We narrowed to 11,627 results for: VARS
-
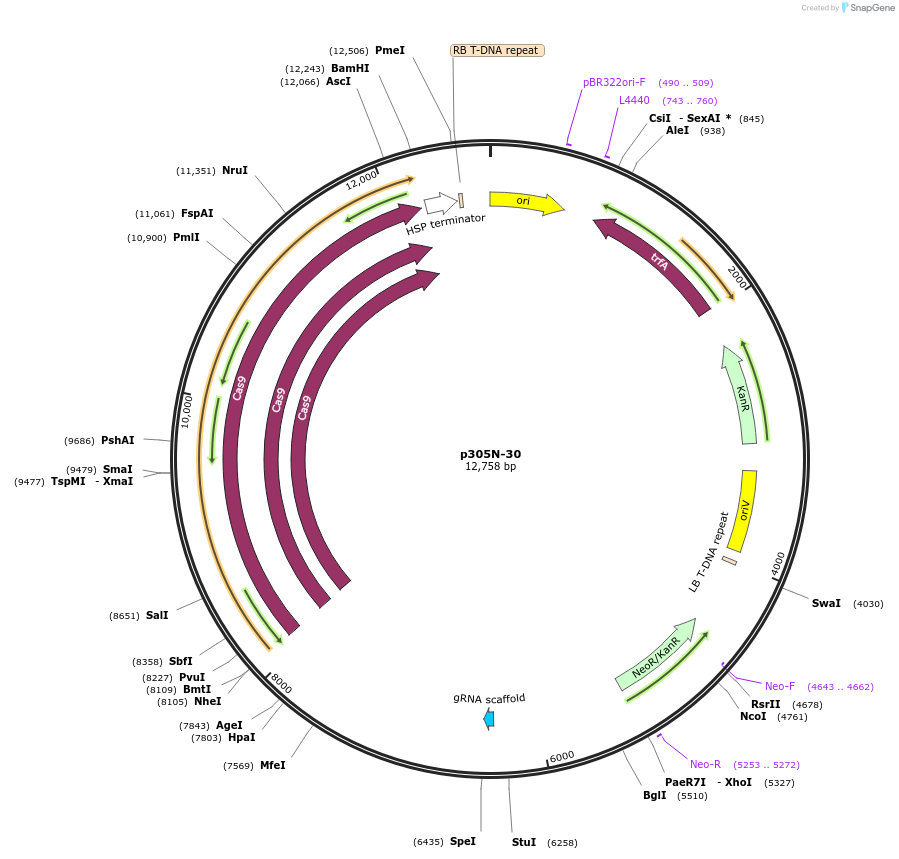
Plasmid#246313PurposeEvaluation of PtU6.2c3 promoter (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertPtU6.2c3 promoter
UseCRISPRExpressionPlantMutationdeletion: -T (-29 from TSS) within TATAPromoterPtU6.2c3 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
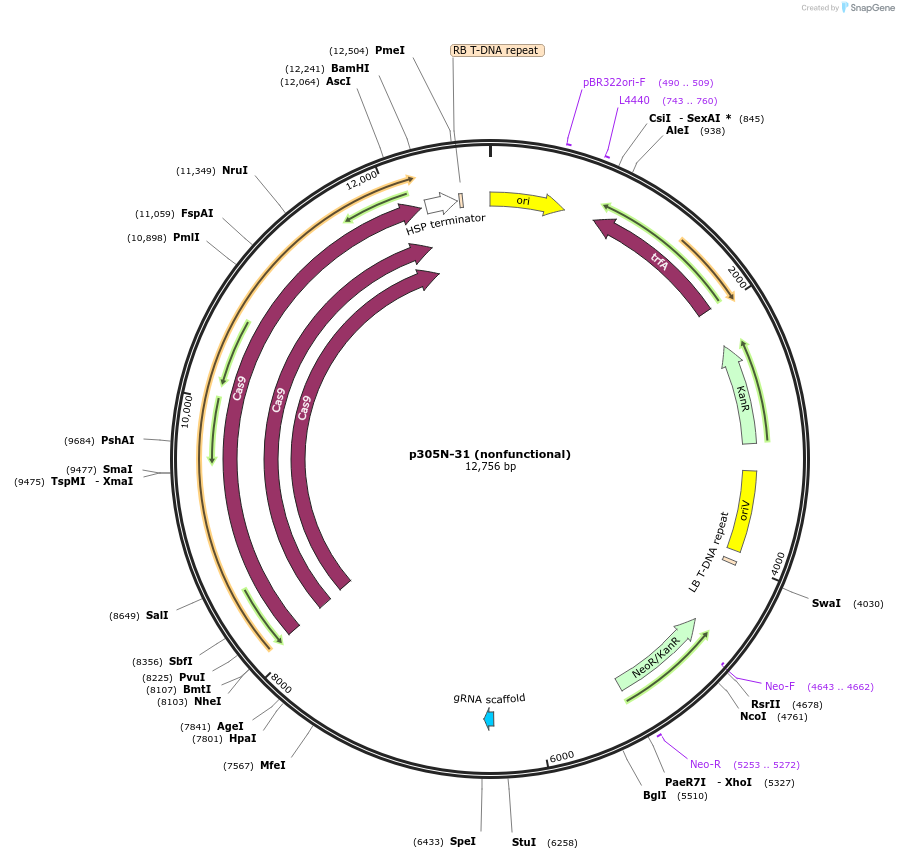
p305N-31 (nonfunctional)
Plasmid#246314PurposeEvaluation of PtU6.2c4 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertPtU6.2c4 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationdeletions: -A (-41 from TSS), -T (-30), -T (-29);…PromoterPtU6.2c4 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
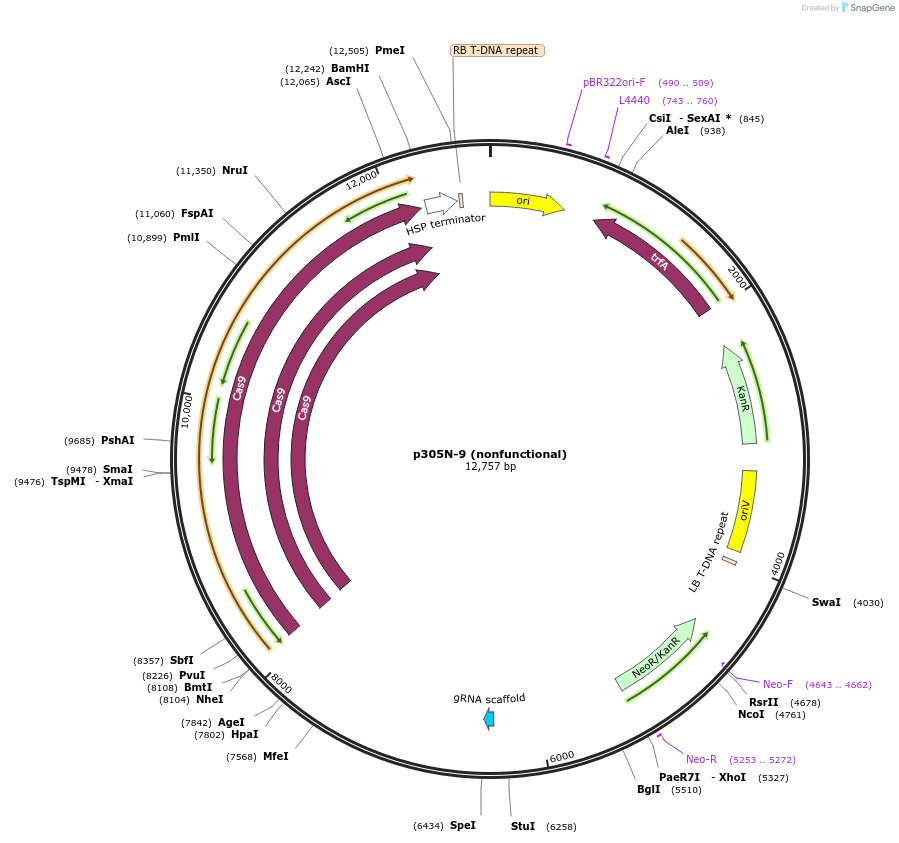
p305N-9 (nonfunctional)
