We narrowed to 20,061 results for: INO
-
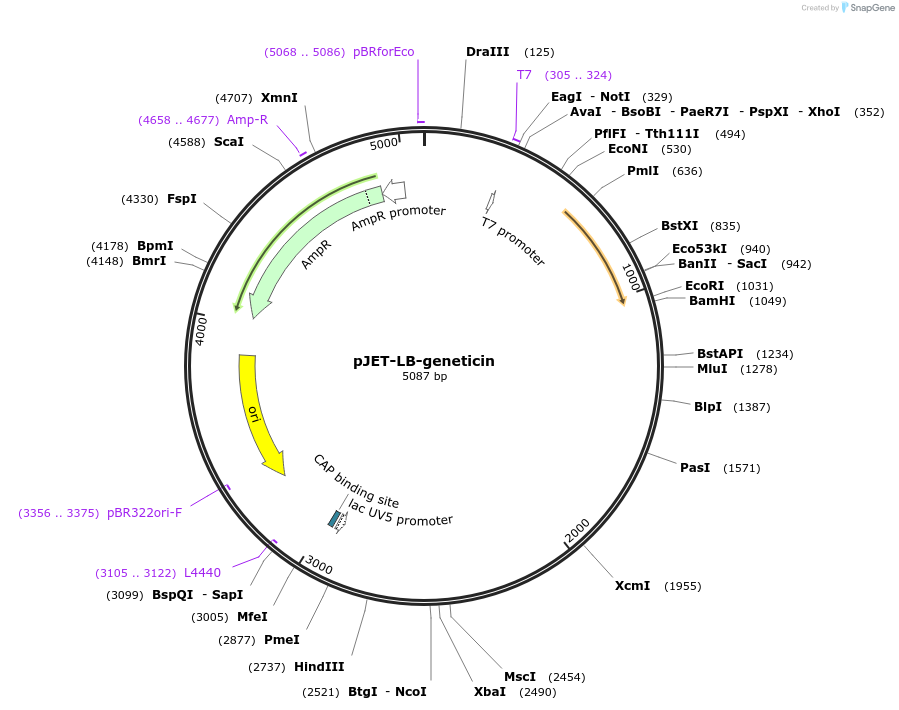
Plasmid#213468PurposeVector to amplify the LB-geneticin fragment for transformation into TSI locus 1DepositorInsertLeft border of the TSI locus 1-split fragment of geneticin cassette
ExpressionBacterialAvailable SinceApril 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
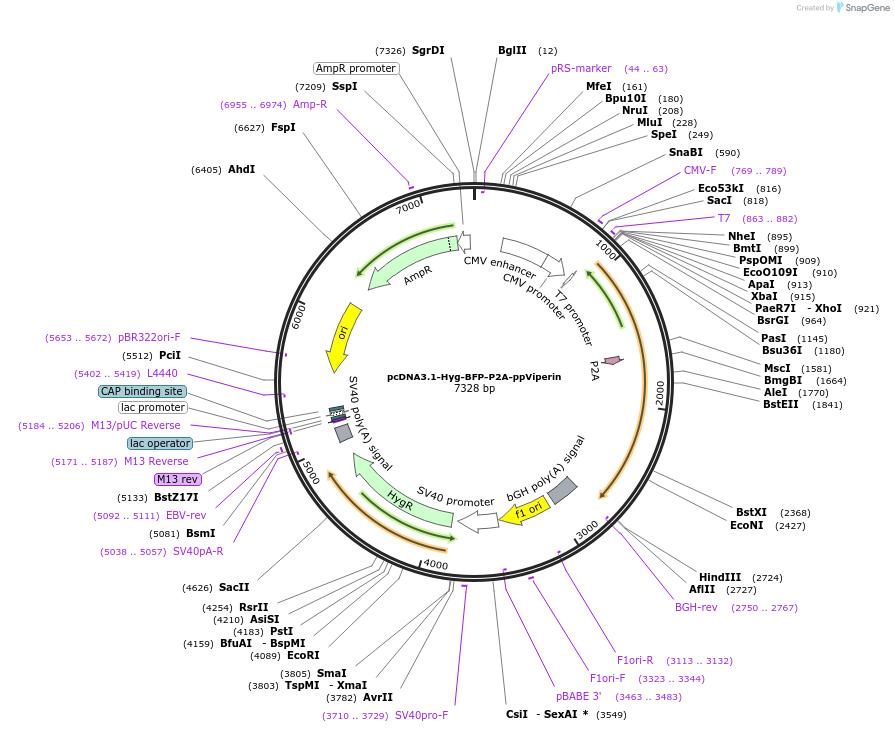
pcDNA3.1-Hyg-BFP-P2A-ppViperin
Plasmid#217481PurposeExpression of Blue Fluorescent Protein - P2A - full length protein kinase R from chinook salmonDepositorInsertBFP-P2A-otPKRFL
ExpressionMammalianPromoterCMVAvailable SinceApril 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
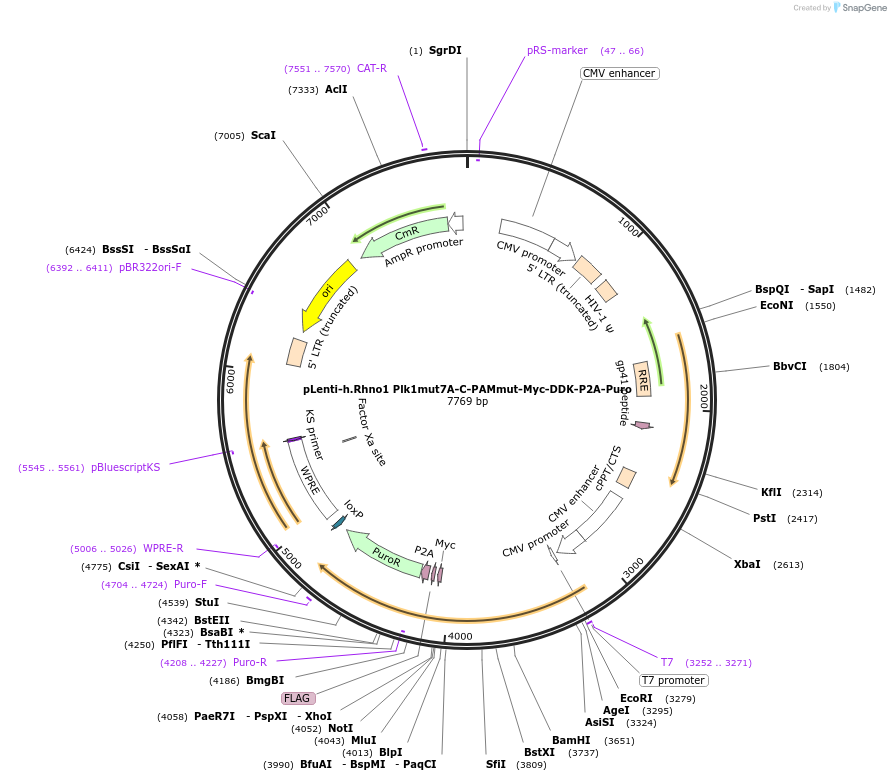
pLenti-h.Rhno1 Plk1mut7A-C-PAMmut-Myc-DDK-P2A-Puro
Plasmid#211616PurposeRhno1 Plk1mut7ADepositorInserthRHNO1 (RHNO1 Human)
