We narrowed to 41,179 results for: ACE
-
Plasmid#204592PurposeGST-lamin A (551-646)DepositorInserthuman lamin A (551-646)
TagsGST - lamin A (551-646)ExpressionBacterialAvailable SinceJan. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
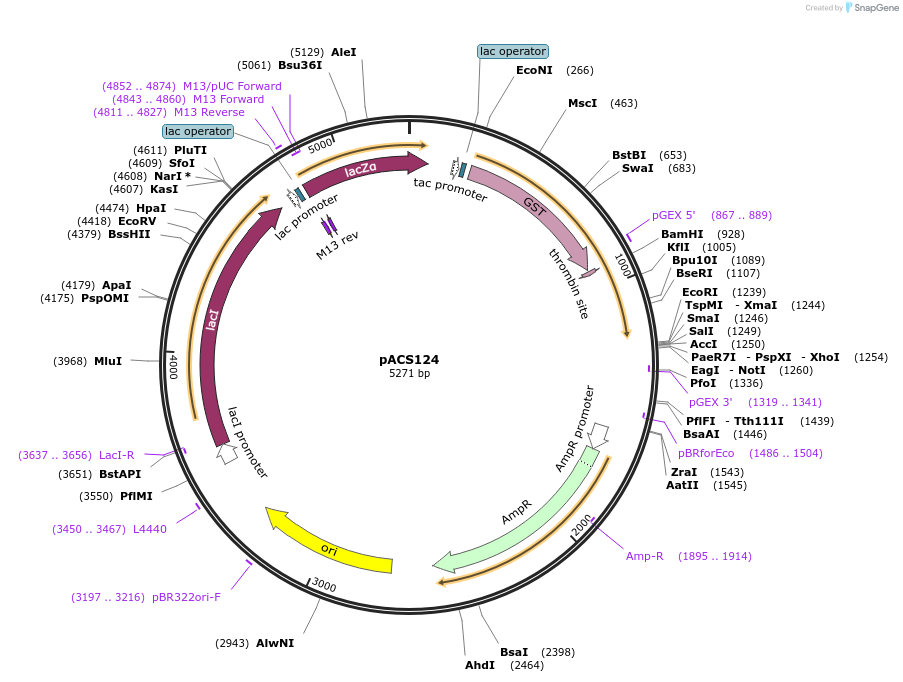
pACS124
Plasmid#204592PurposeGST-lamin A (551-646)DepositorInserthuman lamin A (551-646)
TagsGST - lamin A (551-646)ExpressionBacterialAvailable SinceJan. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
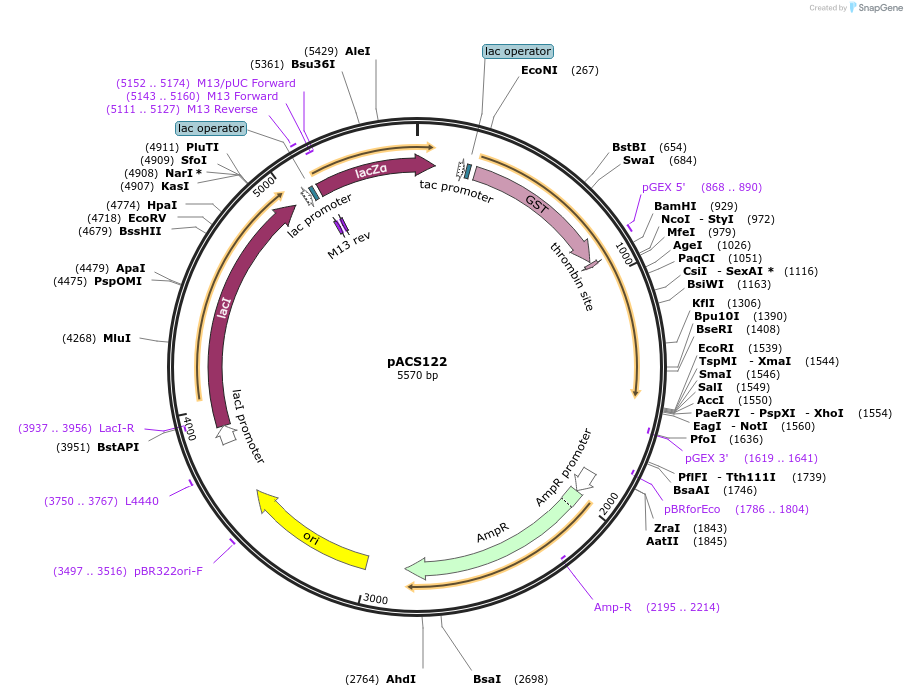
pACS122
Plasmid#204591PurposeGST-lamin A (451-646)DepositorInserthuman lamin A (451-646)
TagsGST - lamin A (451-646)ExpressionBacterialAvailable SinceJan. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
pACS122
Plasmid#204591PurposeGST-lamin A (451-646)DepositorInserthuman lamin A (451-646)
TagsGST - lamin A (451-646)ExpressionBacterialAvailable SinceJan. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
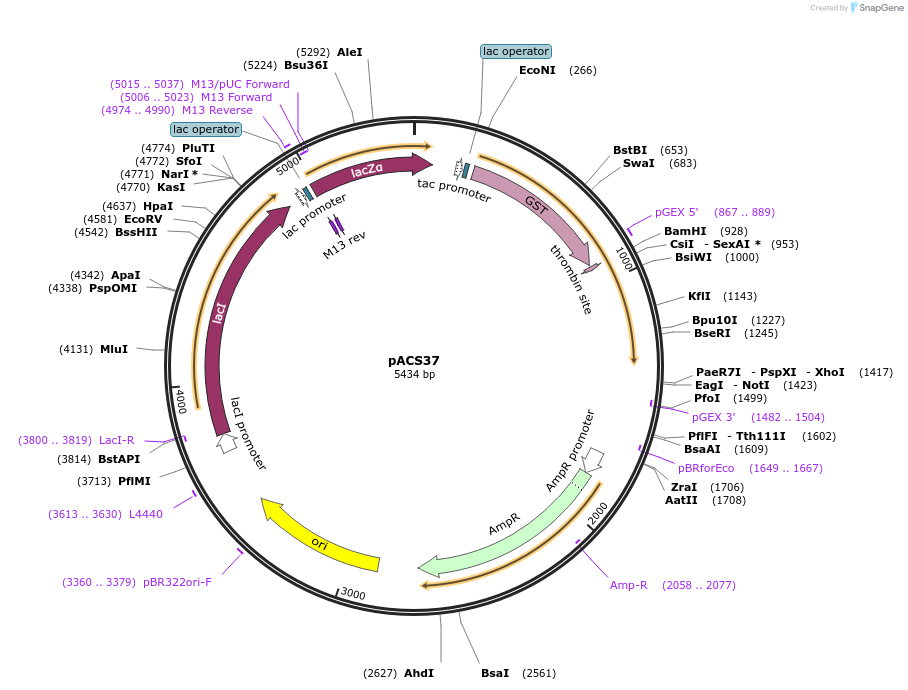
pACS37
Plasmid#204589PurposeGST-lamin A (506-646)DepositorInserthuman lamin A (506-646)
TagsGST - lamin A (506-646)ExpressionBacterialAvailable SinceJan. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
pACS37
Plasmid#204589PurposeGST-lamin A (506-646)DepositorInserthuman lamin A (506-646)