Plasmid#246292PurposeEvaluation of AtU6.29c13 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertAtU6.29c13 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationdeletions: -A (-58 from TSS), -T (-57) within USEPromoterAtU6.29c13 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
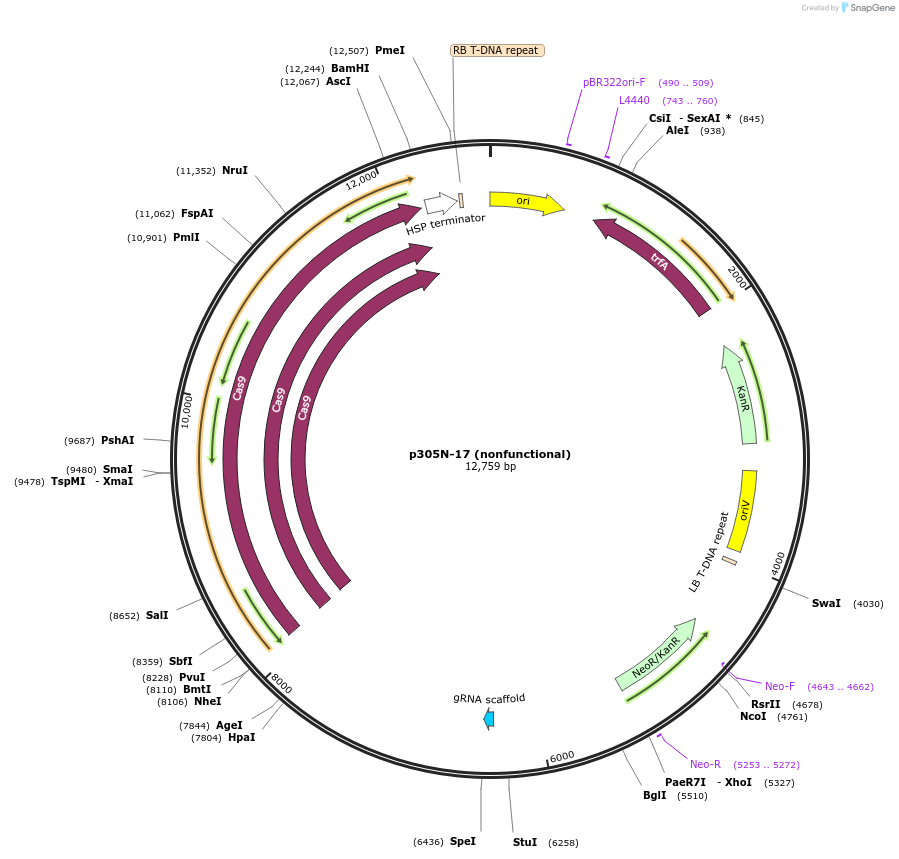
p305N-17 (nonfunctional)
Plasmid#246300PurposeEvaluation of HbU6.2m3 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertHbU6.2m3 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationC-to-T (-29 from TSS), G-to-A (-24)PromoterHbU6.2m3 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
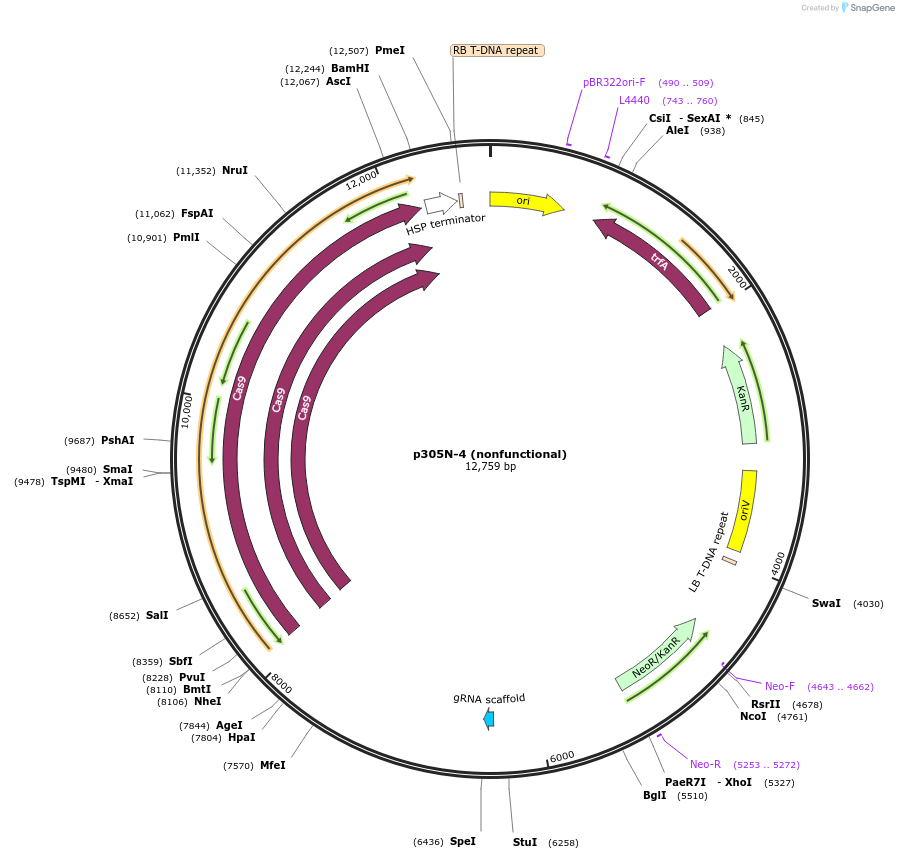
p305N-4 (nonfunctional)
Plasmid#246287PurposeEvaluation of AtU6.1c1 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertAtU6.1c1 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationG-to-C at -15 from TSSPromoterAtU6.1c1 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