TagsmycExpressionMammalianMutationS51A, T52A, S141A, S162A, S163A, S164A, S178APromoterCMVAvailable SinceApril 18, 2024AvailabilityAcademic Institutions and Nonprofits only -
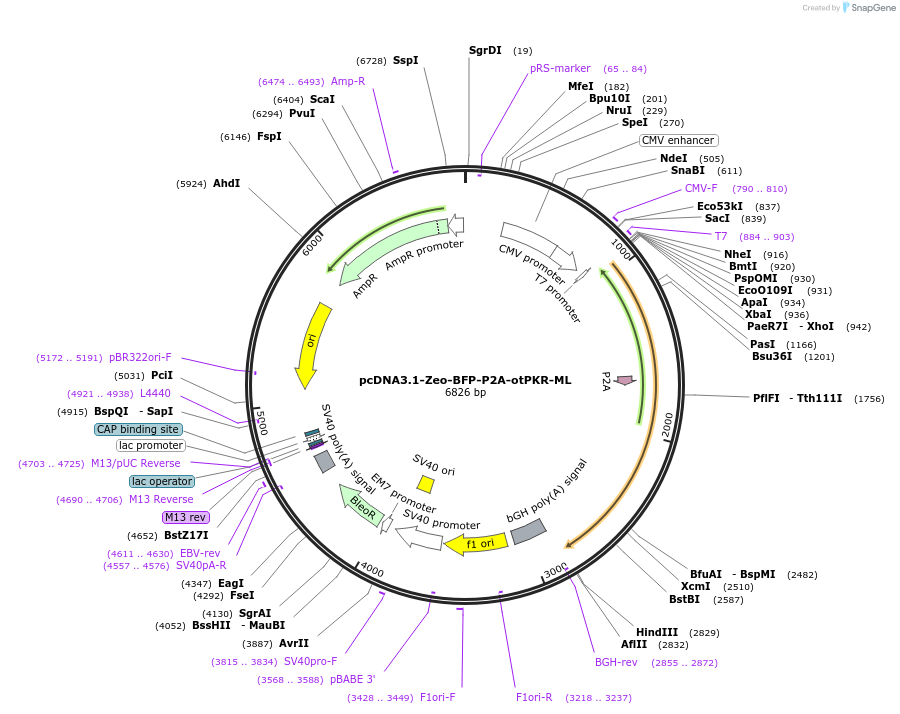
pcDNA3.1-Zeo-BFP-P2A-otPKR-ML
Plasmid#214365PurposeExpression of the Blue Fluorescent Protein - P2A - medium length protein kinase R from chinook salmonDepositorInsertBFP-P2A-otPKRML
ExpressionMammalianPromoterCMVAvailable SinceFeb. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
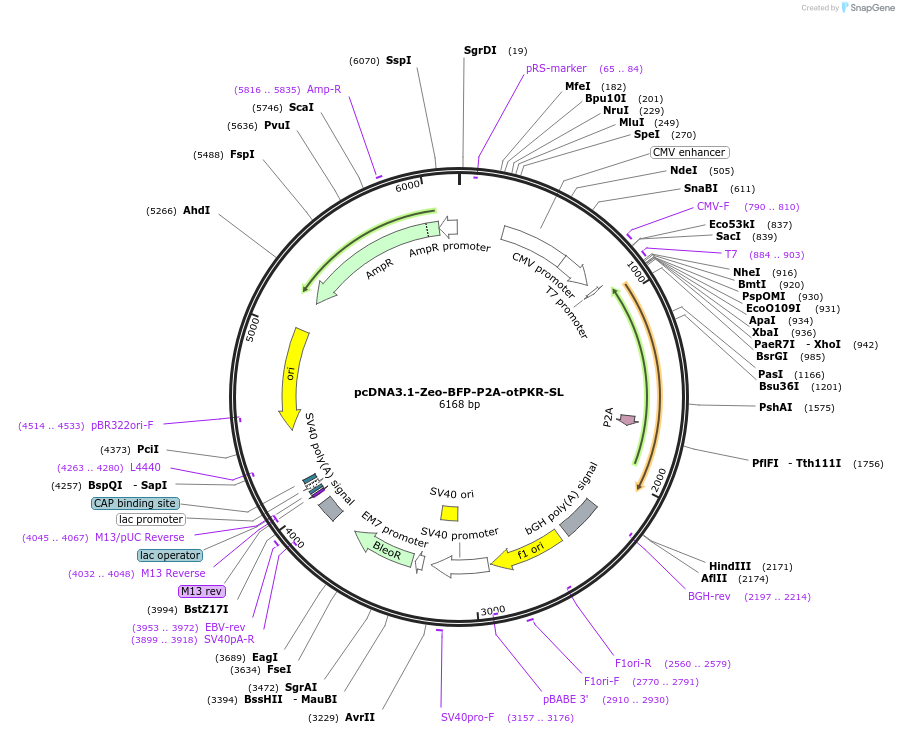
pcDNA3.1-Zeo-BFP-P2A-otPKR-SL
Plasmid#214366PurposeExpression of the Blue Fluorescent Protein - P2A - short length protein kinase R from chinook salmonDepositorInsertBFP-P2A-otPKRSL
ExpressionMammalianPromoterCMVAvailable SinceFeb. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
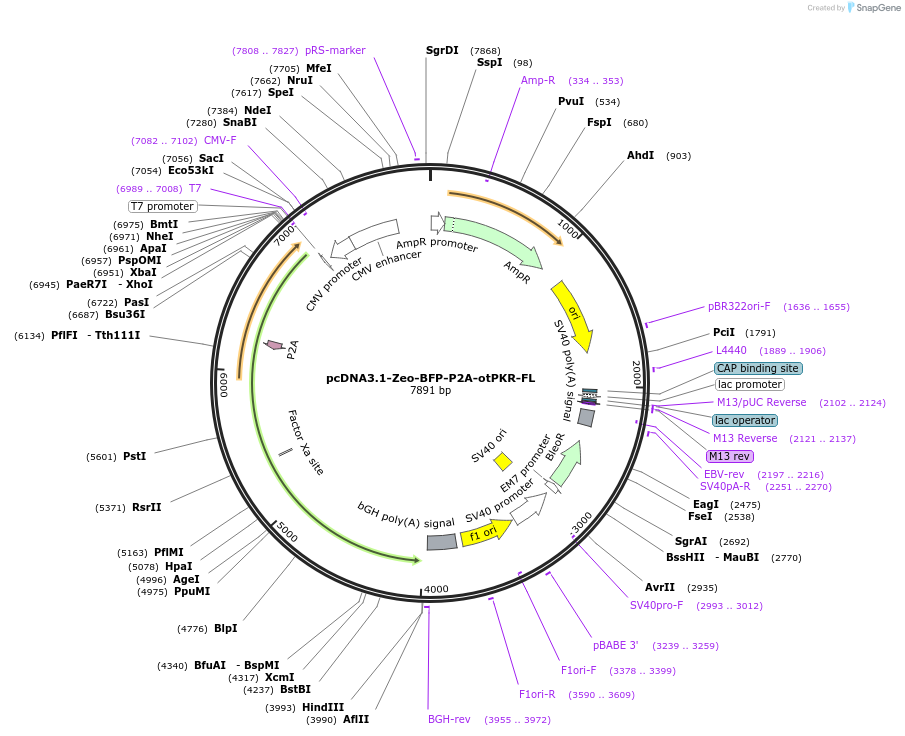