TagsGST - lamin A (506-646)ExpressionBacterialAvailable SinceJan. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
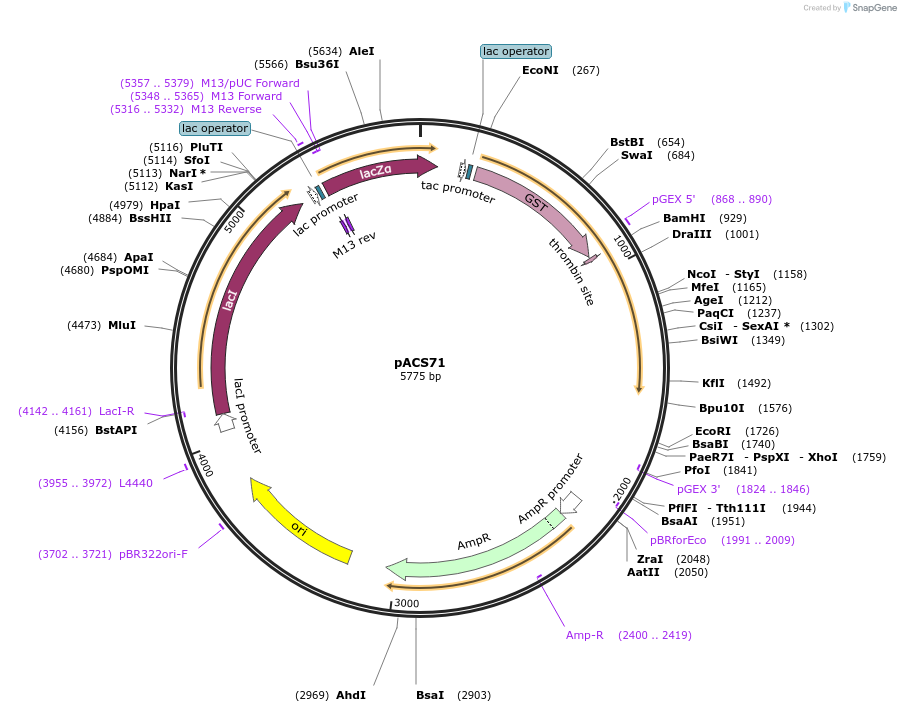
pACS71
Plasmid#204588PurposeGST-lamin A (389-597)DepositorInserthuman lamin A (389-597)
TagsGST-lamin A (389-597)ExpressionBacterialAvailable SinceJan. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
pACS71
Plasmid#204588PurposeGST-lamin A (389-597)DepositorInserthuman lamin A (389-597)
TagsGST-lamin A (389-597)ExpressionBacterialAvailable SinceJan. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
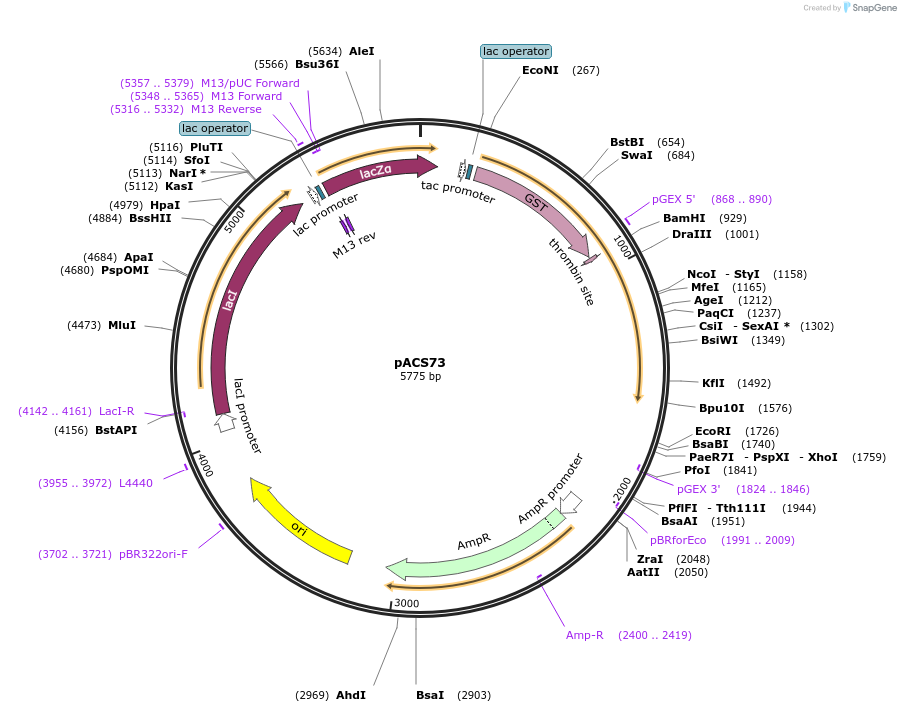
pACS73
Plasmid#204587PurposeGST-lamin A (389-609)DepositorInserthuman lamin A (389-609)
TagsGST-lamin A (389-609)ExpressionBacterialAvailable SinceJan. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
pACS73
Plasmid#204587PurposeGST-lamin A (389-609)DepositorInserthuman lamin A (389-609)
TagsGST-lamin A (389-609)ExpressionBacterialAvailable SinceJan. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
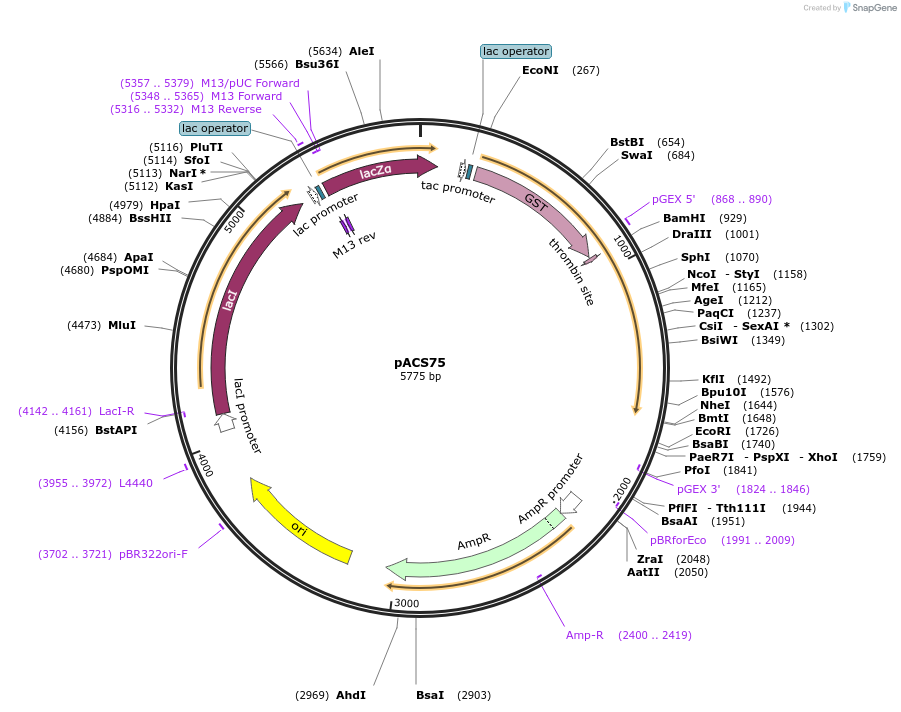
pACS75
Plasmid#204586PurposeGST-lamin A (389-626)DepositorInsertlamin A (389-626)
TagsGST-lamin A (389-626)ExpressionBacterialAvailable SinceJan. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