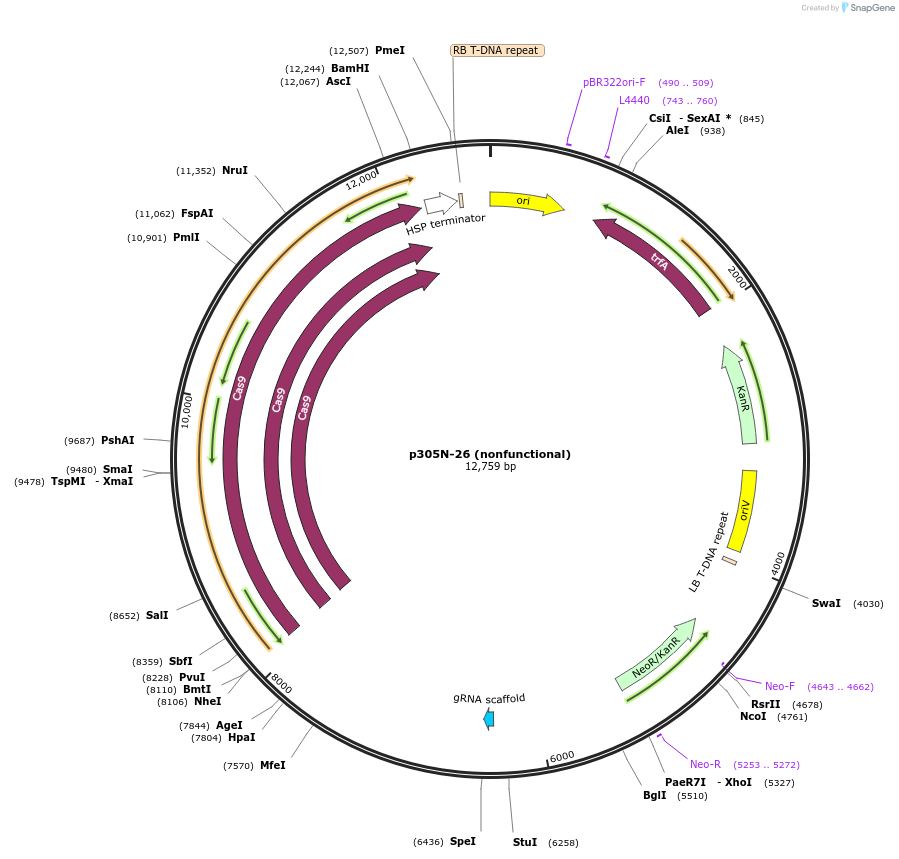
p305N-26 (nonfunctional)
Plasmid#246309PurposeEvaluation of MtU6.6m7 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertMtU6.6m7 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationC-to-A (-63 from TSS), T-to-G (-57)PromoterMtU6.6m7 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
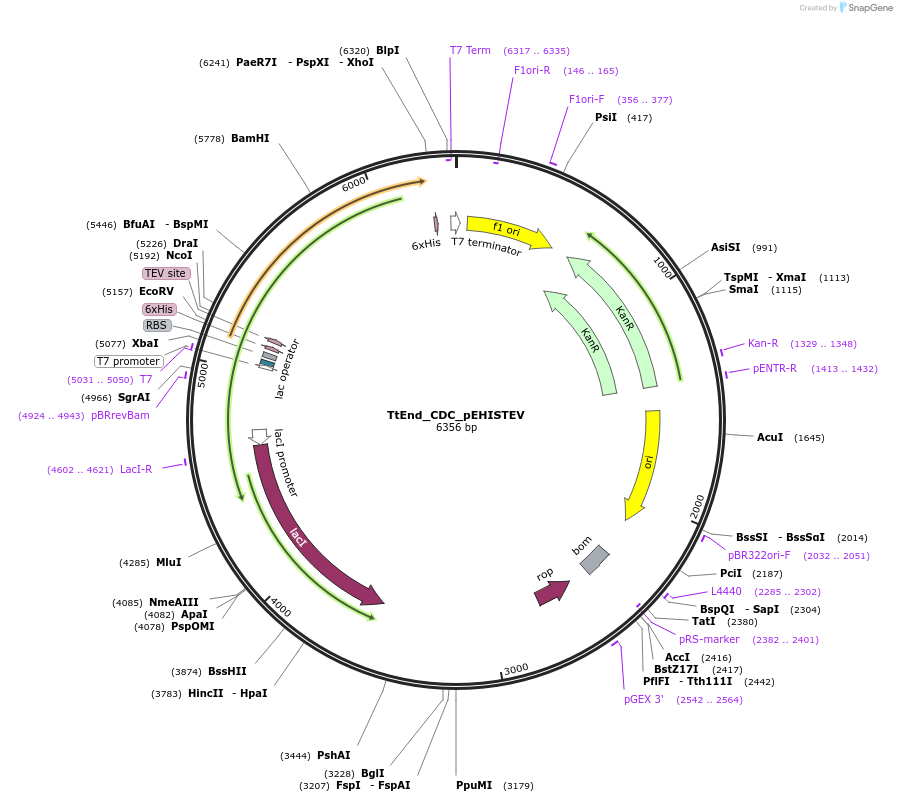
TtEnd_CDC_pEHISTEV
Plasmid#179736PurposeTtEnd_CDC_pEHISTEV expresses tuncated variant of TtEnd5A (186-531 amni caids) comprsisng of cataytic domain and a linker at C-terminal.DepositorInsertTtEnd5A
Tags6xHisExpressionBacterialMutation185 Amino acids from N-terminal were deleted.PromoterT7Available SinceOct. 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