pcDNA3.1-Zeo-BFP-P2A-otPKR-FL
Plasmid#214364PurposeExpression of the Blue Fluorescent Protein - P2A - full length protein kinase R from chinook salmonDepositorInsertBFP-P2A-otPKRFL
ExpressionMammalianPromoterCMVAvailable SinceFeb. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
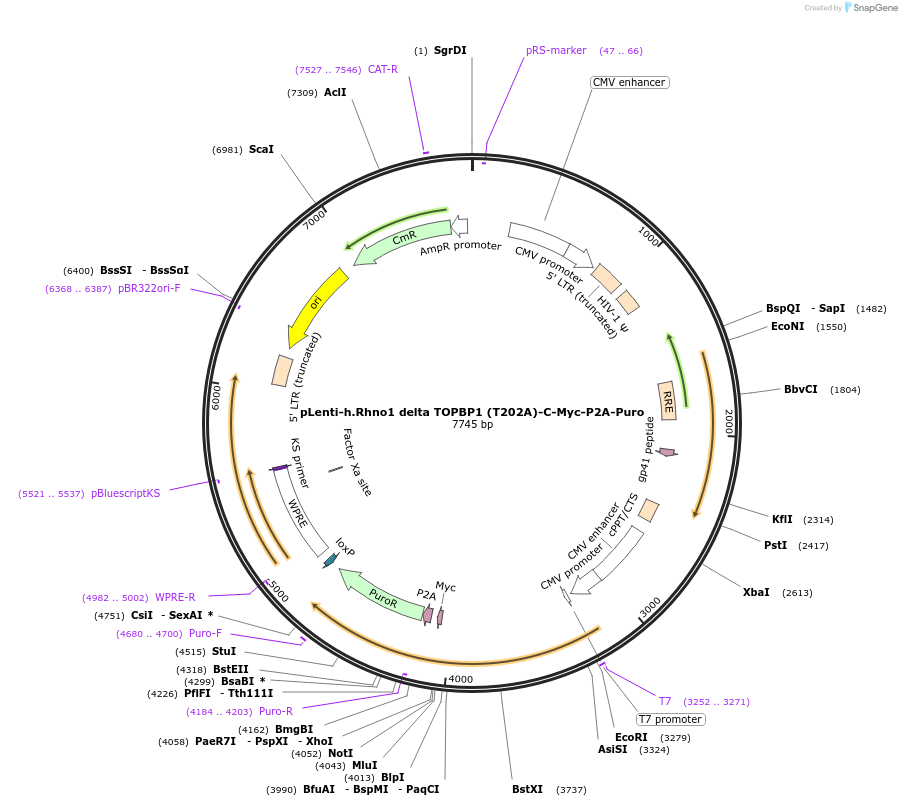
pLenti-h.Rhno1 delta TOPBP1 (T202A)-C-Myc-P2A-Puro
Plasmid#211619PurposeRhno1 delta TOPBP1 (T202A)DepositorAvailable SinceJan. 22, 2024AvailabilityAcademic Institutions and Nonprofits only -
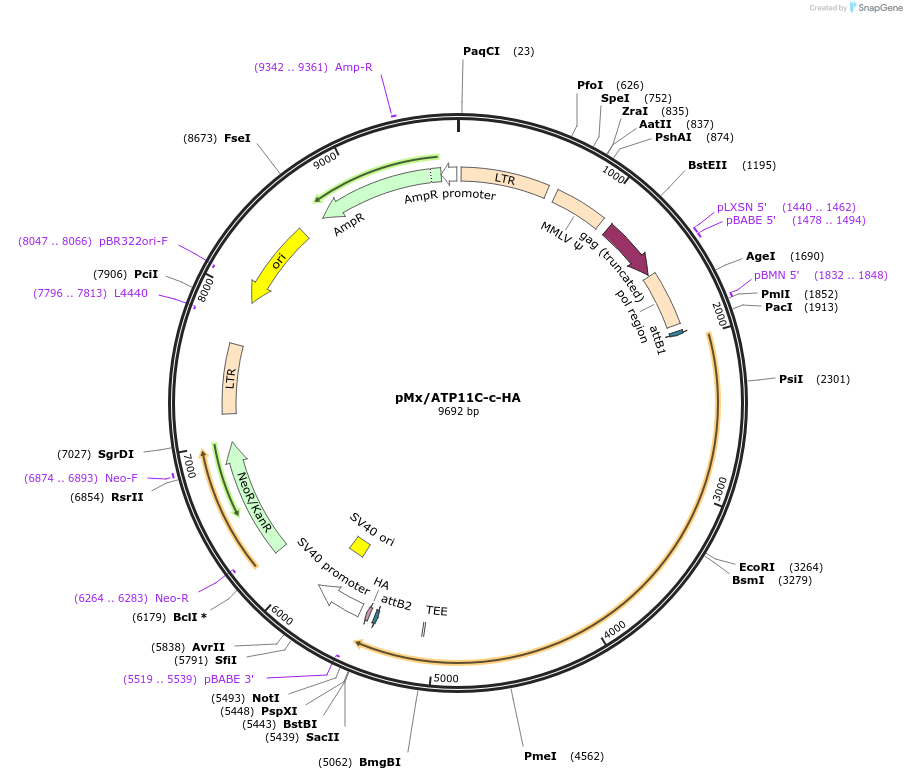
pMx/ATP11C-c-HA
Plasmid#209239PurposeMammalian expression of ATP11C isoform cDepositorAvailable SinceNov. 7, 2023AvailabilityAcademic Institutions and Nonprofits only -
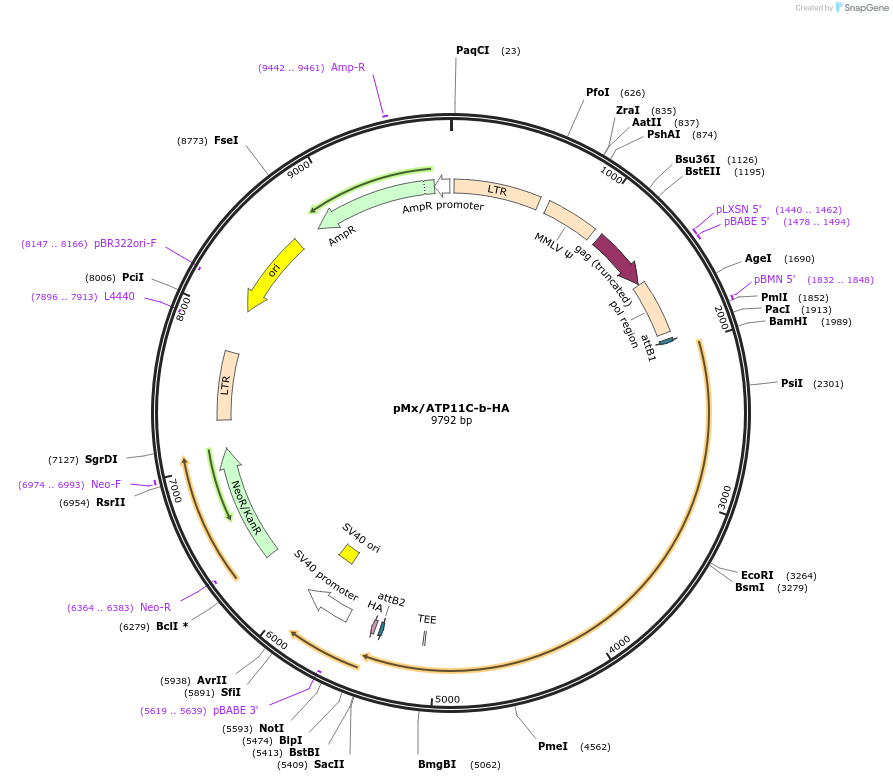
pMx/ATP11C-b-HA