pACS75
Plasmid#204586PurposeGST-lamin A (389-626)DepositorInsertlamin A (389-626)
TagsGST-lamin A (389-626)ExpressionBacterialAvailable SinceJan. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
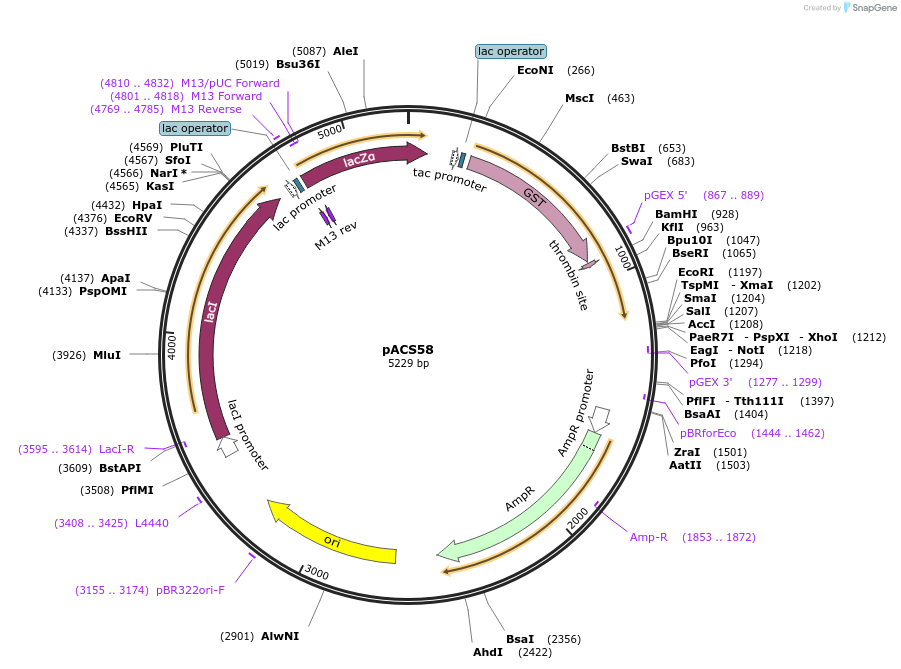
pACS58
Plasmid#204583PurposeGST-lamin A (565-646)DepositorInsertlamin A (565-646)
TagsGST-lamin A (565-646)ExpressionBacterialAvailable SinceJan. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
pACS58
Plasmid#204583PurposeGST-lamin A (565-646)DepositorInsertlamin A (565-646)
TagsGST-lamin A (565-646)ExpressionBacterialAvailable SinceJan. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
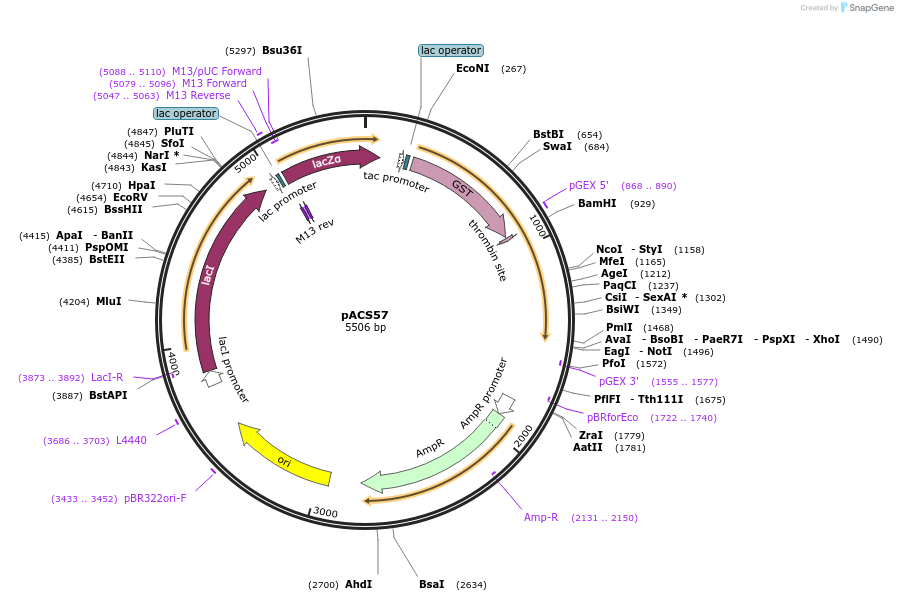
pACS57
Plasmid#204582PurposeGST-lamin C (389-572)DepositorInsertlamin C (389-572)
TagsGST-lamin C (389-572)ExpressionBacterialAvailable SinceJan. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
pACS57
Plasmid#204582PurposeGST-lamin C (389-572)DepositorInsertlamin C (389-572)
TagsGST-lamin C (389-572)ExpressionBacterialAvailable SinceJan. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
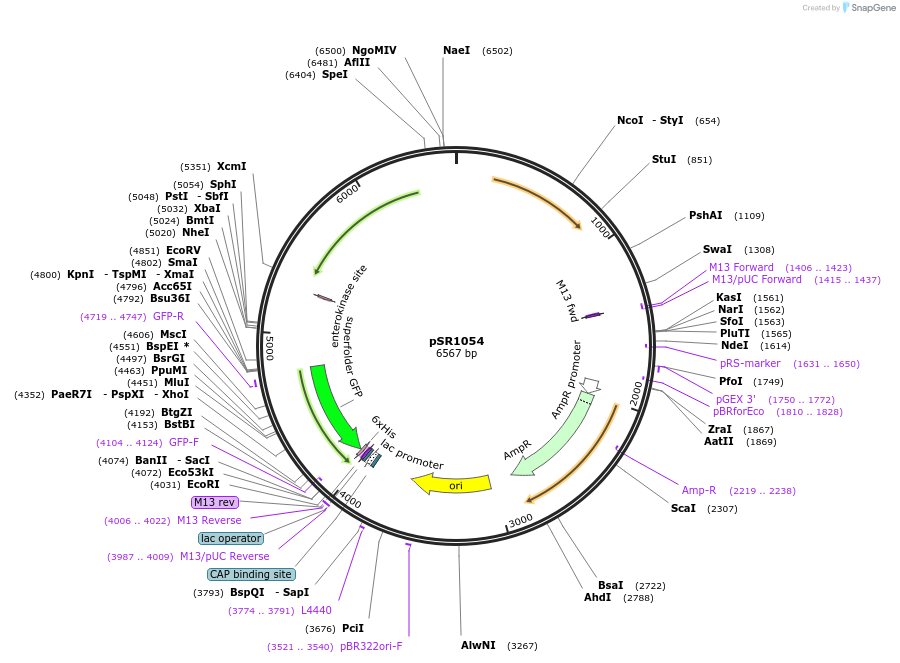
pSR1054