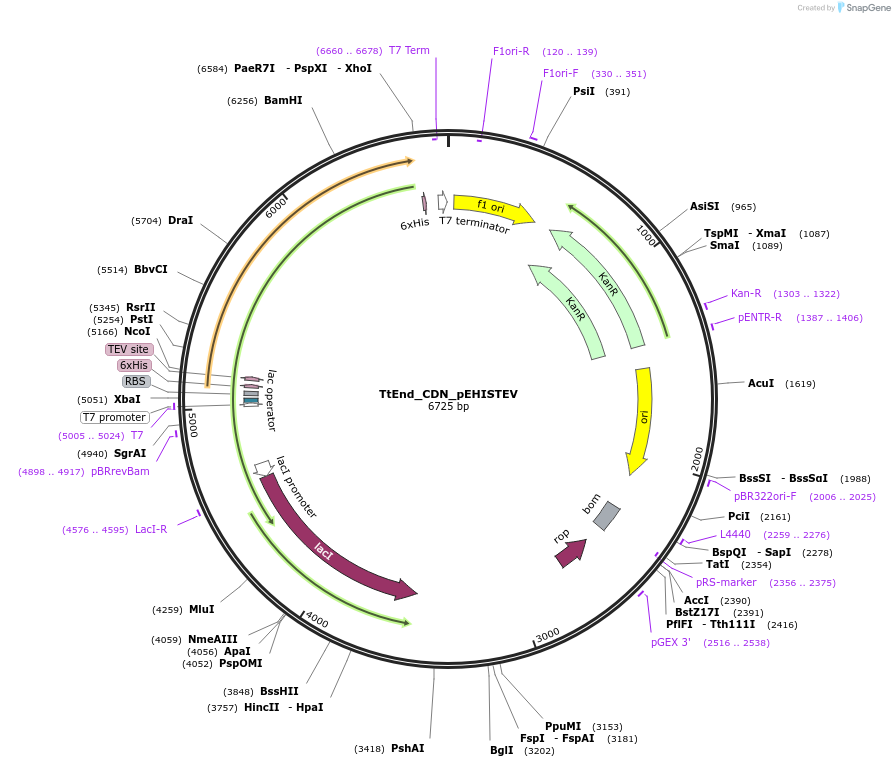
TtEnd_CDN_pEHISTEV
Plasmid#179738PurposeTtEnd_CDN_pEHISTEV expresses tuncated variant of TtEnd5A (19-486 amnino acids) comprsisng of cataytic domain and a linker at N-terminal.DepositorInsertTtEnd5A
Tags6xHisExpressionBacterialMutation45 Amino acids from C-terminal were deleted.PromoterT7Available SinceOct. 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
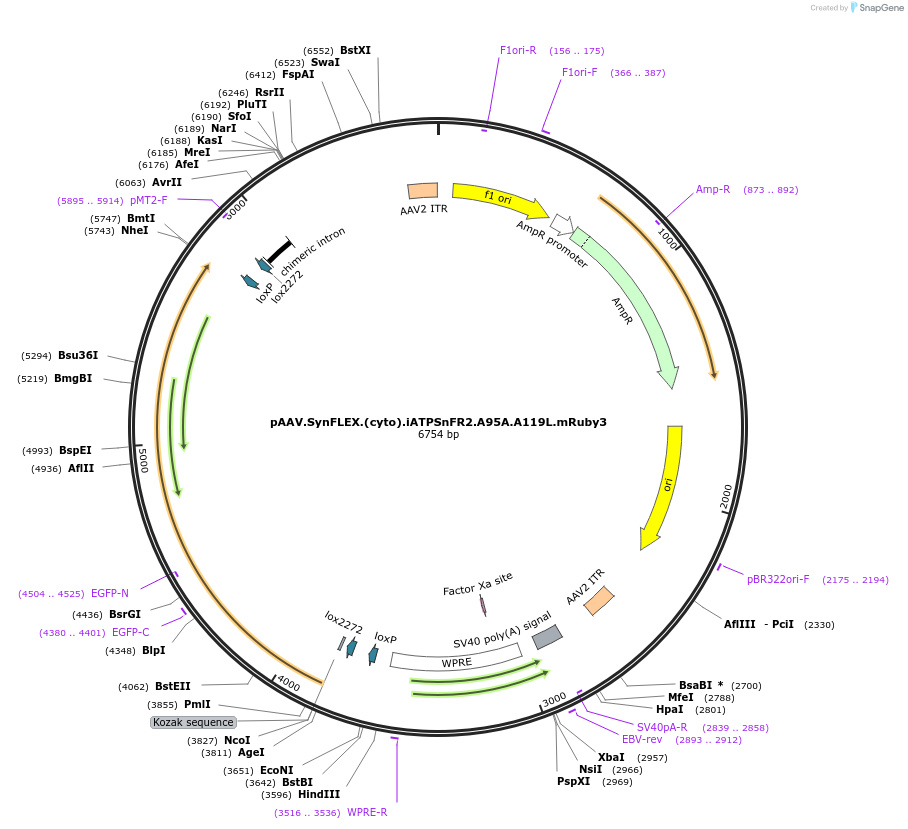
pAAV.SynFLEX.(cyto).iATPSnFR2.A95A.A119L.mRuby3
Plasmid#244913PurposeiATPSnFR2.A95A.A119L variant with non-responsive mRuby3DepositorInsertiATPSnFR2.A95A.A119L.mRuby3
UseAAVMutationA95A.A119LAvailable SinceOct. 8, 2025AvailabilityAcademic Institutions and Nonprofits only -
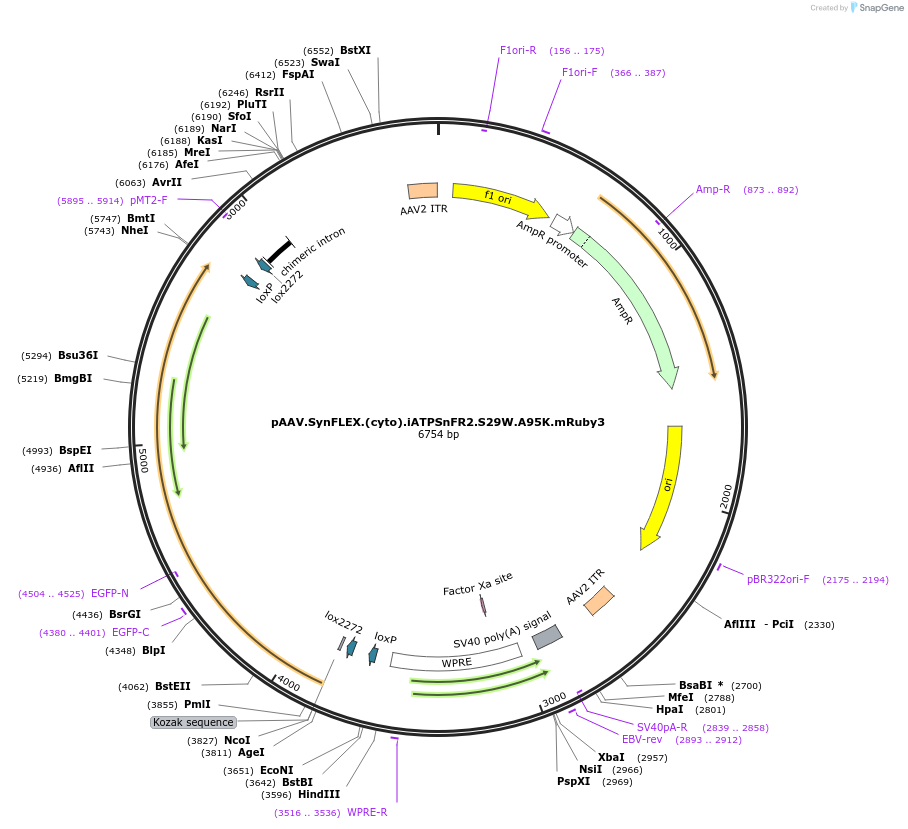
pAAV.SynFLEX.(cyto).iATPSnFR2.S29W.A95K.mRuby3