Plasmid#209238PurposeMammalian expression of ATP11C isoform bDepositorAvailable SinceNov. 7, 2023AvailabilityAcademic Institutions and Nonprofits only -
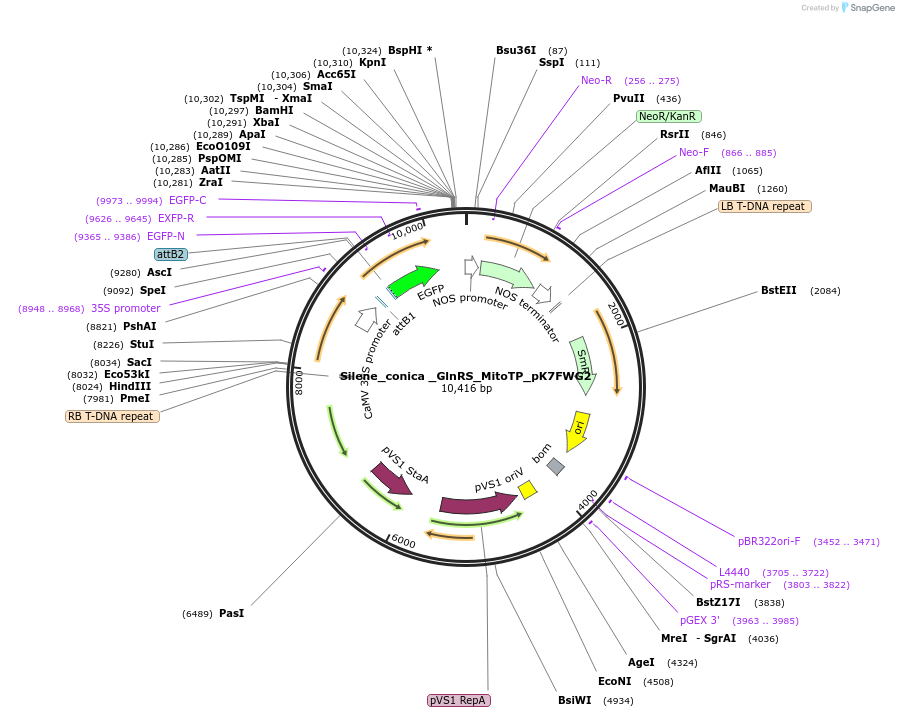
Silene_conica _GlnRS_MitoTP_pK7FWG2
Plasmid#202654PurposeDetermine where transit peptide targets GFP in N. benthamianaDepositorInsertN-terminal transit peptide from Silene conica cytosolic GlnRS predicted to be re-targeted to mitochondria
TagsGreen Florescent ProtienExpressionPlantAvailable SinceAug. 10, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
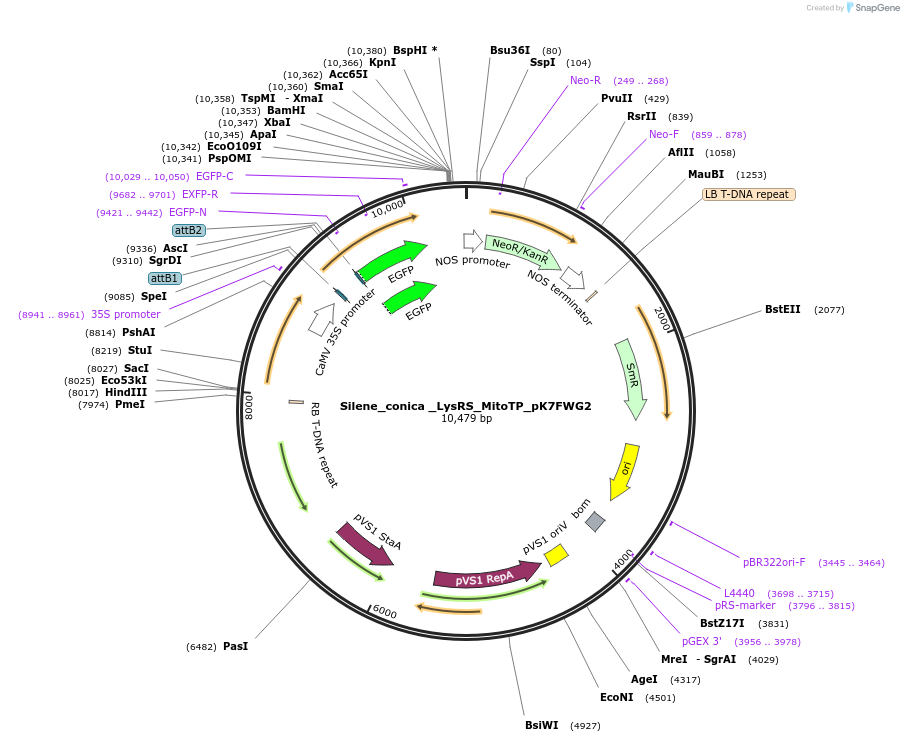
Silene_conica _LysRS_MitoTP_pK7FWG2
Plasmid#202655PurposeDetermine where transit peptide targets GFP in N. benthamianaDepositorInsertN-terminal transit peptide from Silene conica cytosolic LysRS predicted to be re-targeted to mitochondria
TagsGreen Florescent ProtienExpressionPlantAvailable SinceAug. 10, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
