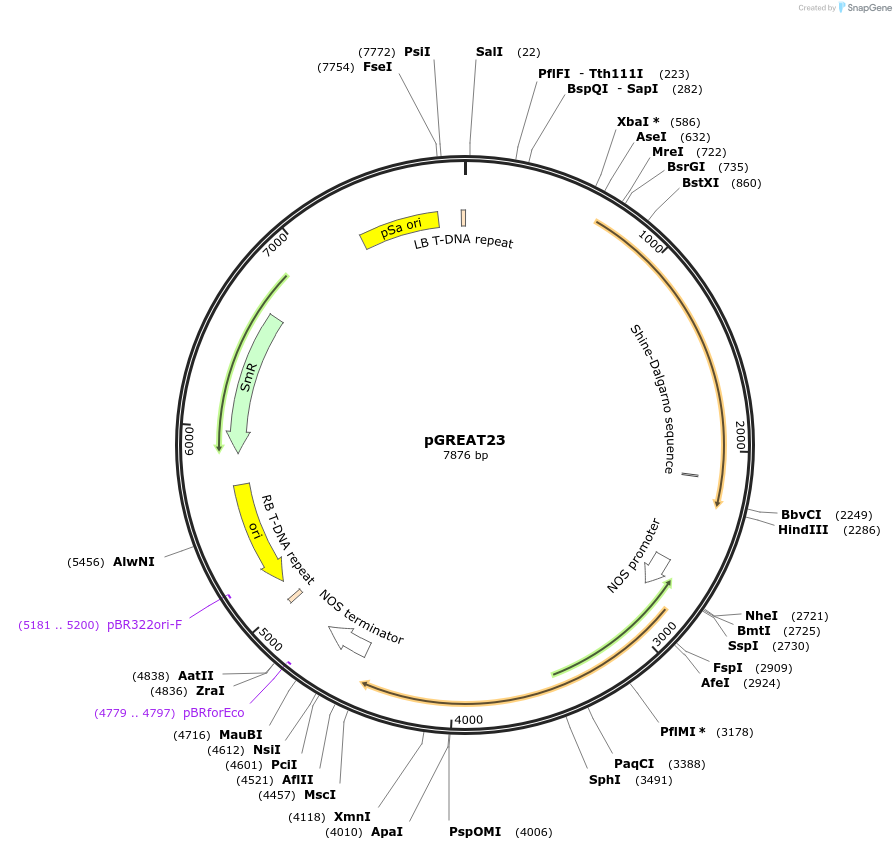
pGREAT23
Plasmid#170911PurposeGoldenBraid Double Transcriptional Unit - pAtUBQ10_E-Luc_35St+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…PromoterpAtUBQ10Available SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
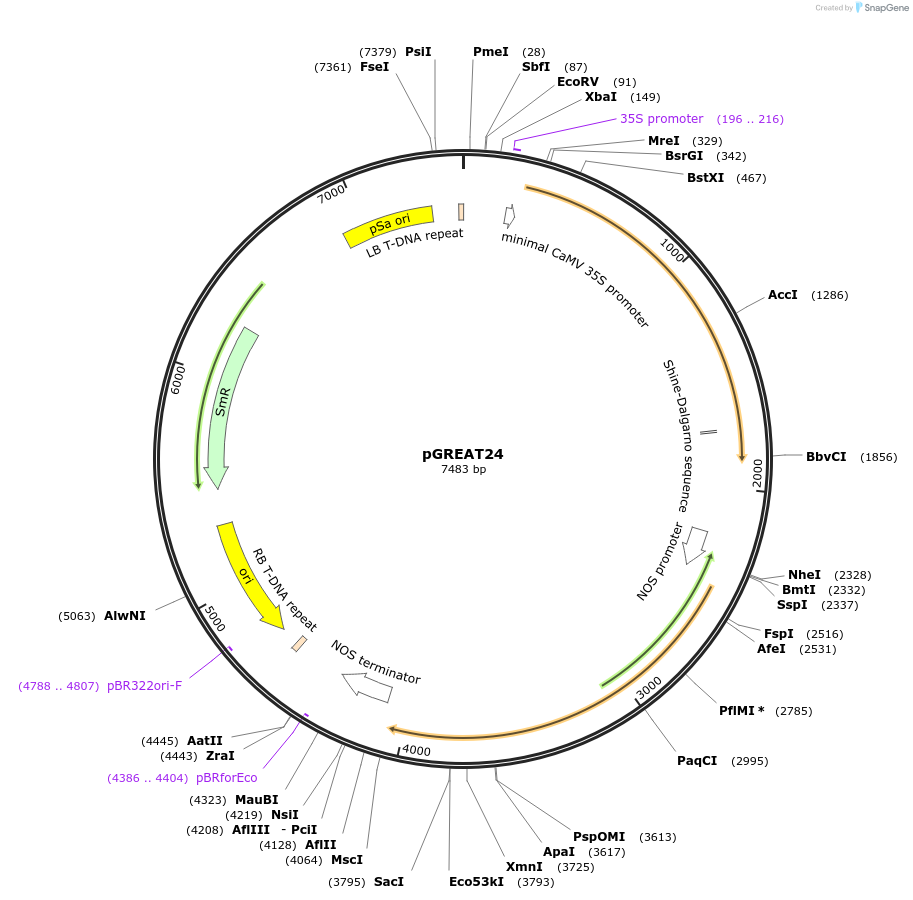
pGREAT24
Plasmid#170912PurposeGoldenBraid Double Transcriptional Unit - pG10-90_E-Luc_35St+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…PromoterpG10-90Available SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
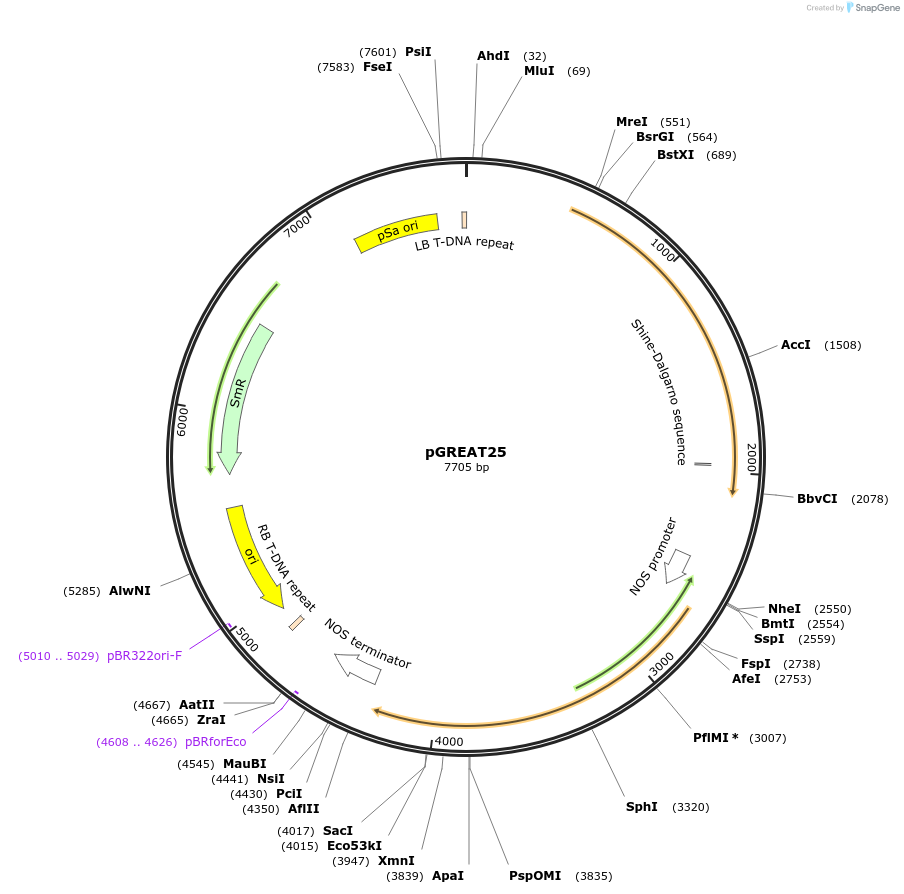
pGREAT25
Plasmid#170913PurposeGoldenBraid Double Transcriptional Unit - pCmYLCV911_E-Luc_35St+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…PromoterpCmYLCV911Available SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
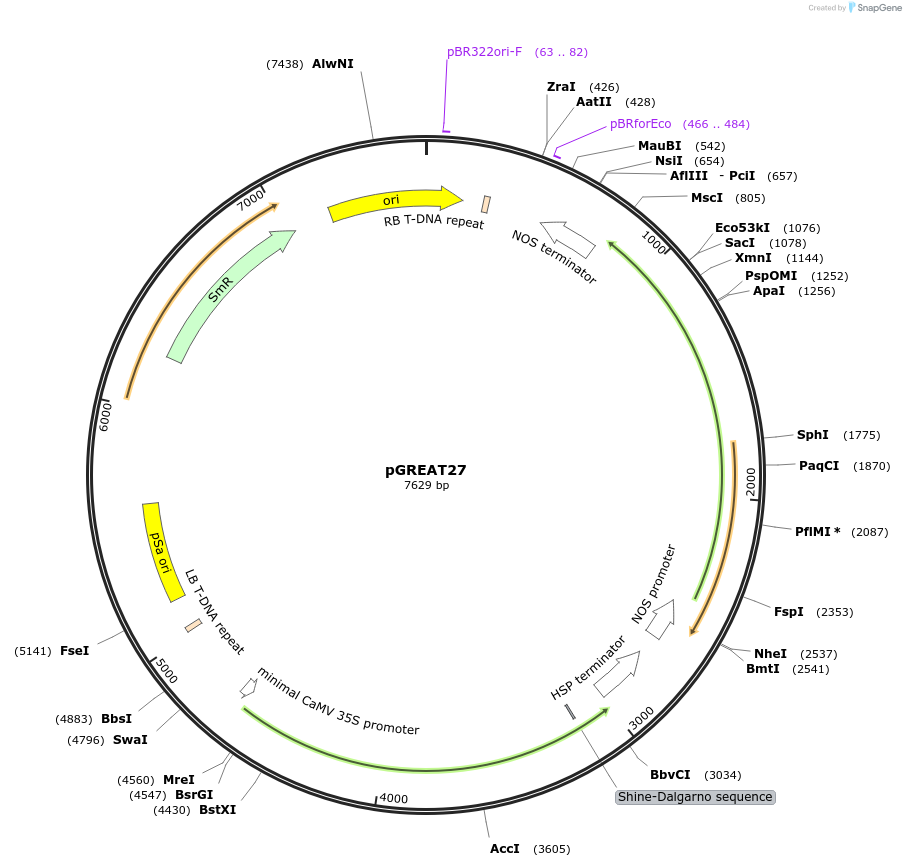
pGREAT27
Plasmid#170915PurposeGoldenBraid Double Transcriptional Unit - pAtNR1_E-Luc_AtHSP18.2t+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…PromoterpAtNR1Available SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