Plasmid#244911PurposeiATPSnFR2.S29W.A95K variant with non-responsive mRuby3DepositorInsertiATPSnFR2.S29W.A95K.mRuby3
UseAAVMutationS29W.A95KAvailable SinceOct. 8, 2025AvailabilityAcademic Institutions and Nonprofits only -
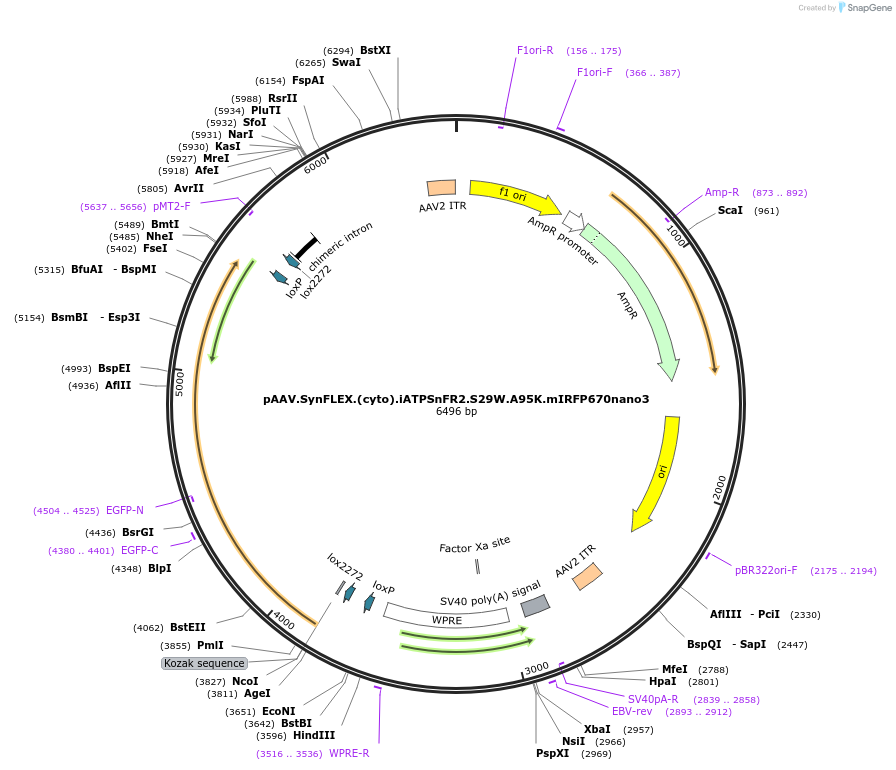
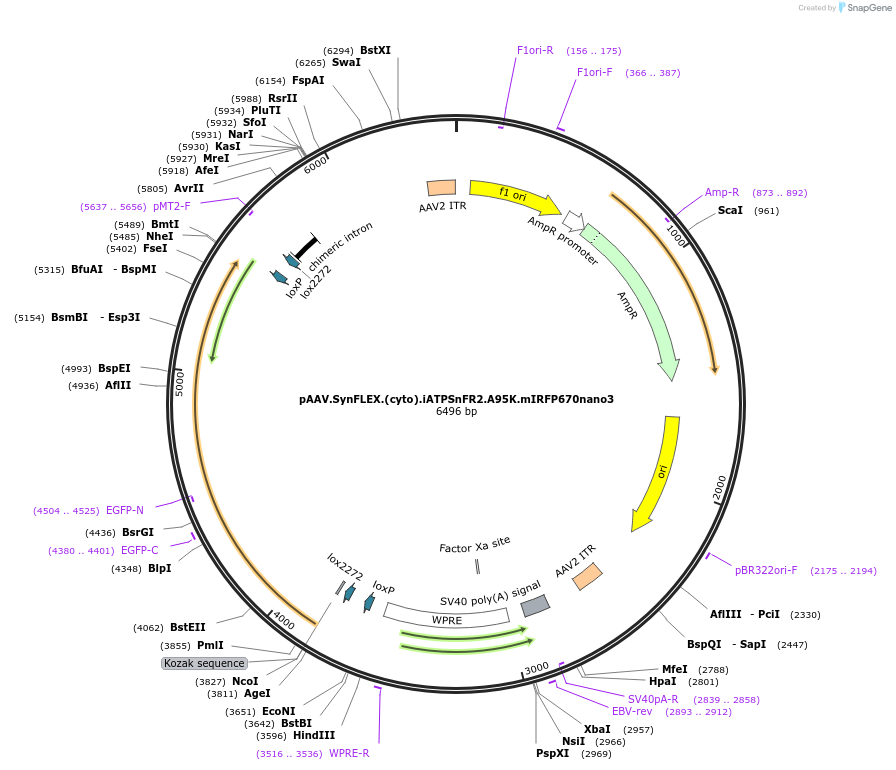
pAAV.SynFLEX.(cyto).iATPSnFR2.S29W.A95K.mIRFP670nano3
Plasmid#244915PurposeiATPSnFR2.S29W.A95K variant with non-responsive mIRFP670nano3DepositorInsertiATPSnFR2.S29W.A95K.mIRFP670nano3
UseAAVMutationS29W.A95KAvailable SinceOct. 8, 2025AvailabilityAcademic Institutions and Nonprofits only -
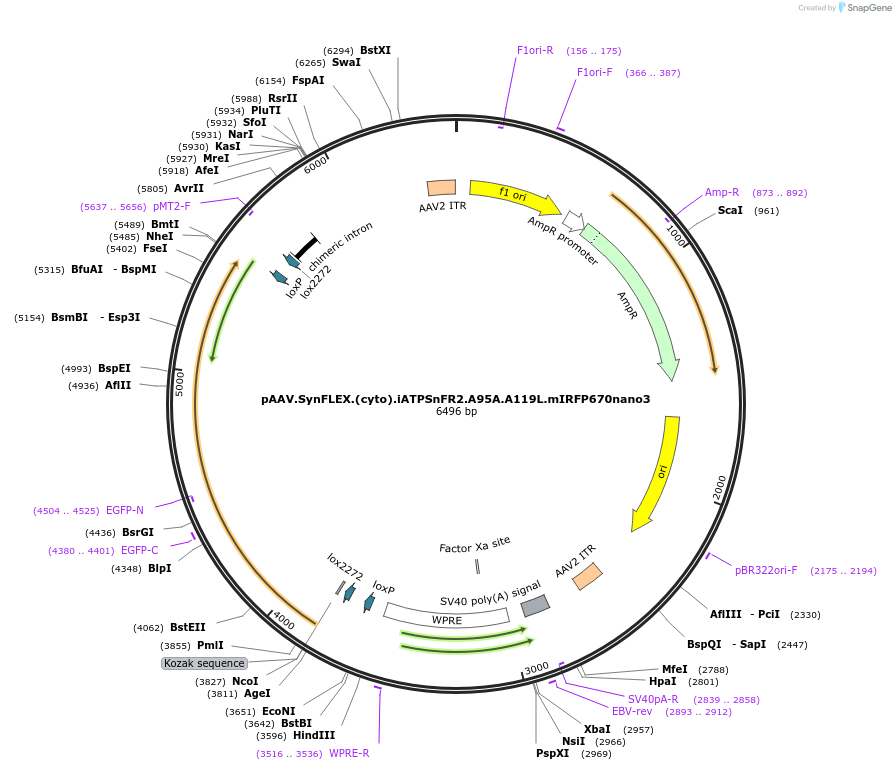
pAAV.SynFLEX.(cyto).iATPSnFR2.A95A.A119L.mIRFP670nano3
Plasmid#244917PurposeiATPSnFR2.A95A.A119L variant with non-responsive mIRFP670nano3DepositorInsertiATPSnFR2.A95A.A119L.mIRFP670nano3