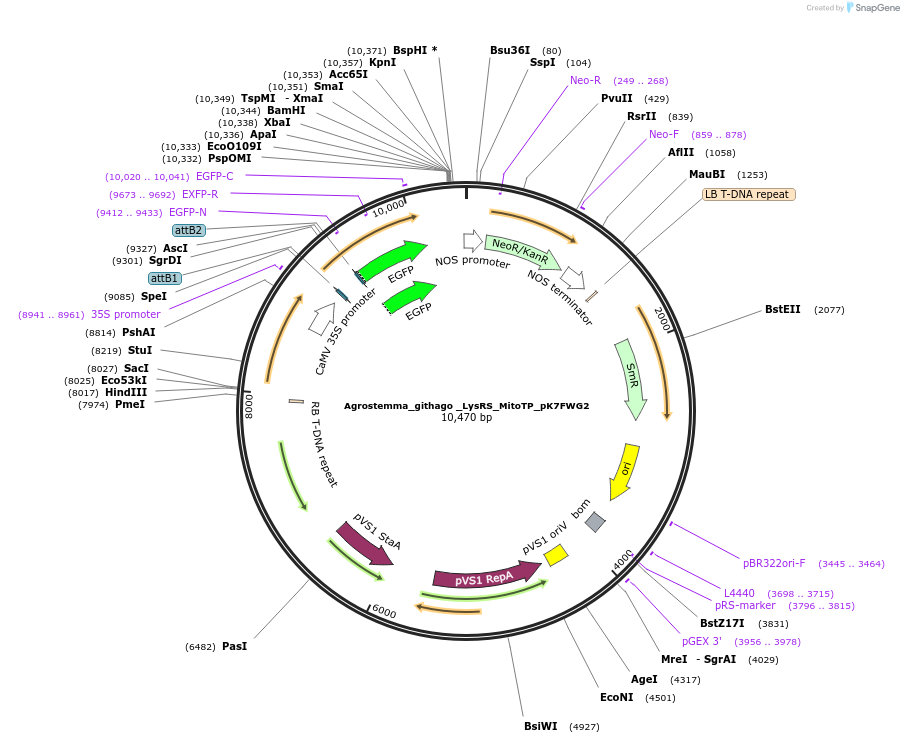
Agrostemma_githago _LysRS_MitoTP_pK7FWG2
Plasmid#202656PurposeDetermine where transit peptide targets GFP in N. benthamianaDepositorInsertN-terminal transit peptide from Agrostemma githago cytosolic LysRS predicted to be re-targeted to mitochondria
TagsGreen Florescent ProtienExpressionPlantAvailable SinceAug. 10, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
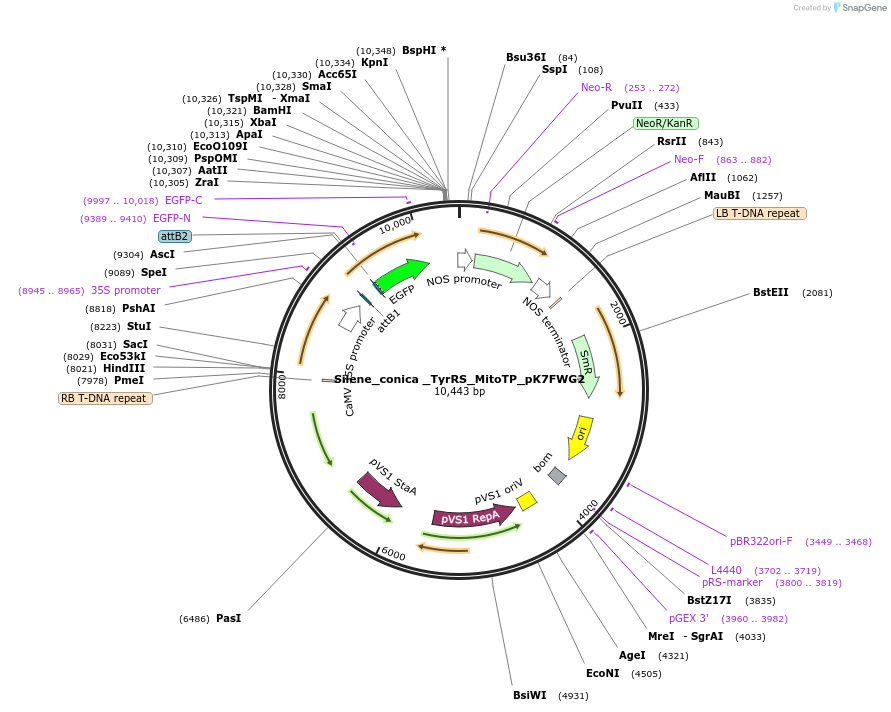
Silene_conica _TyrRS_MitoTP_pK7FWG2
Plasmid#202657PurposeDetermine where transit peptide targets GFP in N. benthamianaDepositorInsertN-terminal transit peptide from Silene conica cytosolic TyrRS predicted to be re-targeted to mitochondria
TagsGreen Florescent ProtienExpressionPlantAvailable SinceAug. 10, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