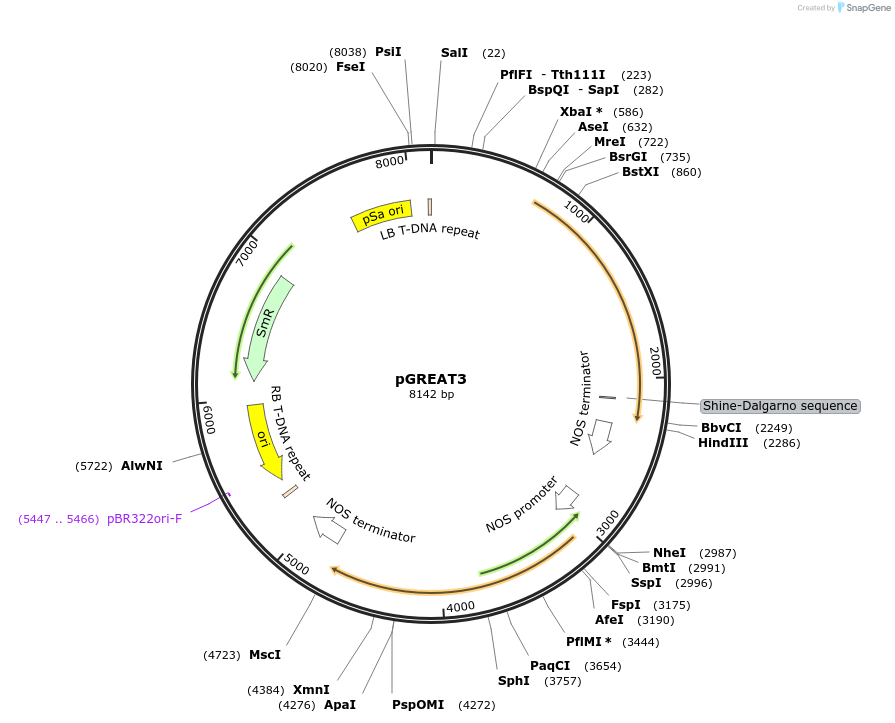
pGREAT3
Plasmid#170891PurposeGoldenBraid Double Transcriptional Unit - pAtUBQ10_E-Luc_NOSt+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…PromoterpAtUBQ10Available SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
pGREAT5
Plasmid#170893PurposeGoldenBraid Double Transcriptional Unit - pCmYLCV911_E-Luc_NOSt+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…PromoterpCmYLCV911Available SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
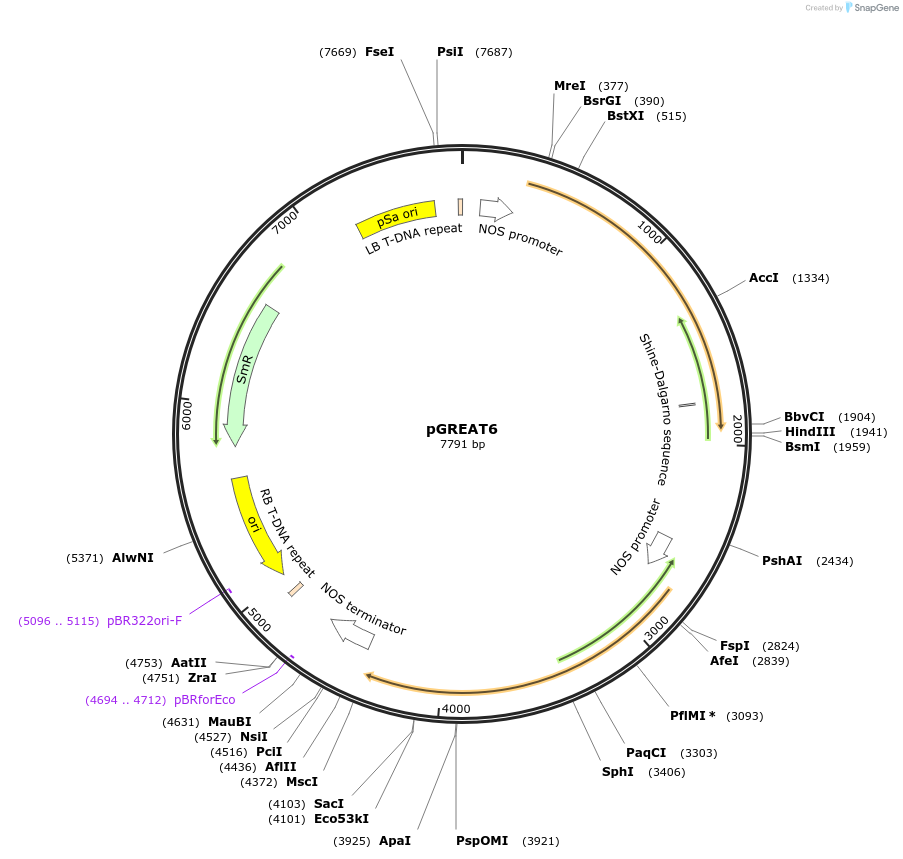
pGREAT6
Plasmid#170894PurposeGoldenBraid Double Transcriptional Unit - pNOS_E-Luc_NtEUt+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…PromoterpNOSAvailable SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
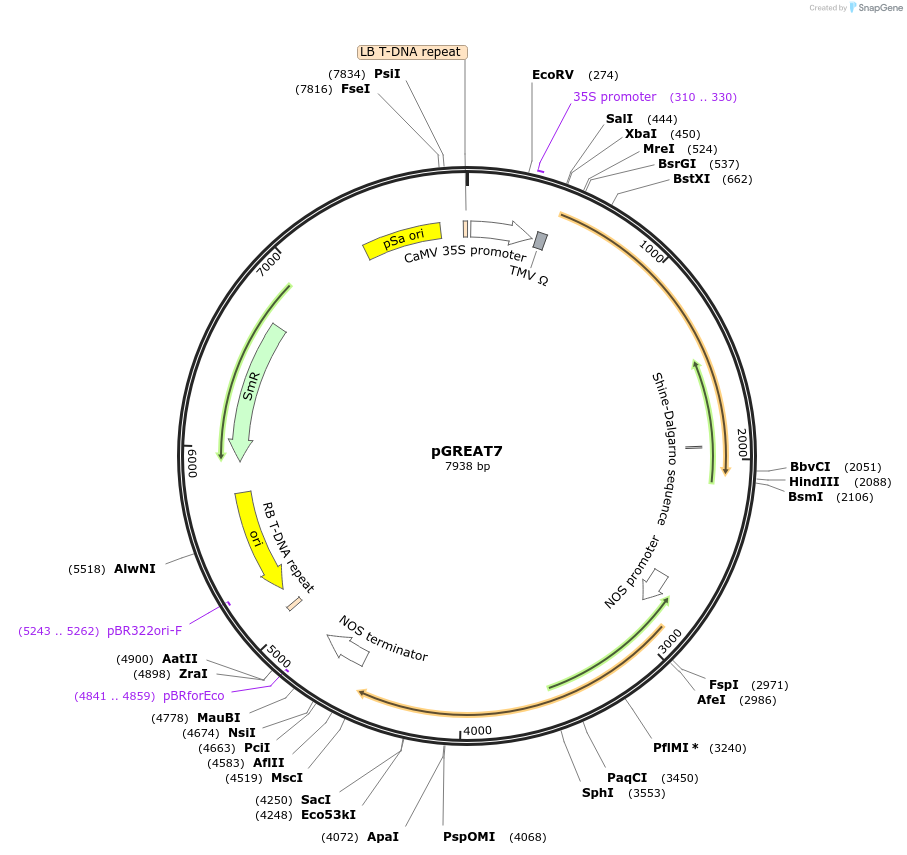
pGREAT7
Plasmid#170895PurposeGoldenBraid Double Transcriptional Unit - p35SoTMV_E-Luc_NtEUt+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…Promoterp35SoTMVAvailable SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