Plasmid#208740PurposeCharacterization plasmid for PAmy (sfgfp expressed from PAmy on the S. aureus pC194 replicon)DepositorInsertPAmy
UseSynthetic BiologyExpressionBacterialPromoterPAmyAvailable SinceDec. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
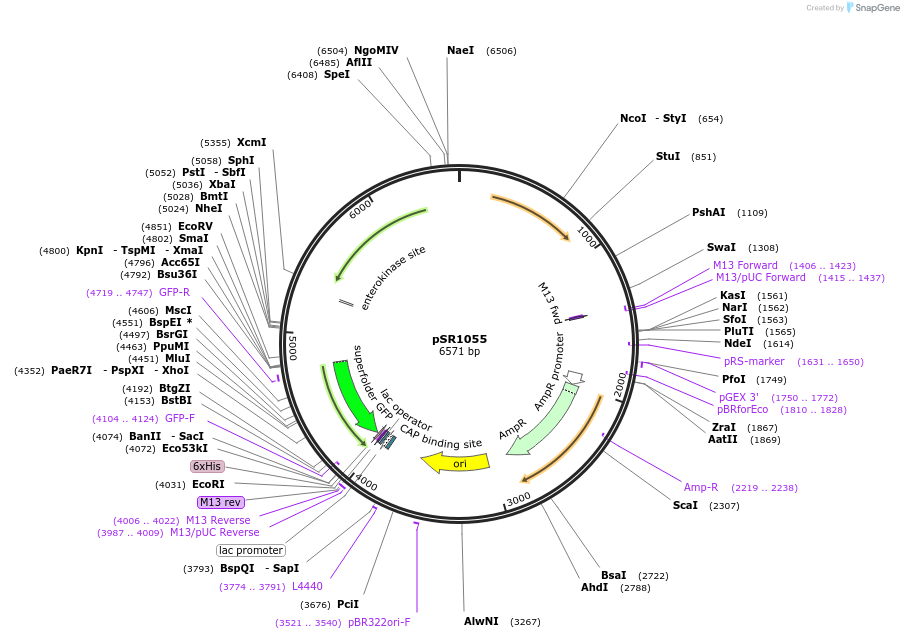
pSR1055
Plasmid#208741PurposeCharacterization plasmid for PHrcA (sfgfp expressed from PHrcA on the S. aureus pC194 replicon)DepositorInsertPHrcA
UseSynthetic BiologyExpressionBacterialPromoterPHrcAAvailable SinceDec. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
pSR1054
Plasmid#208740PurposeCharacterization plasmid for PAmy (sfgfp expressed from PAmy on the S. aureus pC194 replicon)DepositorInsertPAmy
UseSynthetic BiologyExpressionBacterialPromoterPAmyAvailable SinceDec. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
pSR1055
Plasmid#208741PurposeCharacterization plasmid for PHrcA (sfgfp expressed from PHrcA on the S. aureus pC194 replicon)DepositorInsertPHrcA
UseSynthetic BiologyExpressionBacterialPromoterPHrcAAvailable SinceDec. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
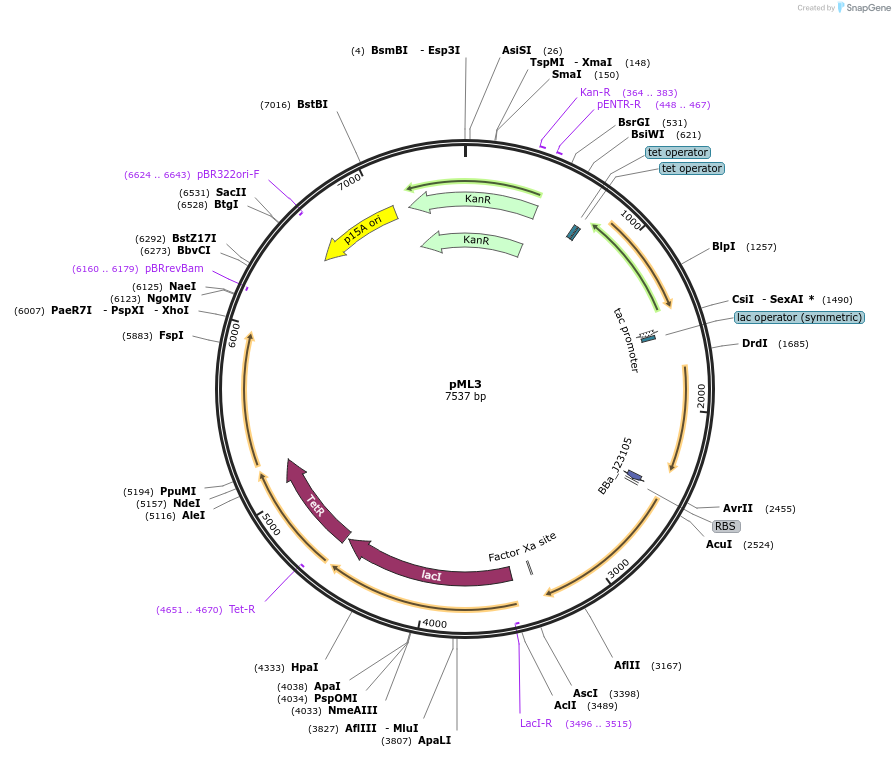
pML3
Plasmid#203337PurposeSR latch circuit on pLW555 backbone: PTet-E1, PTac-P1DepositorInsertsbetI
phlF