UseAAVMutationA95A.A119LAvailable SinceOct. 8, 2025AvailabilityAcademic Institutions and Nonprofits only -
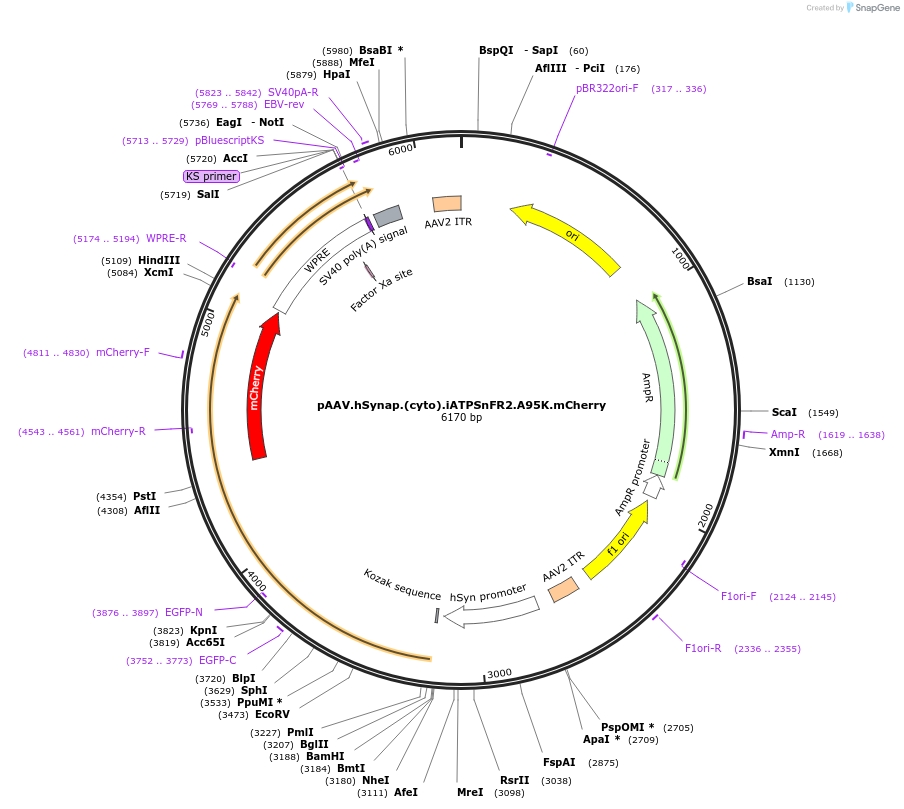
pAAV.hSynap.(cyto).iATPSnFR2.A95K.mCherry
Plasmid#244919PurposeiATPSnFR2.A95K variant with non-responsive mCherryDepositorInsertiATPSnFR2.A95K.mCherry
UseAAVMutationA95KAvailable SinceOct. 8, 2025AvailabilityAcademic Institutions and Nonprofits only -
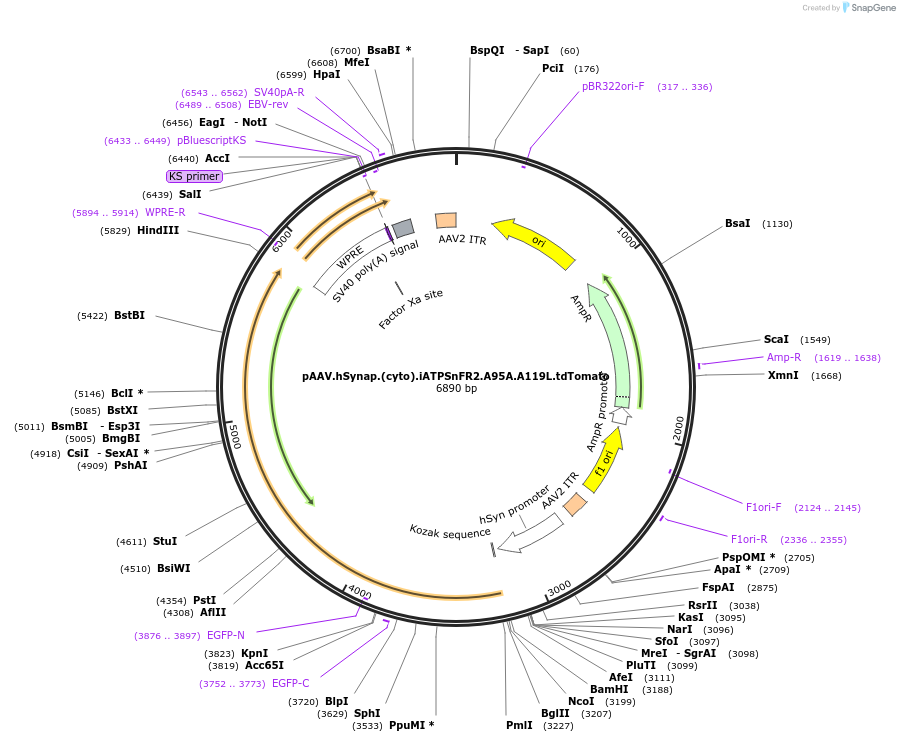
pAAV.hSynap.(cyto).iATPSnFR2.A95A.A119L.tdTomato
Plasmid#244922PurposeiATPSnFR2.A95A.A119L variant with non-responsive tdTomatoDepositorInsertiATPSnFR2.A95A.A119L.tdTomato
UseAAVMutationA95A.A119LAvailable SinceOct. 8, 2025AvailabilityAcademic Institutions and Nonprofits only -
pAAV.SynFLEX.(cyto).iATPSnFR2.A95K.mIRFP670nano3