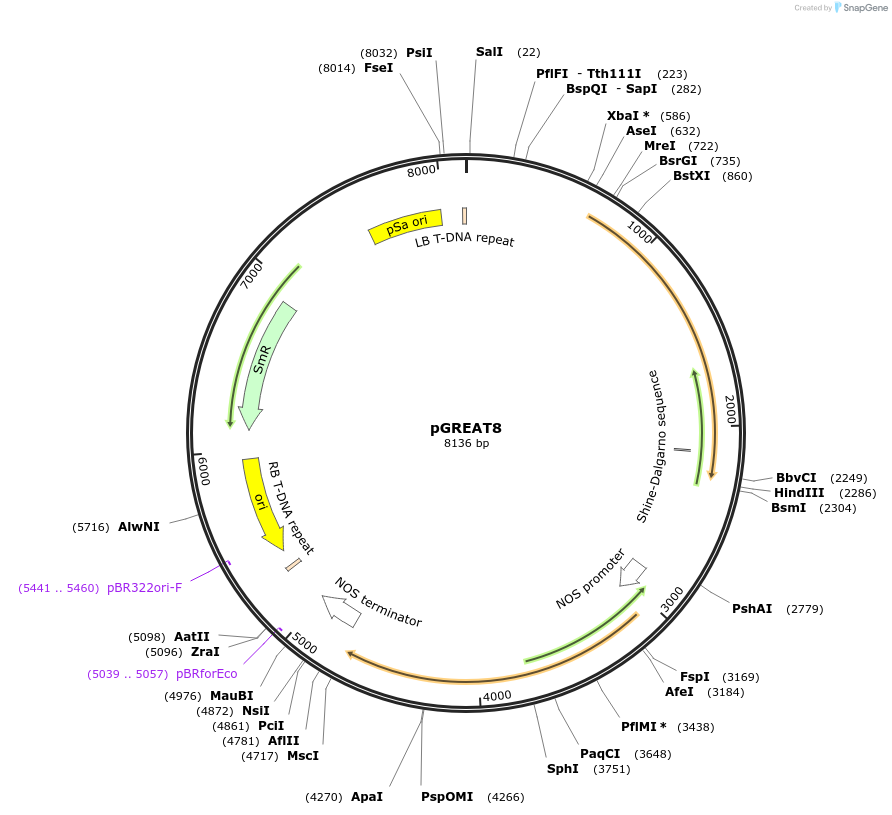
pGREAT8
Plasmid#170896PurposeGoldenBraid Double Transcriptional Unit - pAtUBQ10_E-Luc_NtEUt+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…PromoterpAtUBQ10Available SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
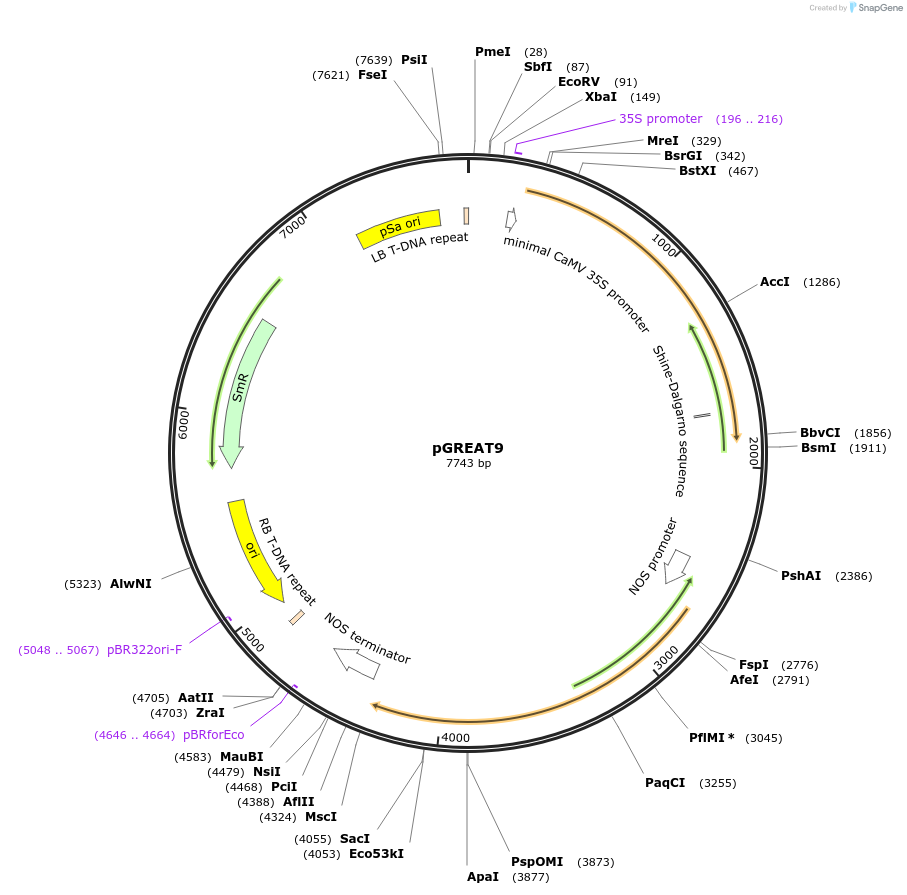
pGREAT9
Plasmid#170897PurposeGoldenBraid Double Transcriptional Unit - pG10-90_E-Luc_NtEUt+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…PromoterpG10-90Available SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
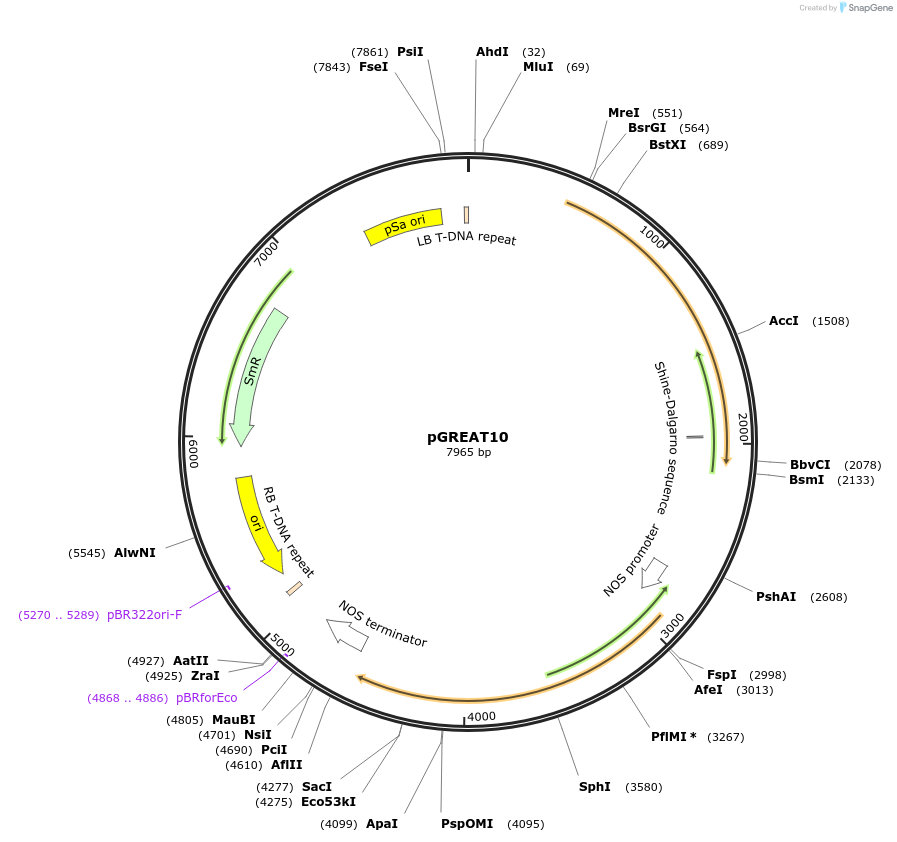
pGREAT10
Plasmid#170898PurposeGoldenBraid Double Transcriptional Unit - pCmYLCV911_E-Luc_NtEUt+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…PromoterpCmYLCV911Available SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
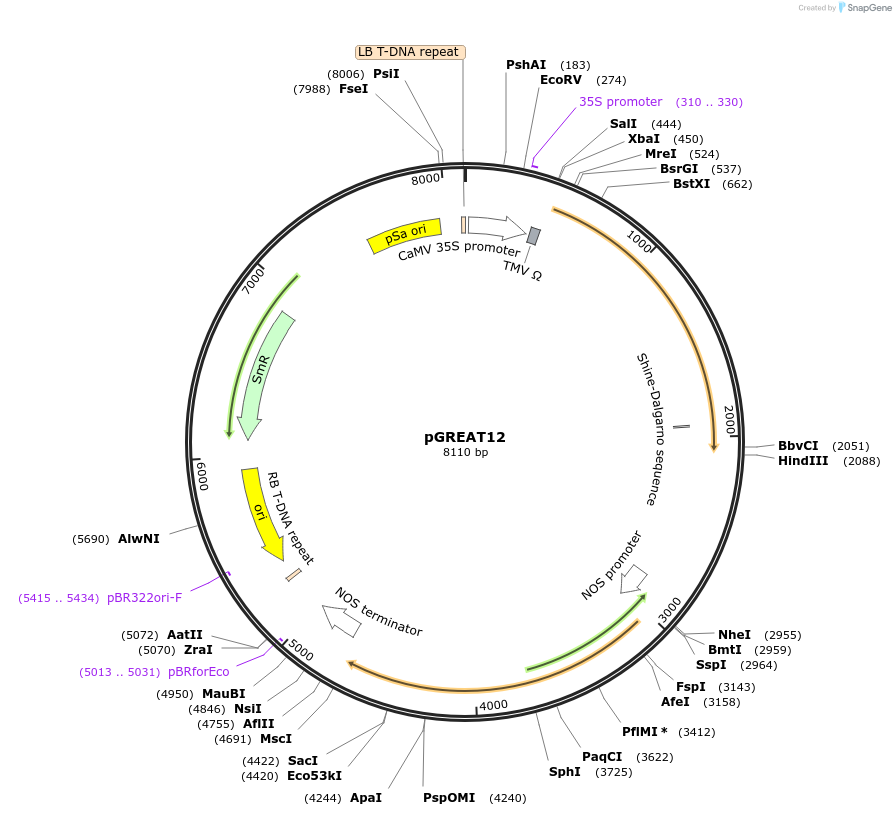
pGREAT12