PromoterPPhlF and PTet and PTac and PBetIAvailable SinceAug. 29, 2023AvailabilityAcademic Institutions and Nonprofits only -
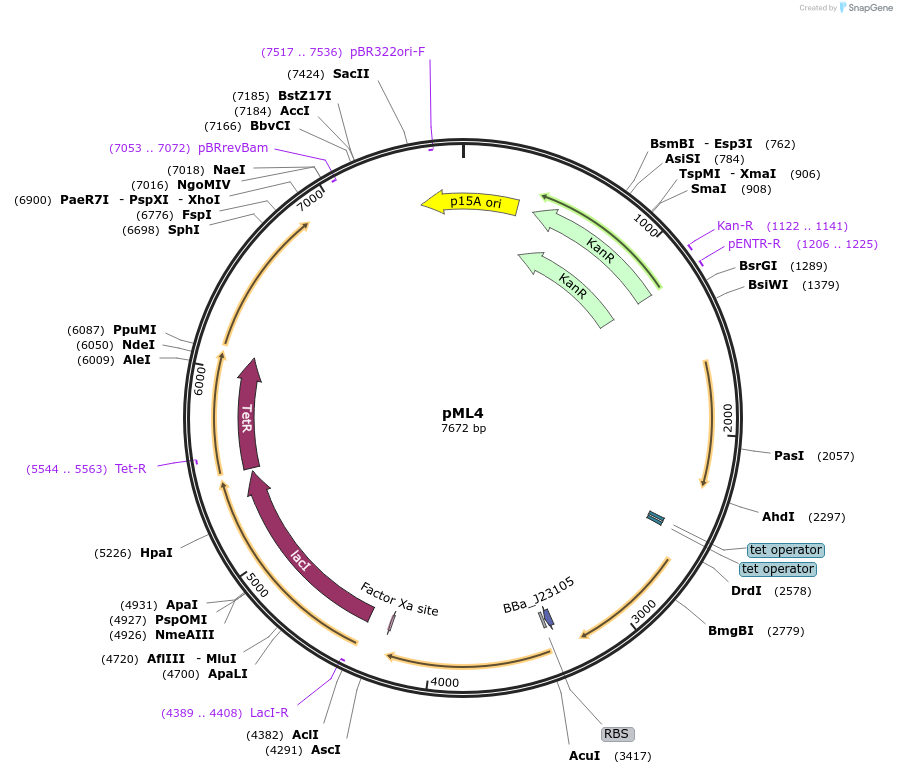
pML4
Plasmid#203338PurposeSR latch circuit on pLW555 backbone: PLux-S3, PTet-P1DepositorInsertssrpR
phlF
PromoterPPhlF and PLux and PSrpR and PTetAvailable SinceAug. 29, 2023AvailabilityAcademic Institutions and Nonprofits only -
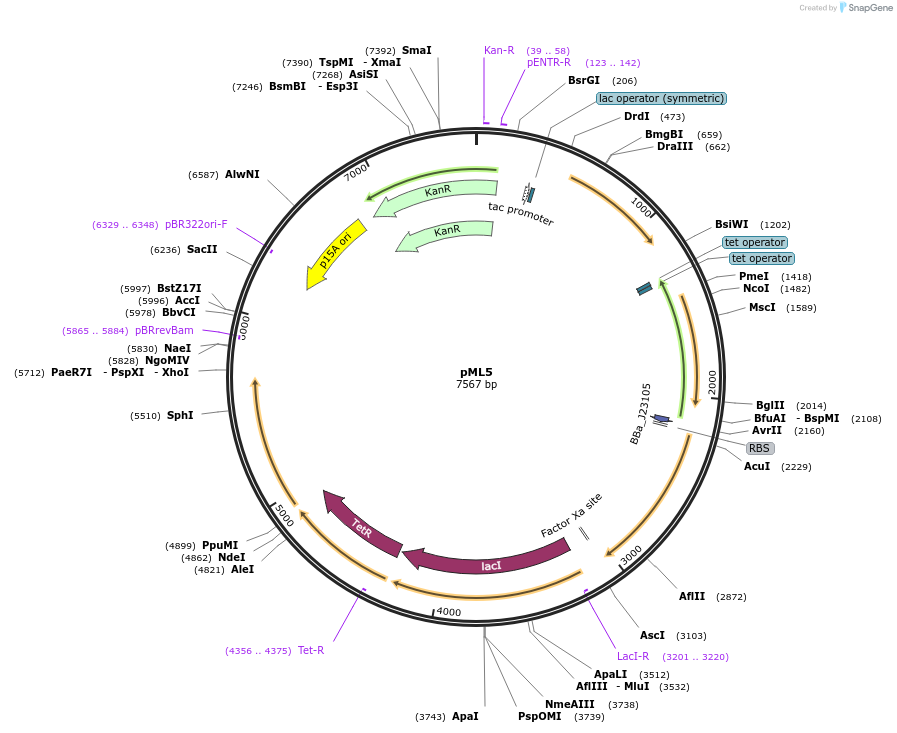
pML5
Plasmid#203339PurposeSR latch circuit on pLW555 backbone: PTac-P3, PTet-H1DepositorInsertsphlF
hlyIIR
PromoterPPhlF and PTet and PTac and PHlyIIRAvailable SinceAug. 29, 2023AvailabilityAcademic Institutions and Nonprofits only -
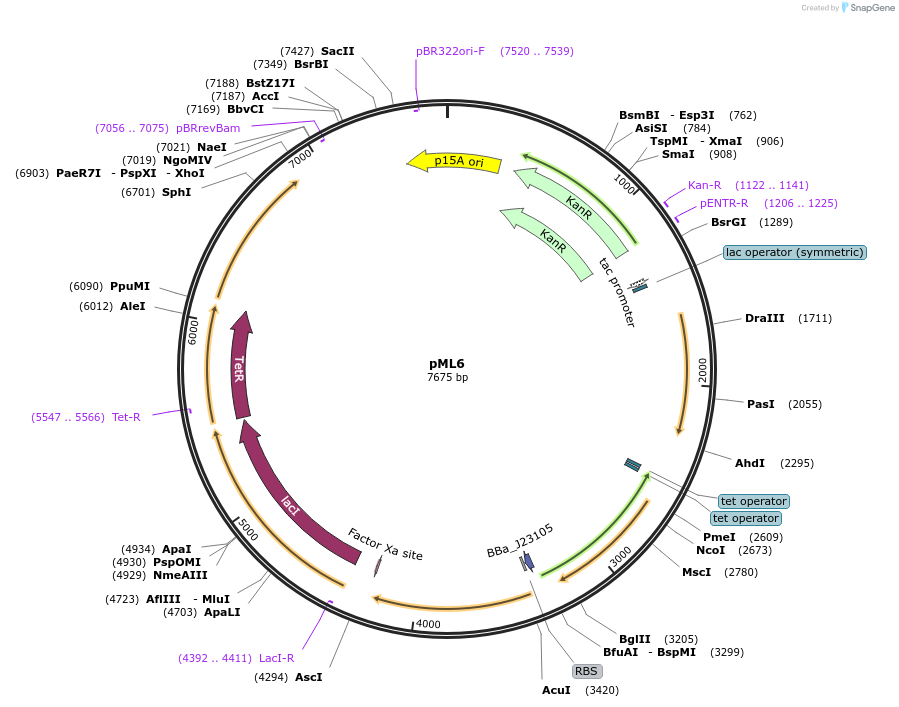
pML6
Plasmid#203340PurposeSR latch circuit on pLW555 backbone: PTac-S2, PTet-H1DepositorInsertssrpR
hlyIIR
PromoterPSrpR and PTet and PTac and PHlyIIRAvailable SinceAug. 29, 2023AvailabilityAcademic Institutions and Nonprofits only -
pML12
Plasmid#203341PurposeSR latch circuit on pLW555 backbone: PTac-S3, PTet-E1DepositorInsertssrpR
betI
PromoterPSrpR and PTet and PTac and PBetIAvailable SinceAug. 29, 2023AvailabilityAcademic Institutions and Nonprofits only -
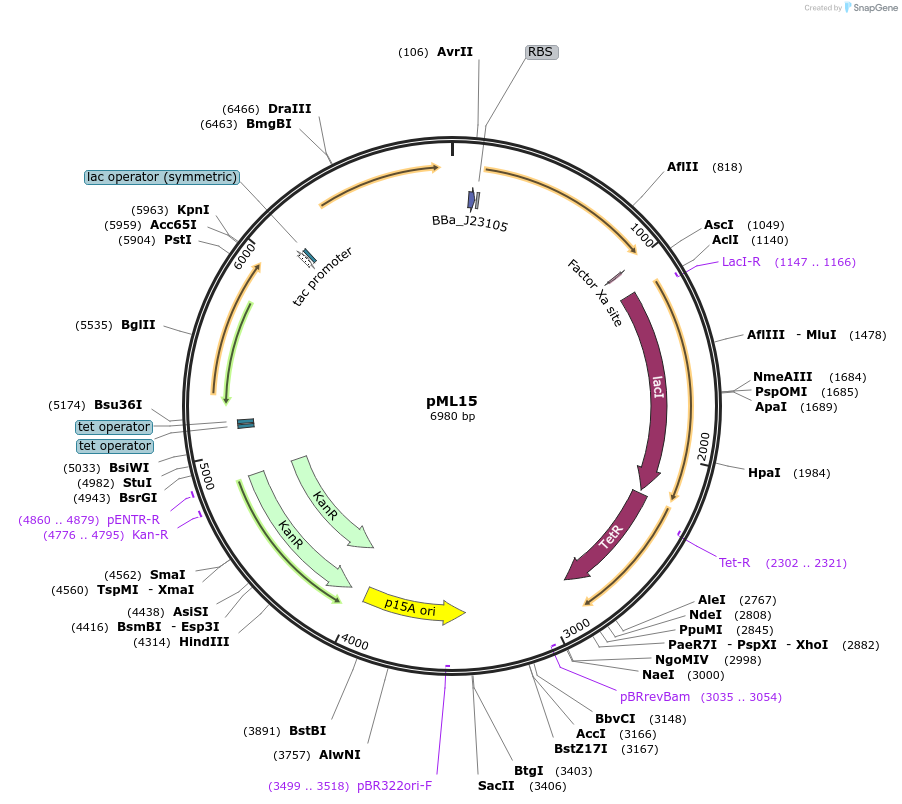
pML15
Plasmid#203342PurposeSR latch circuit on pLW555 backbone: PTet-A1, PTac-P2DepositorInsertsamtR
