We narrowed to 13,573 results for: Mpl
-
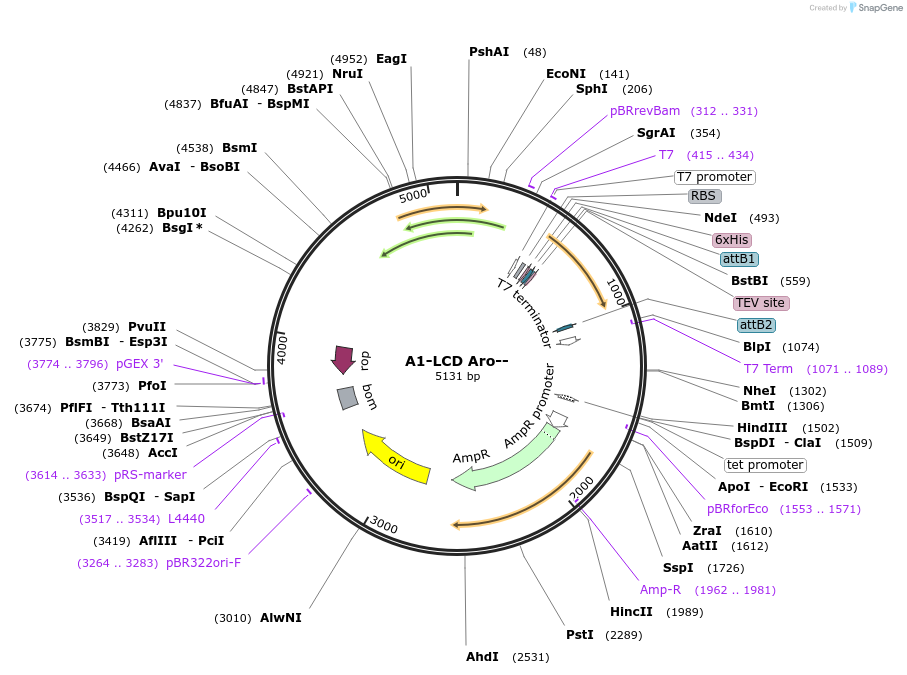
Plasmid#169716PurposeBacterial Expression of human hnRNPA1-A Low complexity domain including amino acids 186-320, aromatic amino acids.DepositorInserthnRNPA1 (HNRNPA1 Human)
Tags6xHisExpressionBacterialMutationhnRNPA1-A Low complexity domain including amino a…PromoterT7Available SinceJune 14, 2021AvailabilityAcademic Institutions and Nonprofits only -
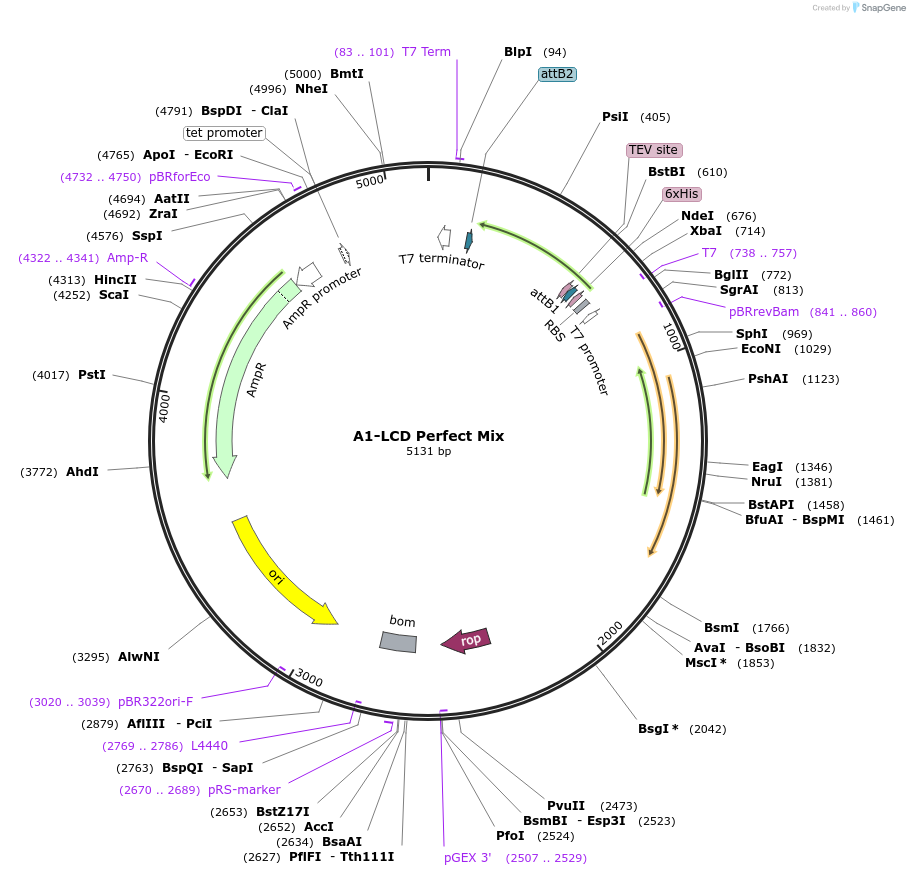
A1-LCD Perfect Mix
Plasmid#169718PurposeBacterial Expression of human hnRNPA1-A Low complexity domain including amino acids 186-320, aromatic amino acids. TYR and PHE shuffled into 5 clusters.DepositorInserthnRNPA1 (HNRNPA1 Human)
Tags6xHisExpressionBacterialMutationhnRNPA1-A Low complexity domain including amino a…PromoterT7Available SinceJune 14, 2021AvailabilityAcademic Institutions and Nonprofits only -