Plasmid#170900PurposeGoldenBraid Double Transcriptional Unit - p35SoTMV_E-Luc_AtACT2t+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…Promoterp35SoTMVAvailable SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
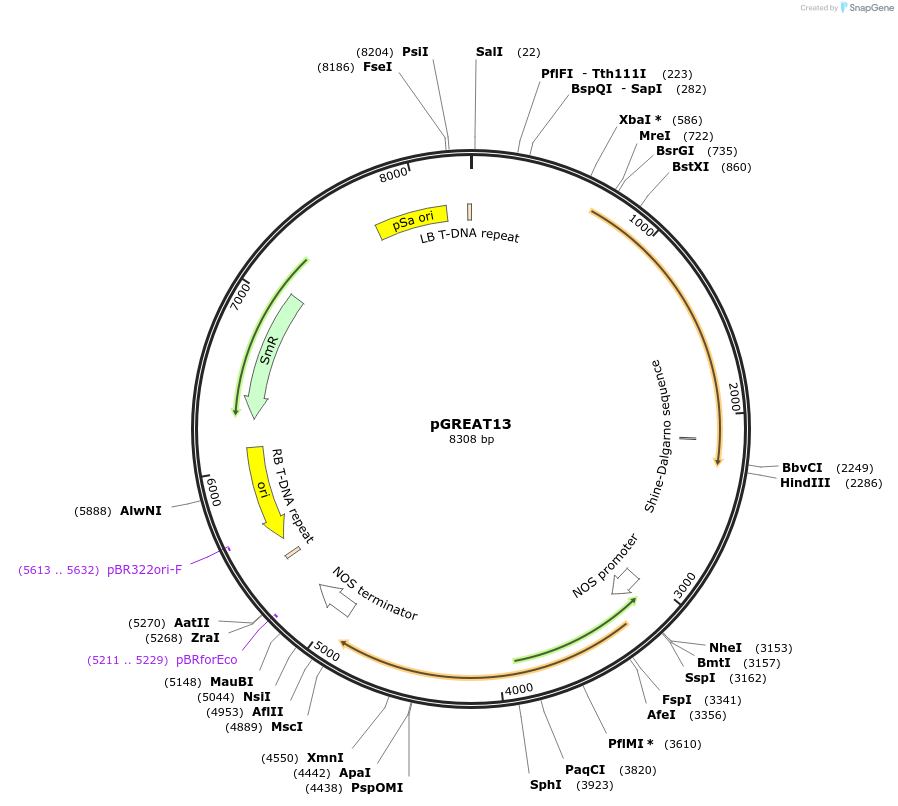
pGREAT13
Plasmid#170901PurposeGoldenBraid Double Transcriptional Unit - pAtUBQ10_E-Luc_AtACT2t+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…PromoterpAtUBQ10Available SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
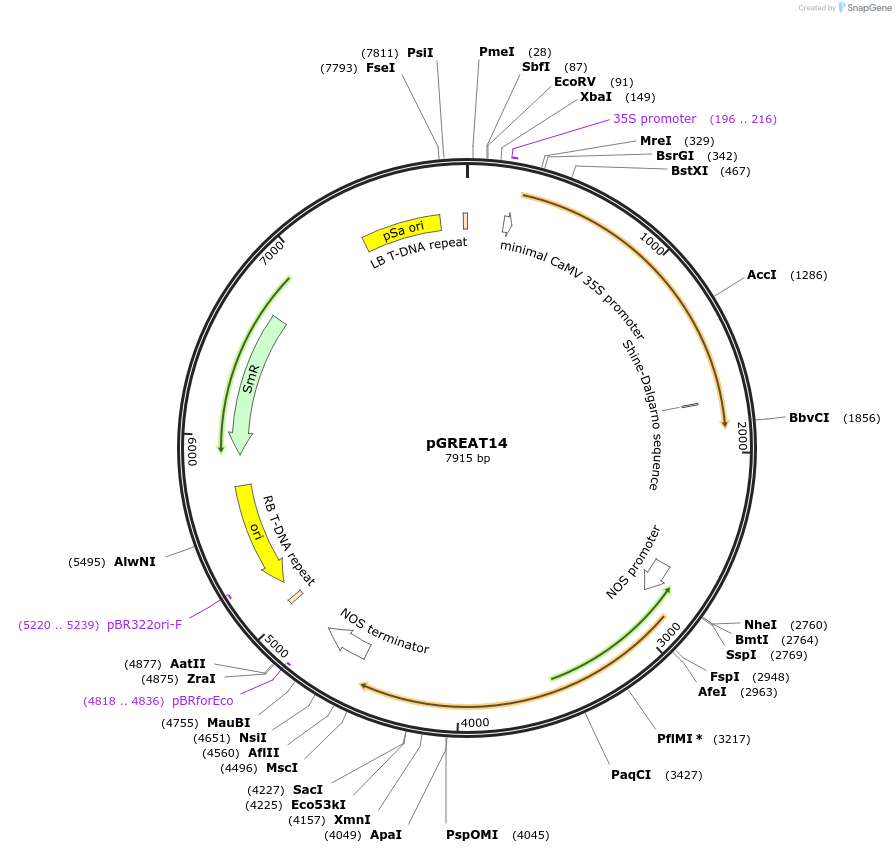
pGREAT14
Plasmid#170902PurposeGoldenBraid Double Transcriptional Unit - pG10-90_E-Luc_AtACT2t+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…PromoterpG10-90Available SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
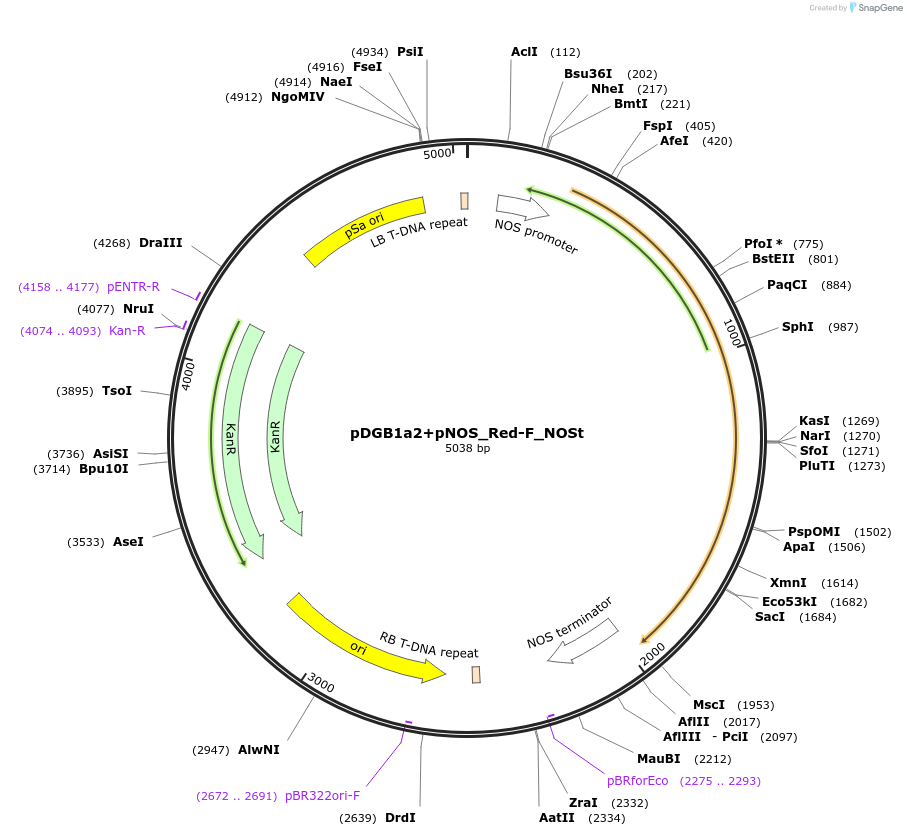
pDGB1a2+pNOS_Red-F_NOSt
Plasmid#170887PurposeGoldenBraid Transcriptional Unit - pNOS_Red-F_NOSt Forward orientationDepositorInsertRed-F
UseLuciferase and Synthetic BiologyExpressionPlantMutationp.S286Y Red mutantPromoterpNOSAvailable SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