phlF
PromoterPPhlF and PTet and PTac and PAmtRAvailable SinceAug. 29, 2023AvailabilityAcademic Institutions and Nonprofits only -
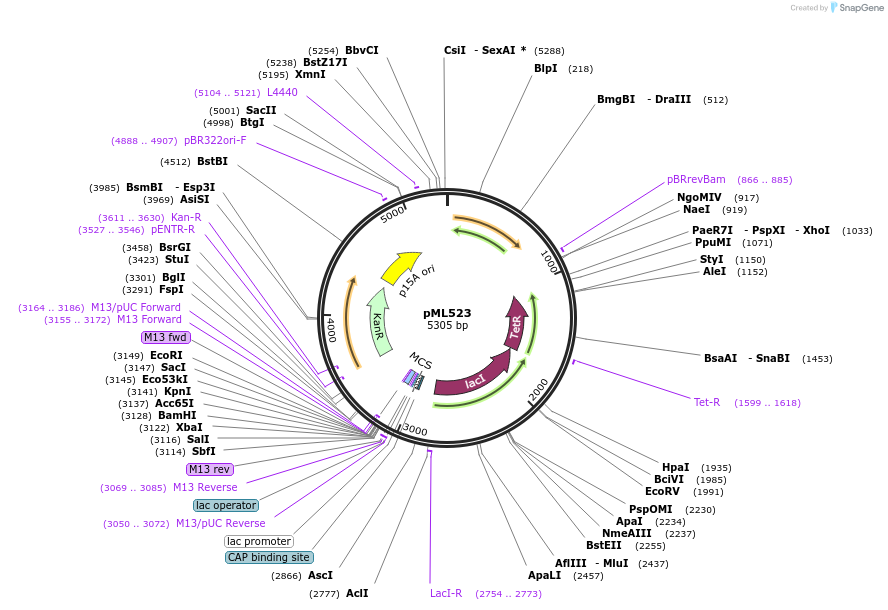
pML523
Plasmid#203345PurposePlasmid backbone for choline sensing circuits. The plasmid contains, lacI and tetR as well as betIM. The plasmid also contains the p15a oriDepositorTypeEmpty backboneAvailable SinceAug. 29, 2023AvailabilityAcademic Institutions and Nonprofits only -
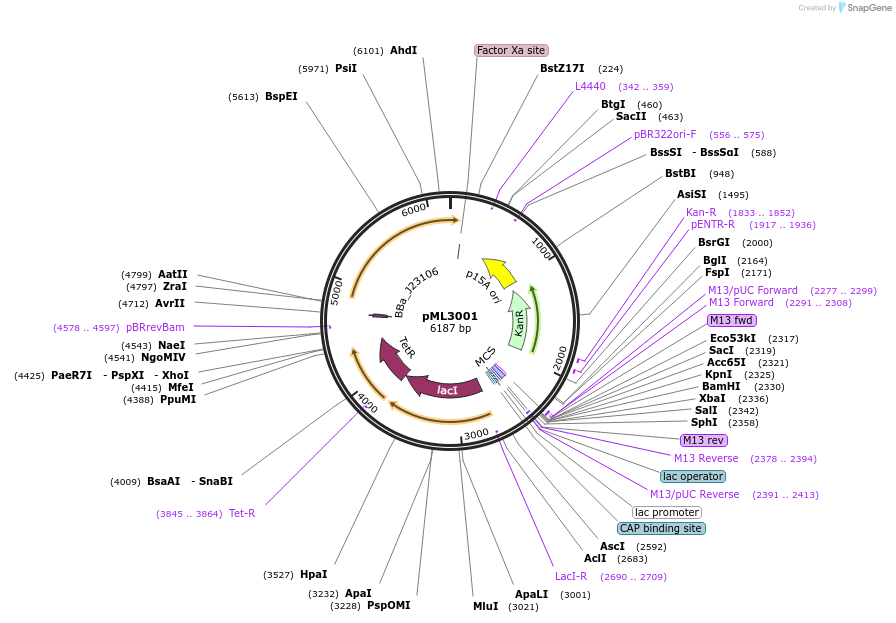
pML3001
Plasmid#203346PurposePlasmid backbone for gamma-aminobutyric acid circuits. The plasmid contains gabR, lacI and tetR as well as the resistance gene kanR and the p15a ori.DepositorTypeEmpty backboneAvailable SinceAug. 29, 2023AvailabilityAcademic Institutions and Nonprofits only -
pML3
Plasmid#203337PurposeSR latch circuit on pLW555 backbone: PTet-E1, PTac-P1DepositorInsertsbetI
phlF
PromoterPPhlF and PTet and PTac and PBetIAvailable SinceAug. 29, 2023AvailabilityAcademic Institutions and Nonprofits only -
pML4
Plasmid#203338PurposeSR latch circuit on pLW555 backbone: PLux-S3, PTet-P1DepositorInsertssrpR
phlF
PromoterPPhlF and PLux and PSrpR and PTetAvailable SinceAug. 29, 2023AvailabilityAcademic Institutions and Nonprofits only -