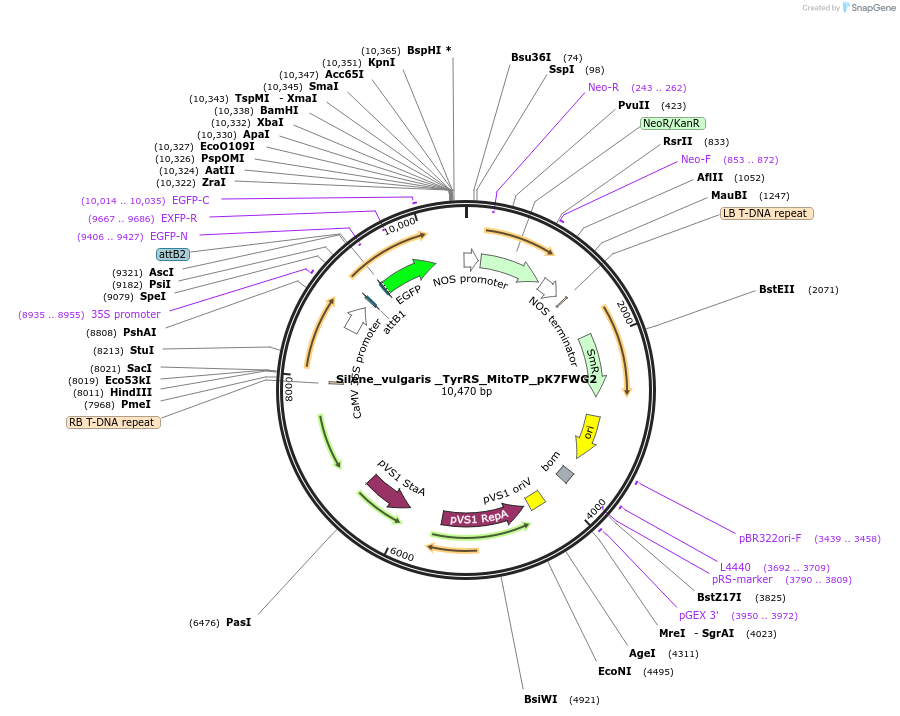
Silene_vulgaris _TyrRS_MitoTP_pK7FWG2
Plasmid#202658PurposeDetermine where transit peptide targets GFP in N. benthamianaDepositorInsertN-terminal transit peptide from Silene vulgaris cytosolic TyrRS predicted to be re-targeted to mitochondria
TagsGreen Florescent ProtienExpressionPlantAvailable SinceAug. 10, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
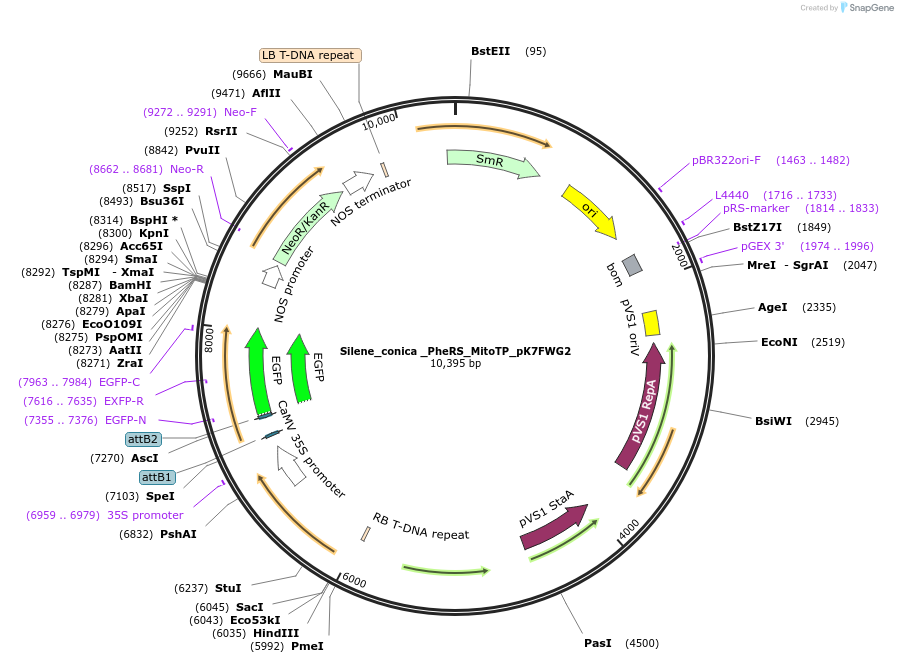
Silene_conica _PheRS_MitoTP_pK7FWG2
Plasmid#202659PurposeDetermine where transit peptide targets GFP in N. benthamianaDepositorInsertN-terminal transit peptide from organellar Silene conica PheRS predicted to be targeted to mito
TagsGreen Florescent ProtienExpressionPlantAvailable SinceAug. 10, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
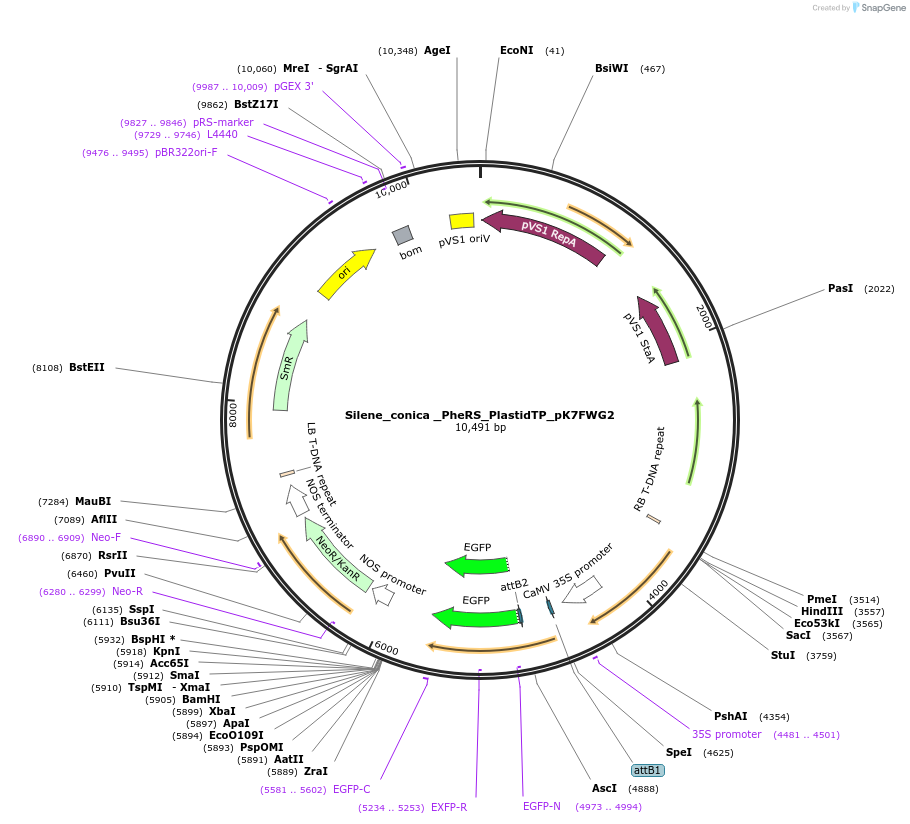