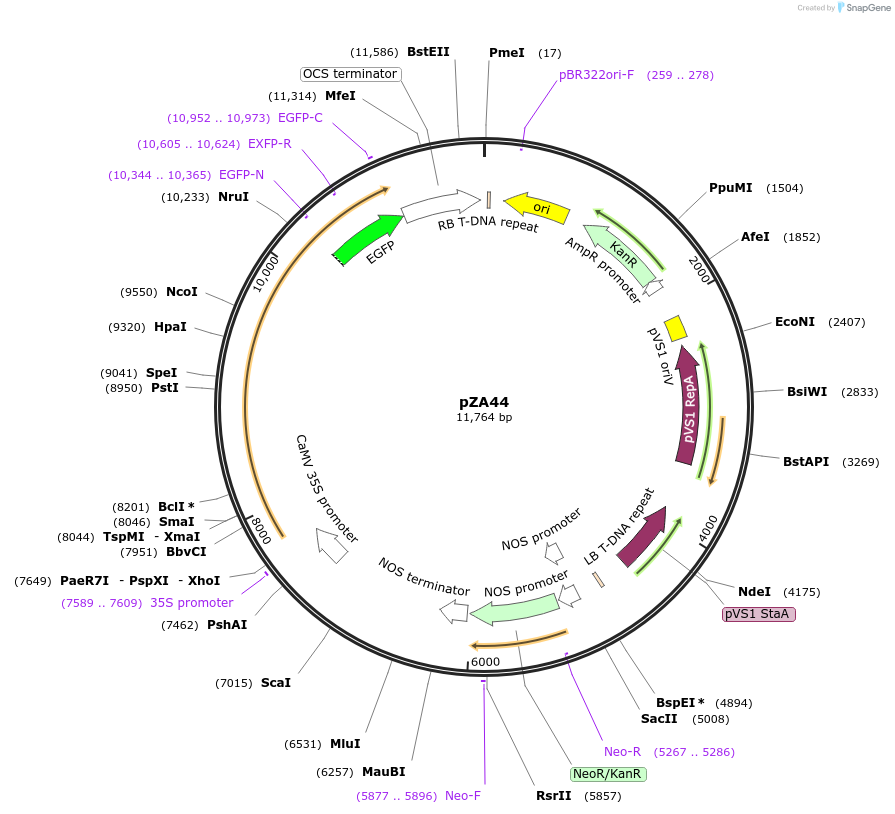
pZA44
Plasmid#158522PurposeIn planta gene expression of Golden gate compatible A. thaliana ZAR1 D498V/L17E-eGFP. Km plant selection marker.DepositorAvailable SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
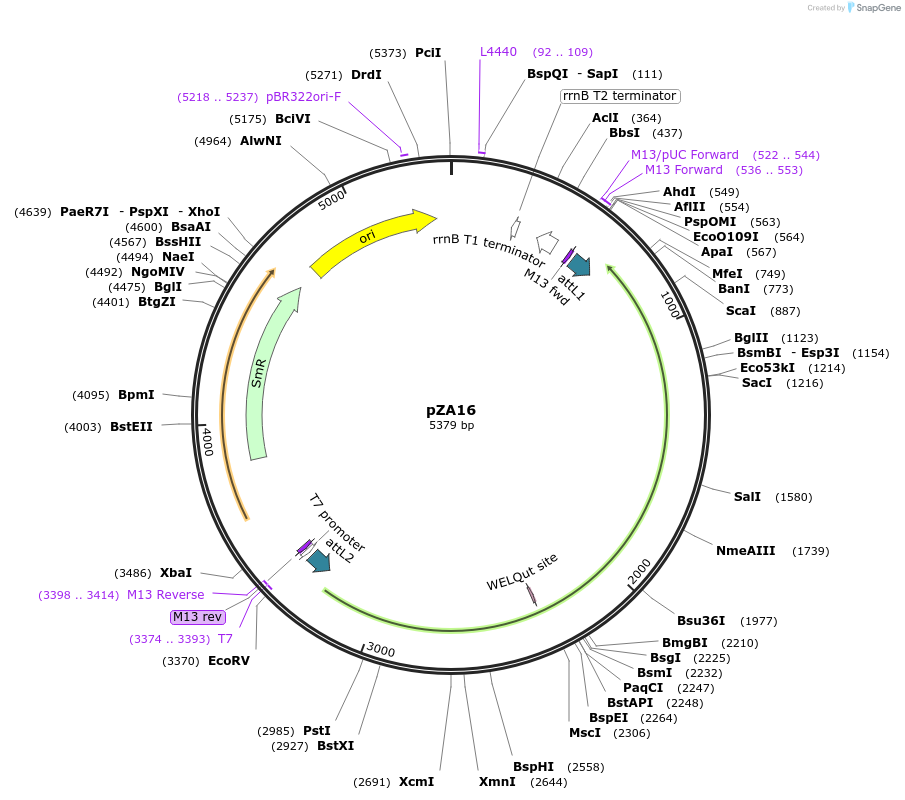
pZA16
Plasmid#158494PurposeEntry clone for Golden Gate assembly. Golden gate compatible N. benthamiana ZAR1 D481V/L17E without STOP codonDepositorInsertZAR1
UseGolden gate entry vectorMutationL17E, D481V, no STOP codonAvailable SinceSept. 17, 2020AvailabilityAcademic Institutions and Nonprofits only -
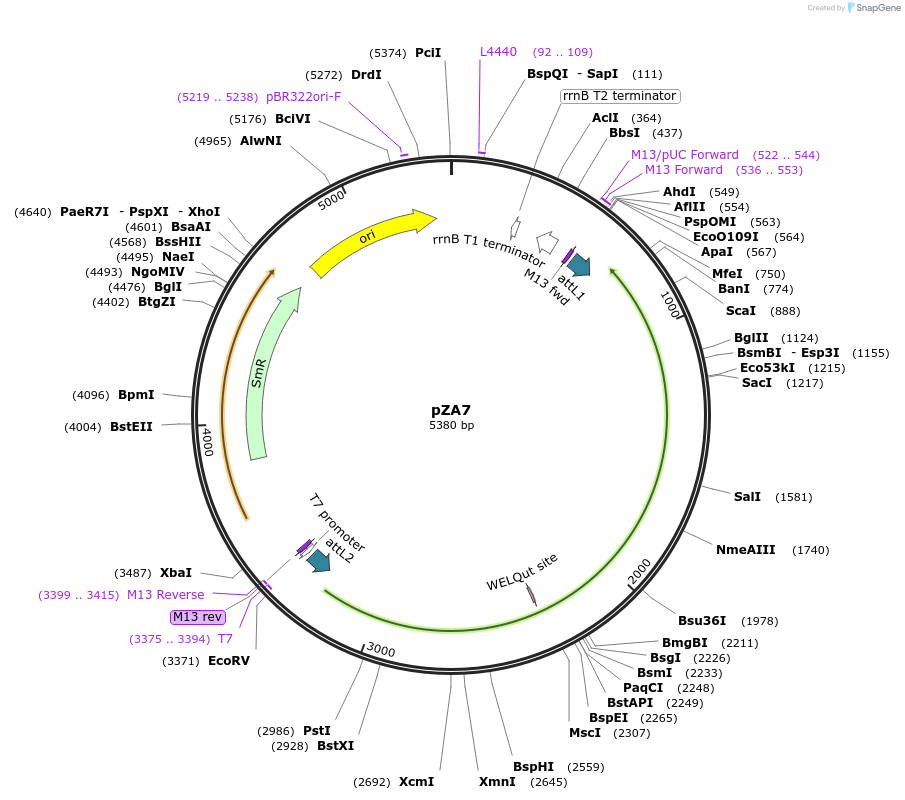
pZA7
Plasmid#158485PurposeEntry clone for Golden Gate assembly. Golden gate compatible N. benthamiana ZAR1 L17E with STOP codonDepositorInsertZAR1
UseGolden gate entry vectorMutationL17E, STOP codonAvailable SinceSept. 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
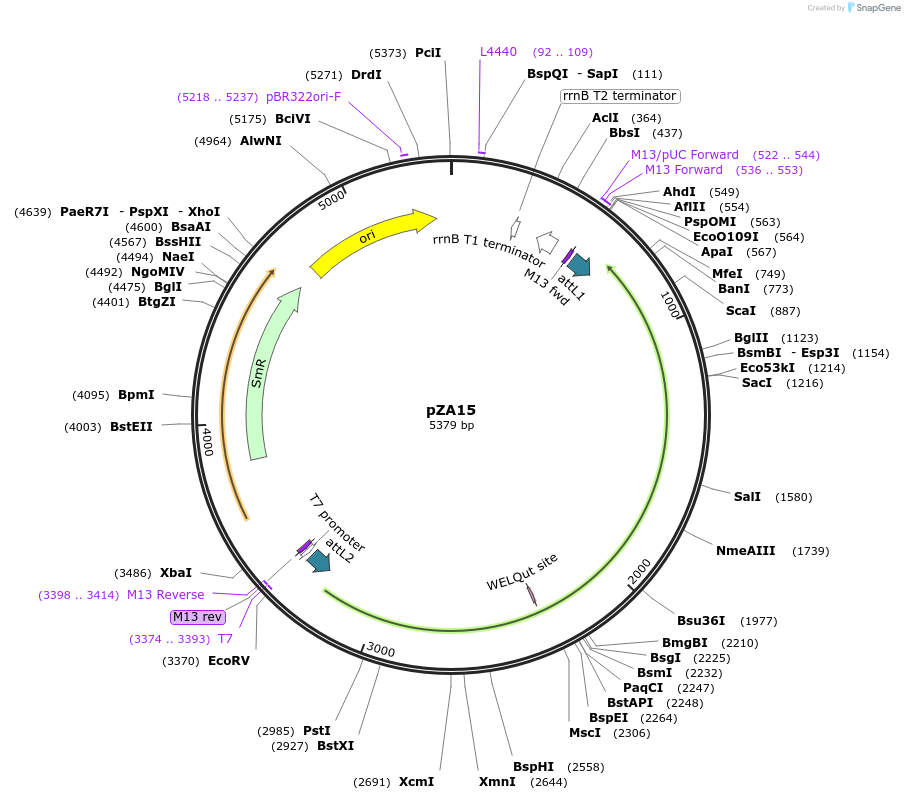
pZA15