pML5
Plasmid#203339PurposeSR latch circuit on pLW555 backbone: PTac-P3, PTet-H1DepositorInsertsphlF
hlyIIR
PromoterPPhlF and PTet and PTac and PHlyIIRAvailable SinceAug. 29, 2023AvailabilityAcademic Institutions and Nonprofits only -
pML6
Plasmid#203340PurposeSR latch circuit on pLW555 backbone: PTac-S2, PTet-H1DepositorInsertssrpR
hlyIIR
PromoterPSrpR and PTet and PTac and PHlyIIRAvailable SinceAug. 29, 2023AvailabilityAcademic Institutions and Nonprofits only -
pML12
Plasmid#203341PurposeSR latch circuit on pLW555 backbone: PTac-S3, PTet-E1DepositorInsertssrpR
betI
PromoterPSrpR and PTet and PTac and PBetIAvailable SinceAug. 29, 2023AvailabilityAcademic Institutions and Nonprofits only -
pML15
Plasmid#203342PurposeSR latch circuit on pLW555 backbone: PTet-A1, PTac-P2DepositorInsertsamtR
phlF
PromoterPPhlF and PTet and PTac and PAmtRAvailable SinceAug. 29, 2023AvailabilityAcademic Institutions and Nonprofits only -
pML523
Plasmid#203345PurposePlasmid backbone for choline sensing circuits. The plasmid contains, lacI and tetR as well as betIM. The plasmid also contains the p15a oriDepositorTypeEmpty backboneAvailable SinceAug. 29, 2023AvailabilityAcademic Institutions and Nonprofits only -
pML3001