Silene_conica _PheRS_PlastidTP_pK7FWG2
Plasmid#202660PurposeDetermine where transit peptide targets GFP in N. benthamianaDepositorInsertN-terminal transit peptide from organellar Silene conica PheRS predicted to be targeted to plastid
TagsGreen Florescent ProtienExpressionPlantAvailable SinceAug. 10, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
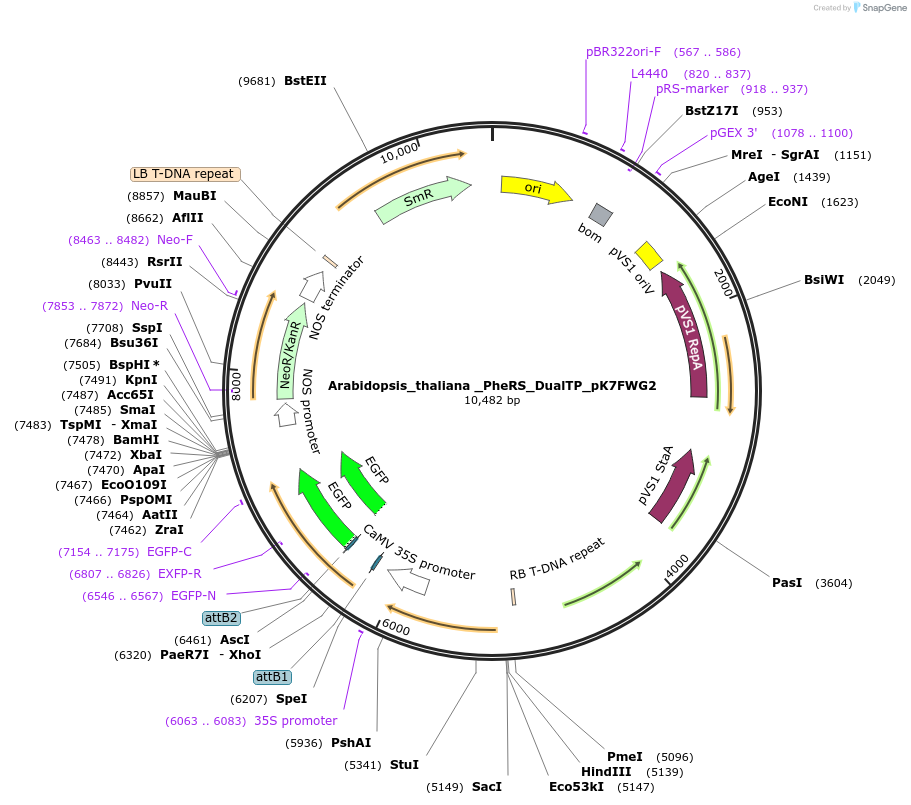
Arabidopsis_thaliana _PheRS_DualTP_pK7FWG2
Plasmid#202661PurposeDetermine where transit peptide targets GFP in N. benthamianaDepositorInsertN-terminal transit peptide from organellar Arabidopsis thaliana PheRS known to be dual targeted to plastid and mitochondria
TagsGreen Florescent ProtienExpressionPlantAvailable SinceAug. 10, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
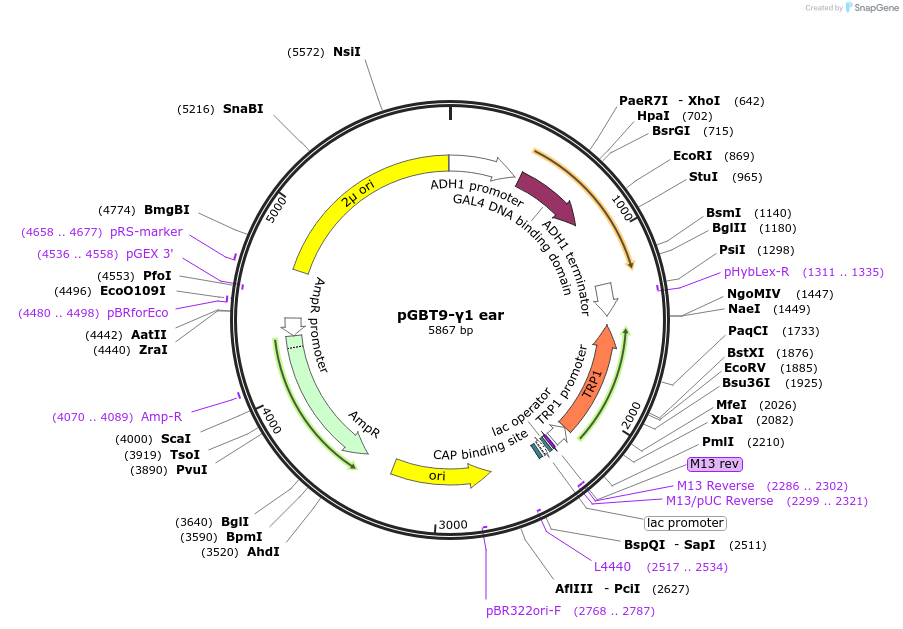
pGBT9-γ1 ear