Plasmid#158493PurposeEntry clone for Golden Gate assembly. Golden gate compatible N. benthamiana ZAR1 L17E without STOP codonDepositorInsertZAR1
UseGolden gate entry vectorMutationL17E, no STOP codonAvailable SinceSept. 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
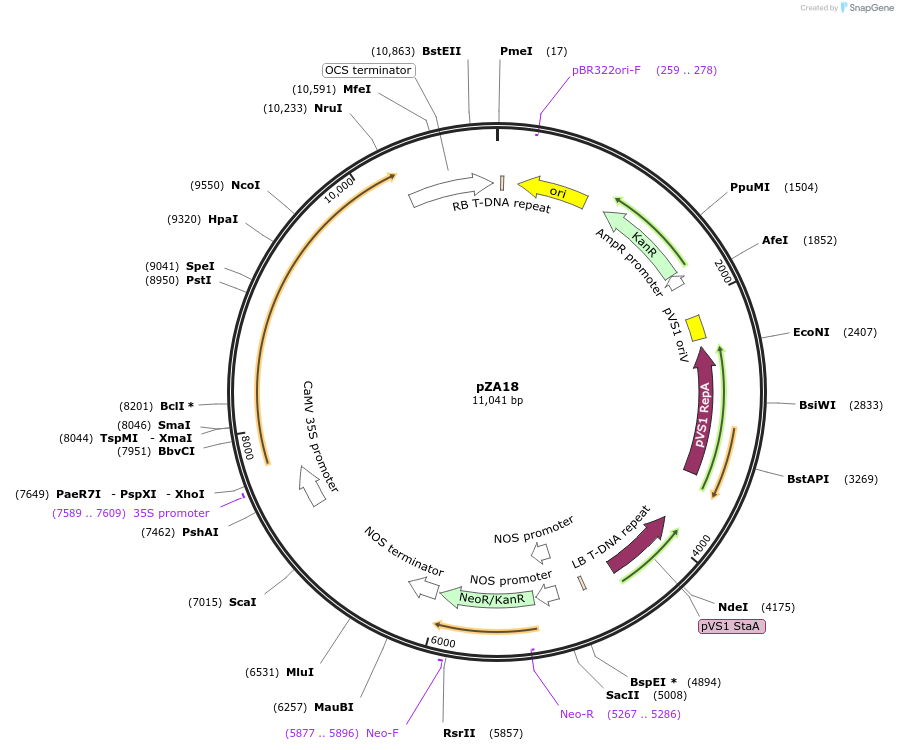
pZA18
Plasmid#158496PurposeIn planta gene expression of Golden gate compatibleA. thaliana ZAR1 D489V. Km plant selection marker.DepositorAvailable SinceSept. 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
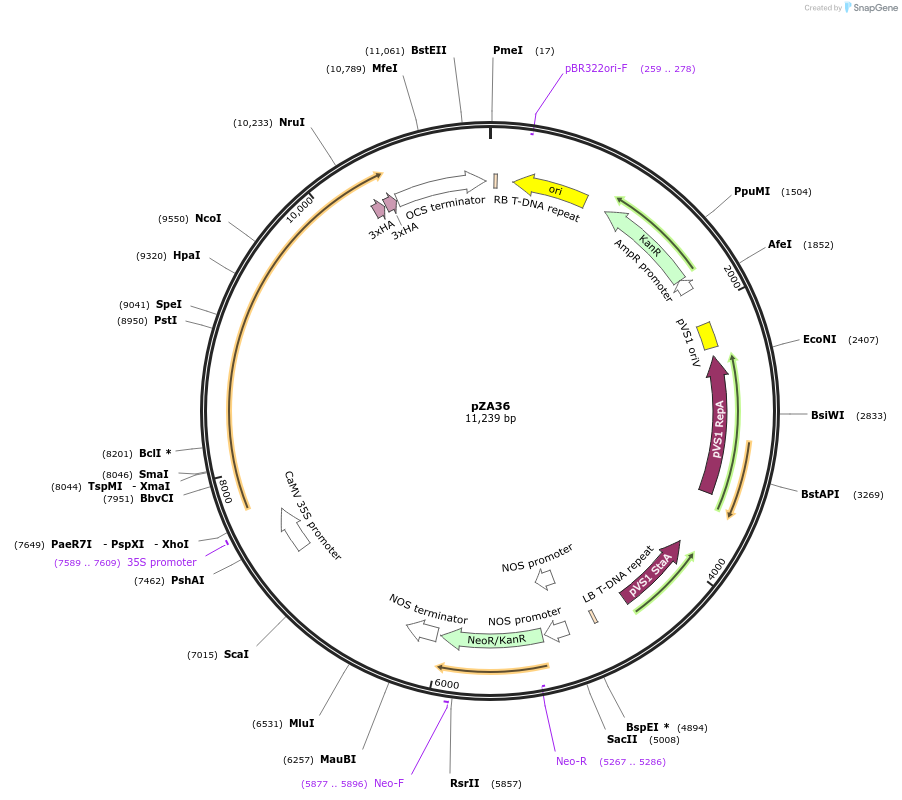
pZA36
Plasmid#158514PurposeIn planta gene expression of Golden gate compatible A. thaliana ZAR1 D498V/L17E-6xHA. Km plant selection marker.DepositorAvailable SinceSept. 4, 2020AvailabilityAcademic Institutions and Nonprofits only -
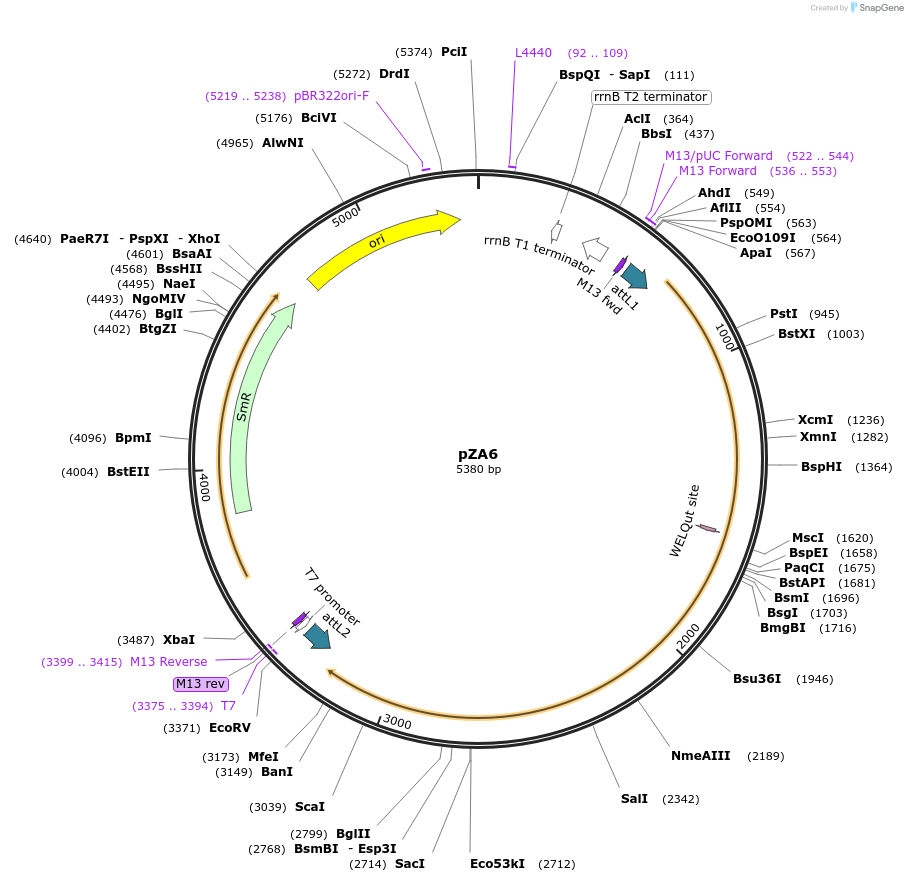
pZA6
Plasmid#158484PurposeEntry clone for Golden Gate assembly. Golden gate compatible N. benthamiana ZAR1 D481V with STOP codonDepositorInsertZAR1
UseGolden gate entry vectorMutationD481V, STOP codonAvailable SinceAug. 27, 2020AvailabilityAcademic Institutions and Nonprofits only -