Plasmid#244916PurposeiATPSnFR2.A95K variant with non-responsive mIRFP670nano3DepositorInsertiATPSnFR2.A95K.mIRFP670nano3
UseAAVMutationA95KAvailable SinceOct. 8, 2025AvailabilityAcademic Institutions and Nonprofits only -
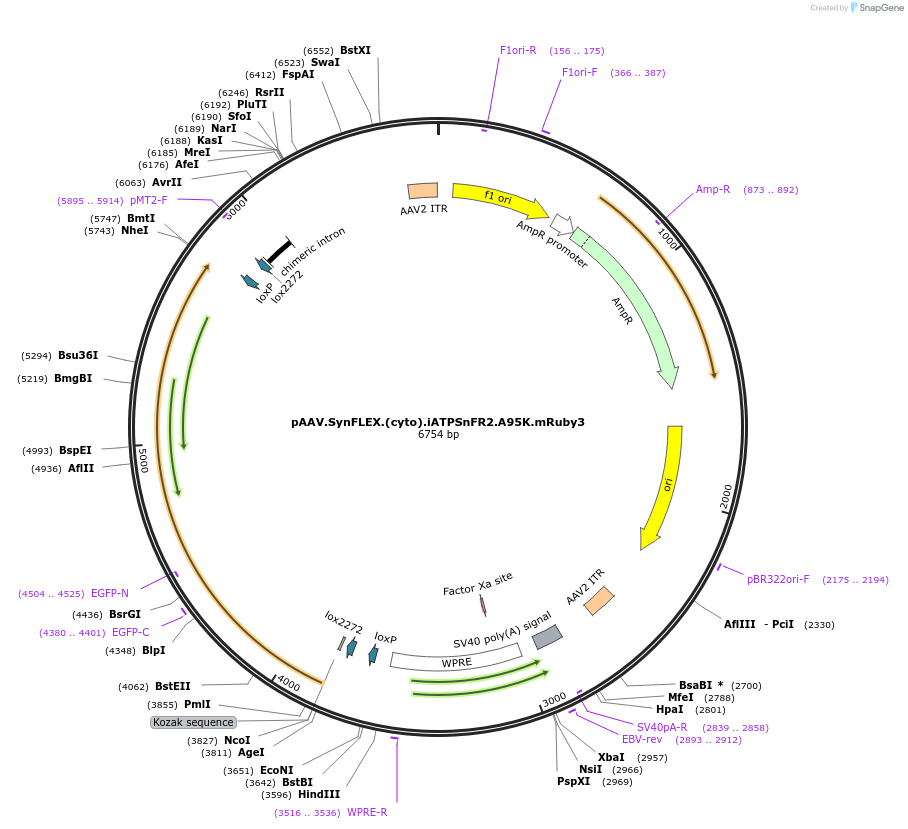
pAAV.SynFLEX.(cyto).iATPSnFR2.A95K.mRuby3
Plasmid#244912PurposeiATPSnFR2.A95K variant with non-responsive mRuby3DepositorInsertiATPSnFR2.A95K.mRuby3
UseAAVMutationA95KAvailable SinceSept. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
pET28a-OuIAD(E502Q)
Plasmid#226973PurposeExpressing Olsenella uli indoleacetate decarboxylase (IAD) E502Q variant in E. coliDepositorInsertOuIAD(E502Q)
TagsHis6 (on backbone)ExpressionBacterialMutationE502QAvailable SinceJan. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
pET28a-OuIAD(W392A)
Plasmid#226980PurposeExpressing Olsenella uli indoleacetate decarboxylase (IAD) W392A variant in E. coliDepositorInsertOuIAD(W392A)
TagsHis6 (on backbone)ExpressionBacterialMutationW392AAvailable SinceNov. 4, 2024AvailabilityAcademic Institutions and Nonprofits only -
pET28a-OuIAD(W392F)
Plasmid#226979PurposeExpressing Olsenella uli indoleacetate decarboxylase (IAD) W392F variant in E. coliDepositorInsertOuIAD(W392F)
TagsHis6 (on backbone)ExpressionBacterialMutationW392FAvailable SinceNov. 4, 2024AvailabilityAcademic Institutions and Nonprofits only -
pET28a-OuIAD(F401A)
Plasmid#226978PurposeExpressing Olsenella uli indoleacetate decarboxylase (IAD) F401A variant in E. coliDepositorInsertOuIAD(F401A)
TagsHis6 (on backbone)ExpressionBacterialMutationF401AAvailable SinceNov. 4, 2024AvailabilityAcademic Institutions and Nonprofits only