Plasmid#203346PurposePlasmid backbone for gamma-aminobutyric acid circuits. The plasmid contains gabR, lacI and tetR as well as the resistance gene kanR and the p15a ori.DepositorTypeEmpty backboneAvailable SinceAug. 29, 2023AvailabilityAcademic Institutions and Nonprofits only -
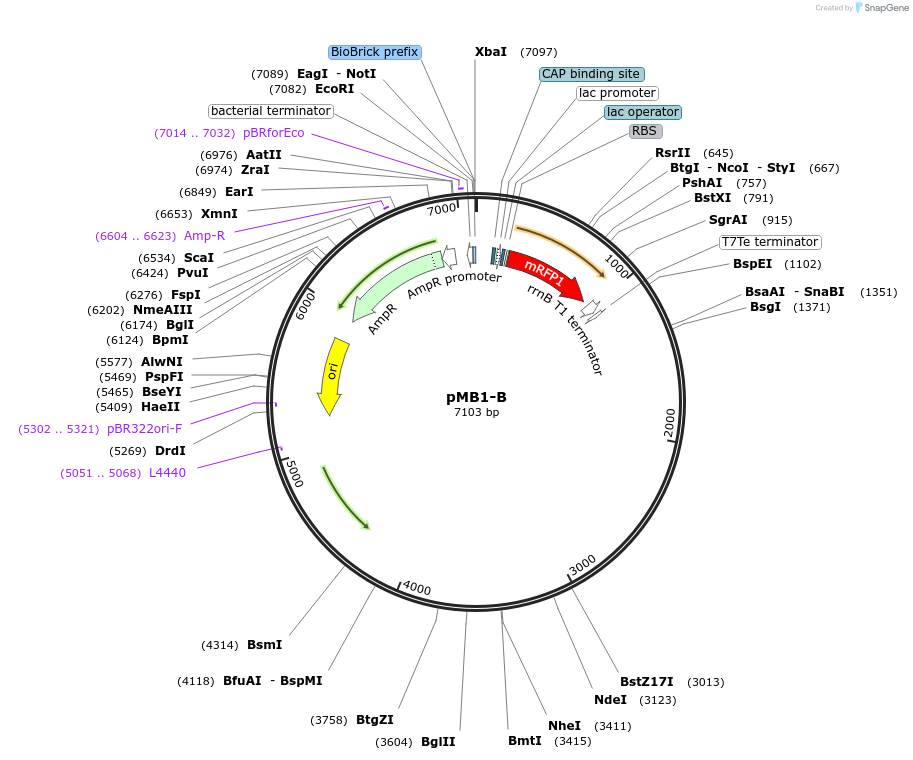
pMB1-B
Plasmid#190115PurposeLevel 1 MoClo plasmid for A. baumanniiDepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterialAvailable SinceAug. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
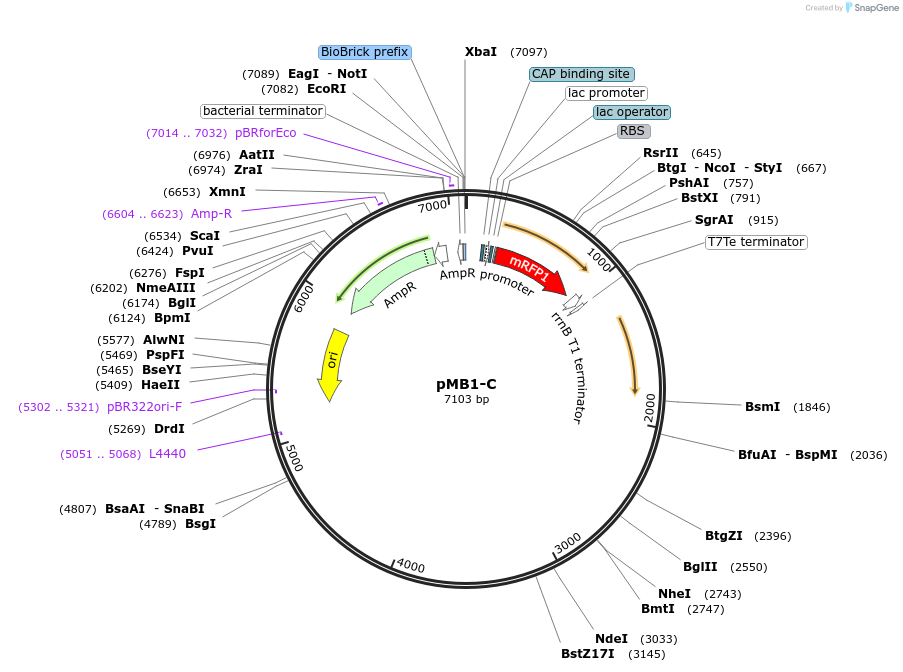
pMB1-C
Plasmid#190116PurposeLevel 1 MoClo plasmid for A. baumanniiDepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterialAvailable SinceAug. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
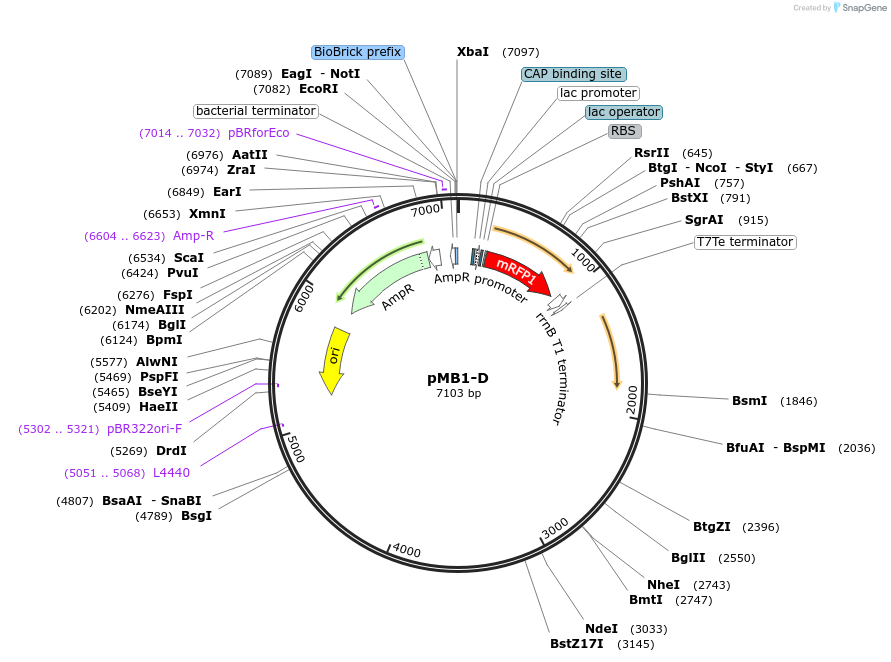
pMB1-D
Plasmid#190117PurposeLevel 1 MoClo plasmid for A. baumanniiDepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterialAvailable SinceAug. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
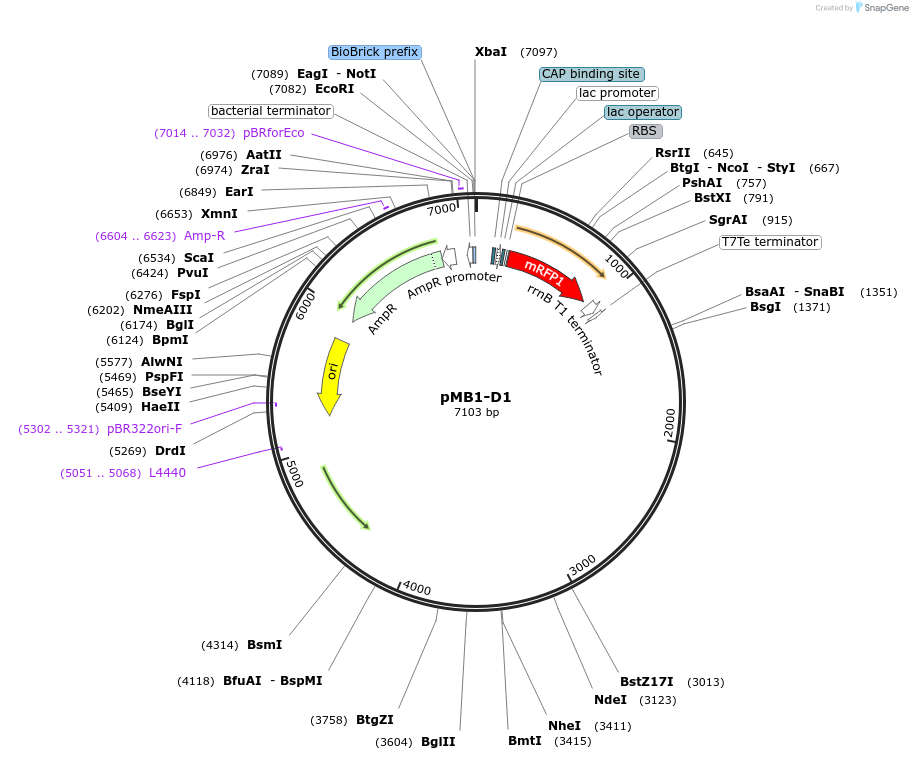
pMB1-D1
Plasmid#190118PurposeLevel 1 MoClo plasmid for A. baumanniiDepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterialAvailable SinceAug. 24, 2023AvailabilityAcademic Institutions and Nonprofits only