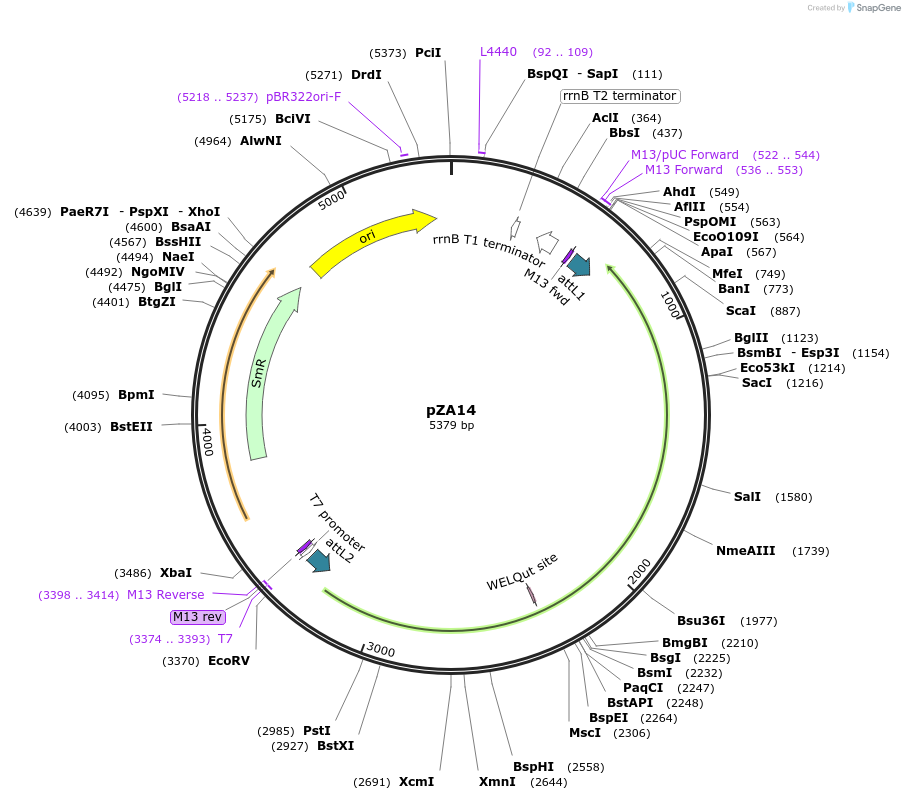
pZA14
Plasmid#158492PurposeEntry clone for Golden Gate assembly. Golden gate compatible N. benthamiana ZAR1 D481V without STOP codonDepositorInsertZAR1
UseGolden gate entry vectorMutationD481V, no STOP codonAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
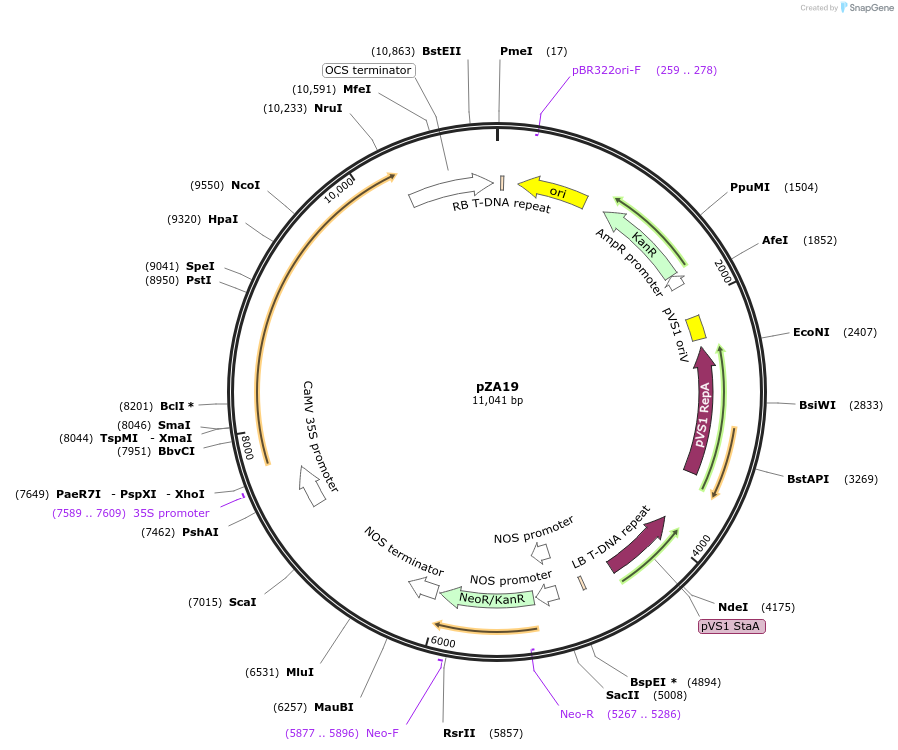
pZA19
Plasmid#158497PurposeIn planta gene expression of Golden gate compatlible A. thaliana ZAR1 L17E. Km plant selection marker.DepositorAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
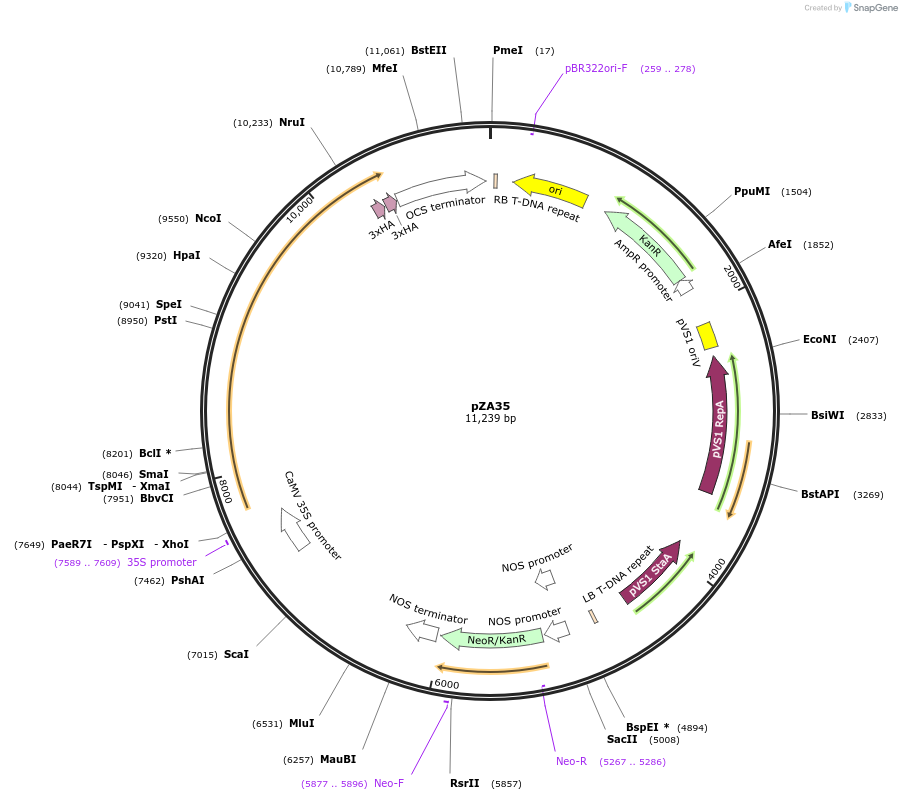
pZA35
Plasmid#158513PurposeIn planta gene expression of Golden gate compatible A. thaliana ZAR1 L17E-6xHA. Km plant selection marker.DepositorAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
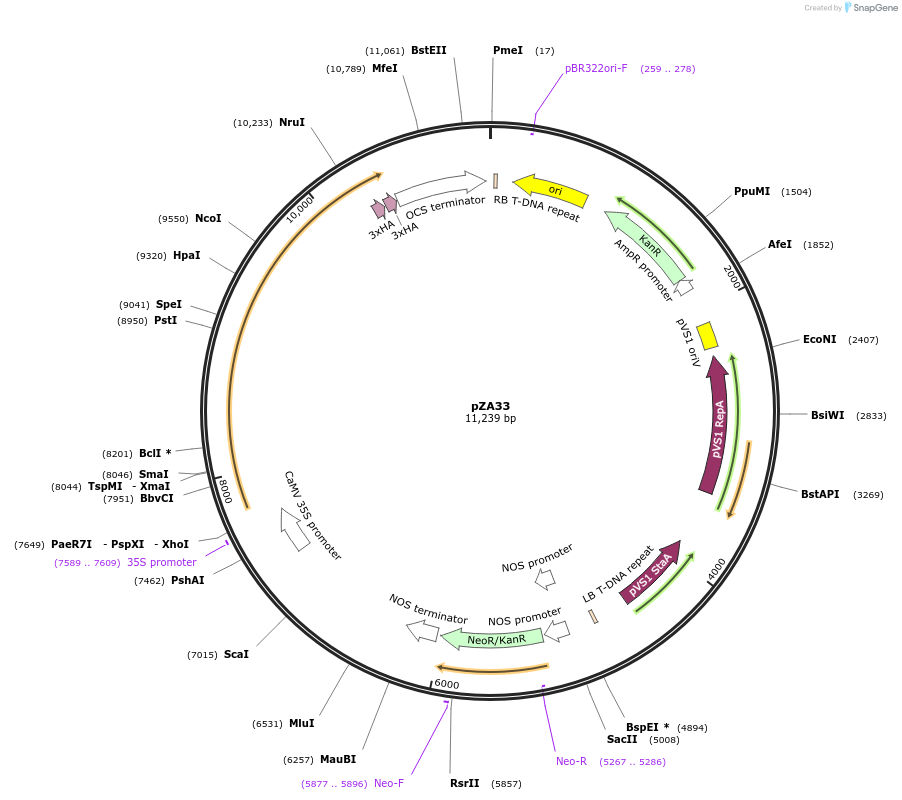
pZA33
Plasmid#158511PurposeIn planta gene expression of Golden gate compatible A. thaliana ZAR1-6xHA. Km plant selection marker.DepositorAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