Plasmid#199177PurposeExpression of GAL4 DNA-binding domain (BD)-AP-1 γ1 ear domain fusion protein in yeast (yeast two-hybrid assays)DepositorInsertAP-1 γ1 ear domain (Ap1g1 Mouse)
TagsGAL4-DNA binding domain fragmentExpressionYeastPromoterADH1Available SinceApril 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
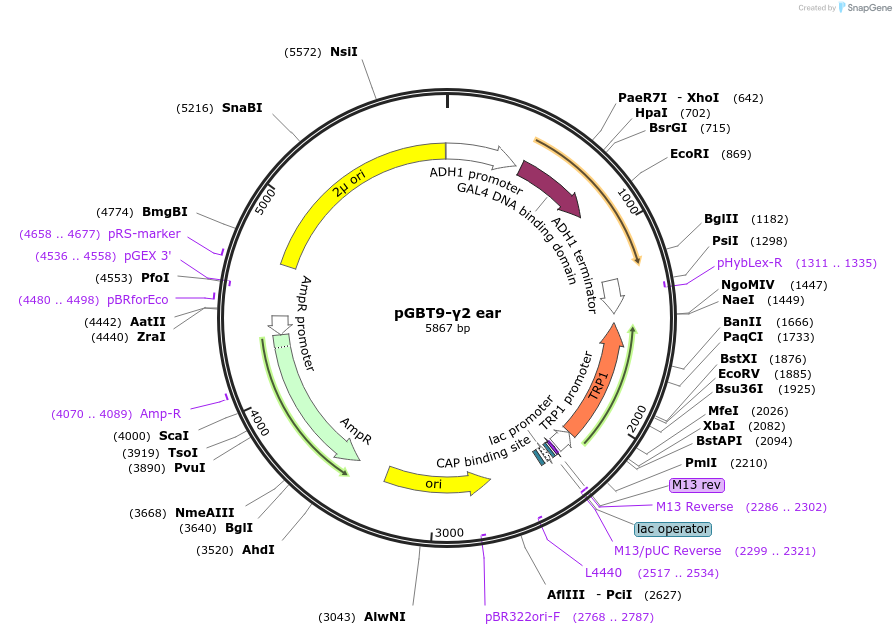
pGBT9-γ2 ear
Plasmid#199178PurposeExpression of GAL4 DNA-binding domain (BD)-AP-1 γ2 ear domain fusion protein in yeast (yeast two-hybrid assays)DepositorInsertAP-1 γ2 ear domain (AP1G2 Human)
TagsGAL4-DNA binding domain fragmentExpressionYeastPromoterADH1Available SinceApril 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
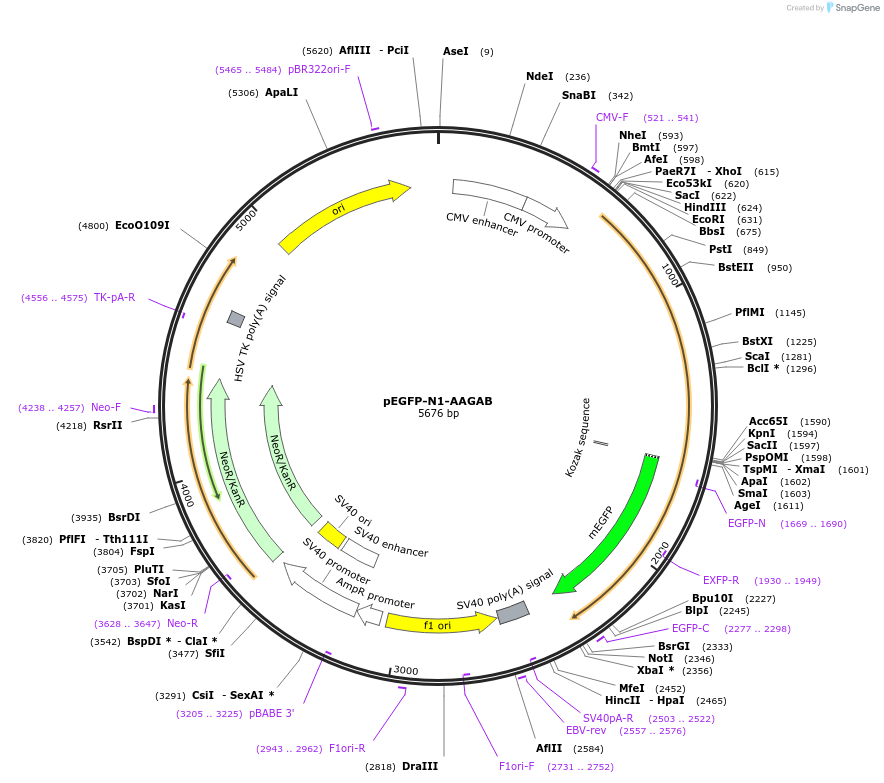
pEGFP-N1-AAGAB
Plasmid#198333PurposeExpresses AAGAB-GFP fusion in mammalian cellsDepositorAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only