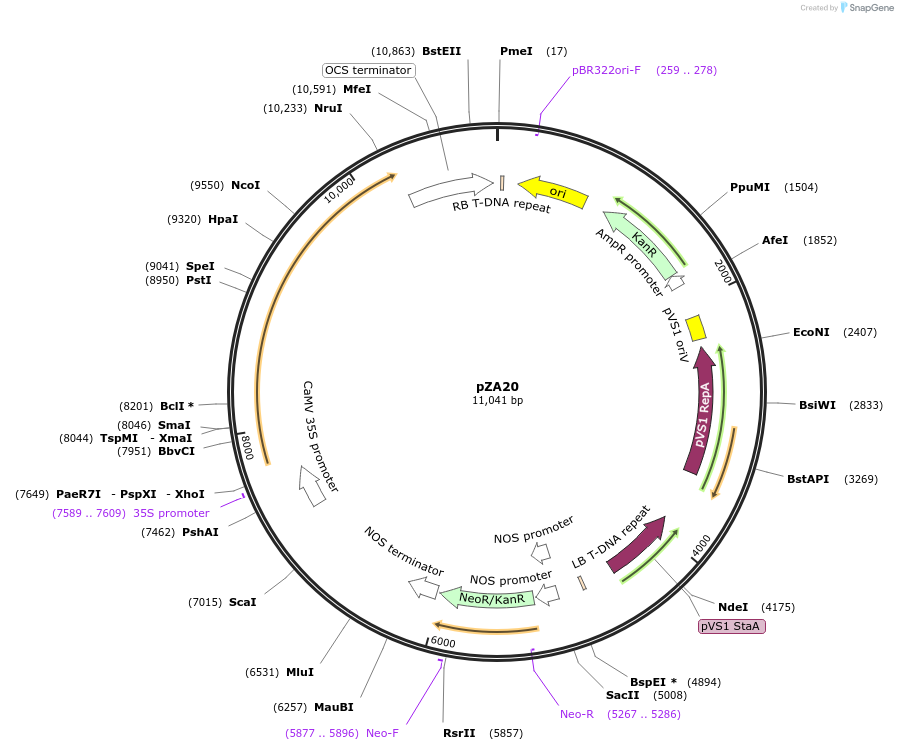
pZA20
Plasmid#158498PurposeIn planta gene expression of Golden gate complatible A. thaliana ZAR1 D498V/L17E. Km plant selection marker.DepositorAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
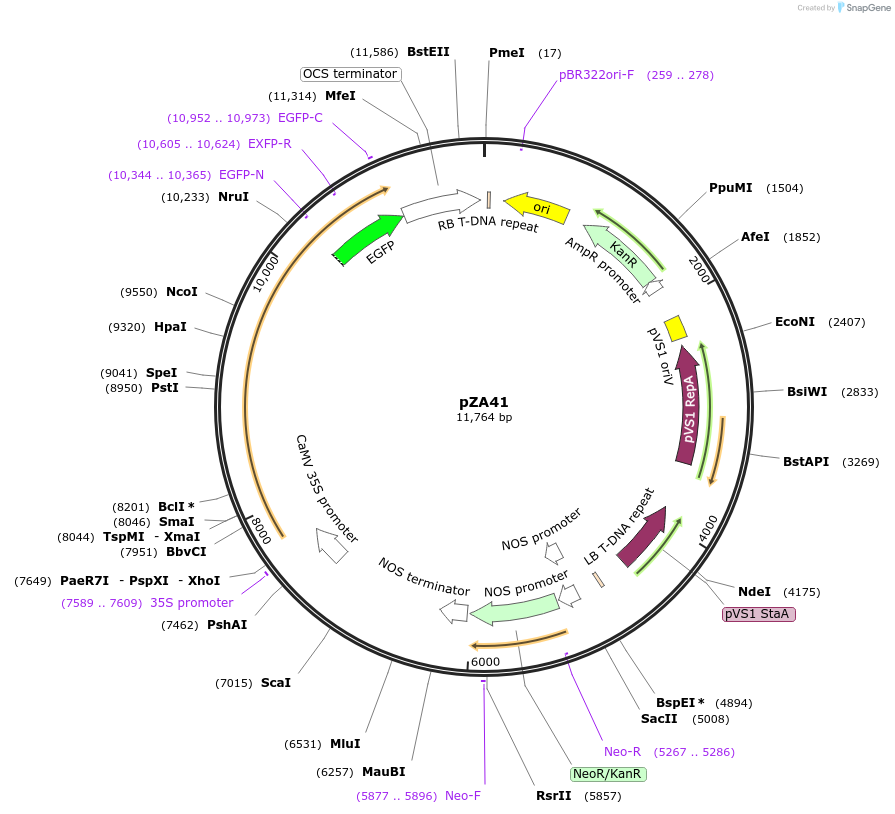
pZA41
Plasmid#158519PurposeIn planta gene expression of Golden gate compatible A. thaliana ZAR1-eGFP. Km plant selection marker.DepositorAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
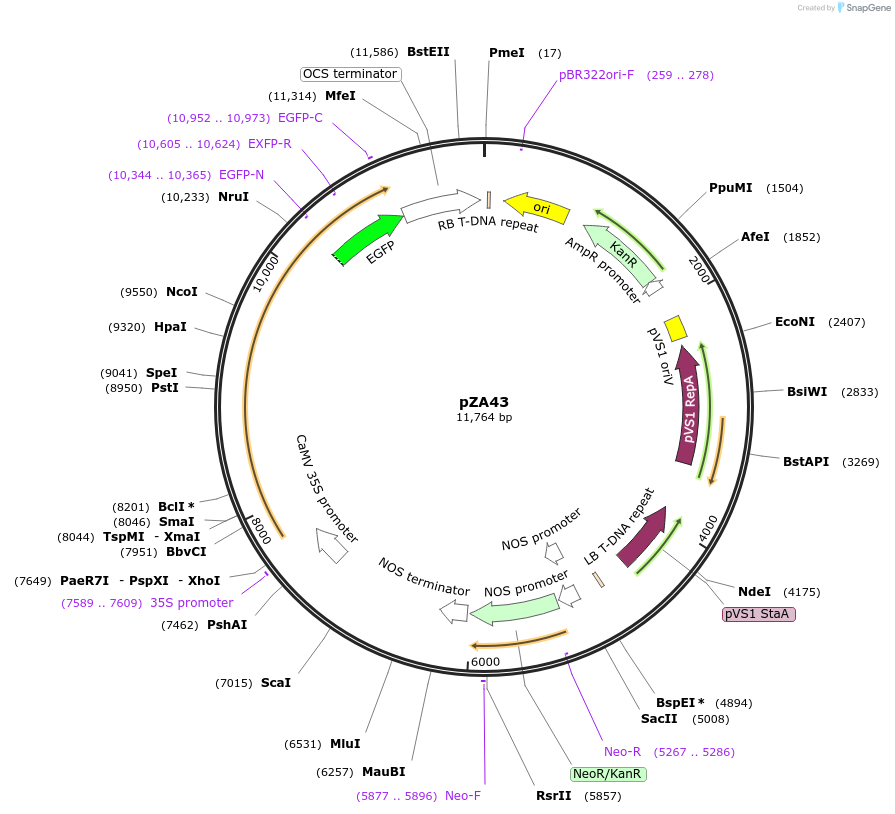
pZA43
Plasmid#158521PurposeIn planta gene expression of Golden gate compatible A. thaliana ZAR1 L17E-eGFP. Km plant selection marker.DepositorAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
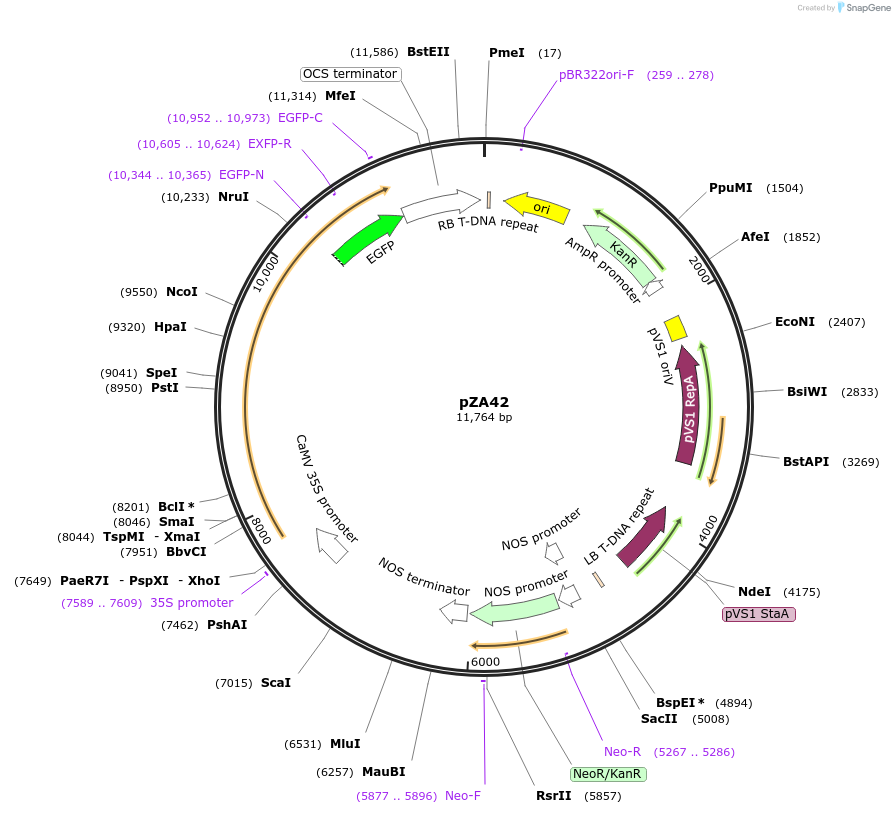
pZA42
Plasmid#158520PurposeIn planta gene expression of Golden gate compatible A. thaliana ZAR1 D498V-eGFP. Km plant selection marker.DepositorAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only