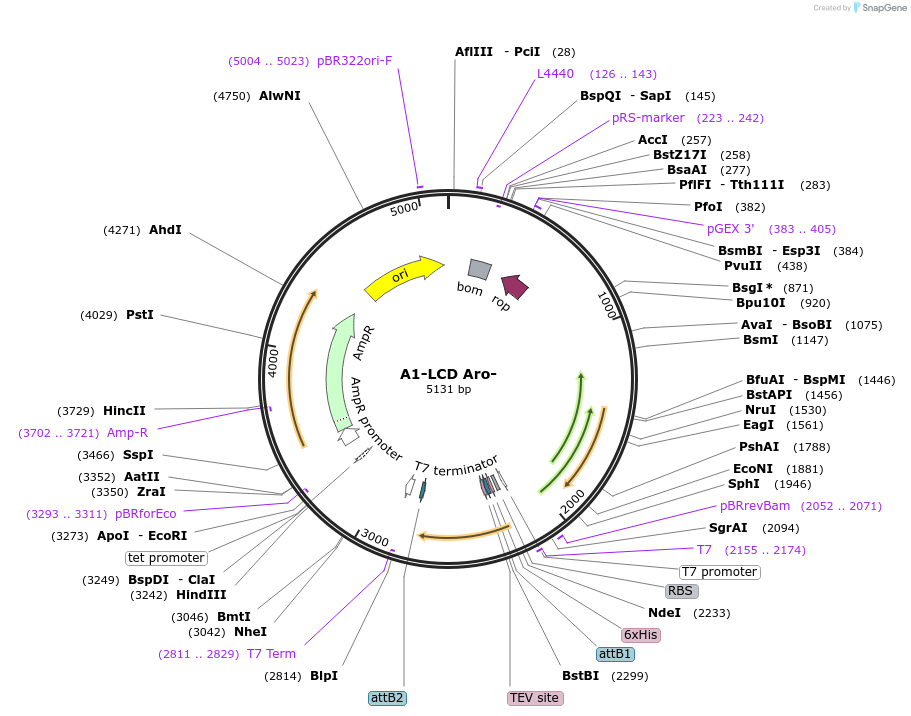
A1-LCD Aro-
Plasmid#169715PurposeBacterial Expression of human hnRNPA1-A Low complexity domain including amino acids 186-320, aromatic amino acids.DepositorInserthnRNPA1 (HNRNPA1 Human)
Tags6xHisExpressionBacterialMutationhnRNPA1-A Low complexity domain including amino a…PromoterT7Available SinceJune 14, 2021AvailabilityAcademic Institutions and Nonprofits only -
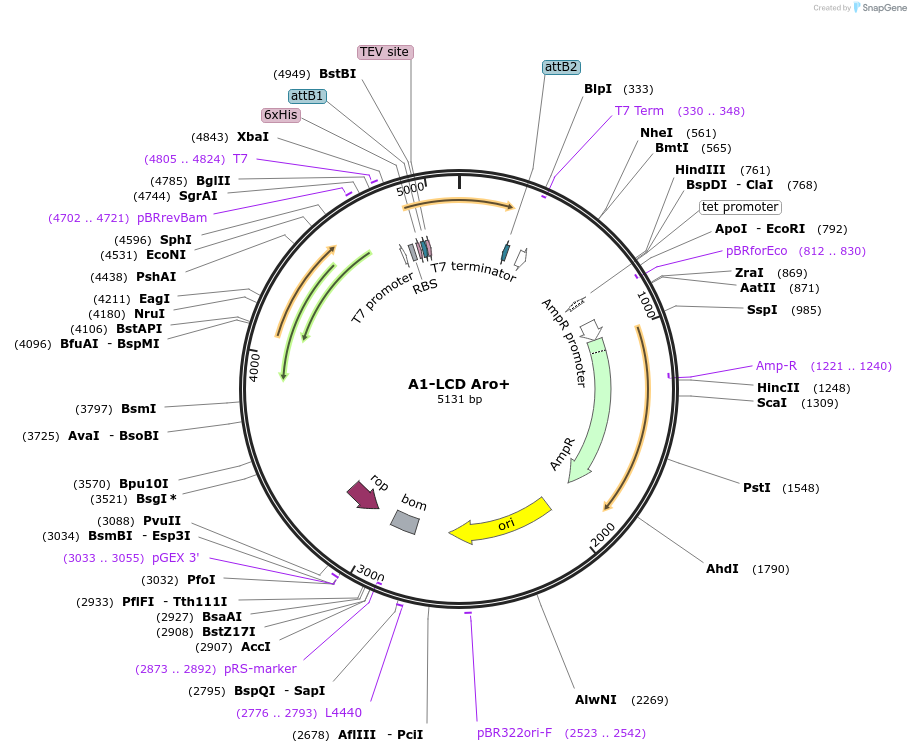
A1-LCD Aro+
Plasmid#169717PurposeBacterial Expression of human hnRNPA1-A Low complexity domain including amino acids 186-320, aromatic amino acids. PHE and TYR amino acids shuffled to optimize mixing.DepositorInserthnRNPA1 (HNRNPA1 Human)
Tags6xHisExpressionBacterialMutationhnRNPA1-A Low complexity domain including amino a…PromoterT7Available SinceJune 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
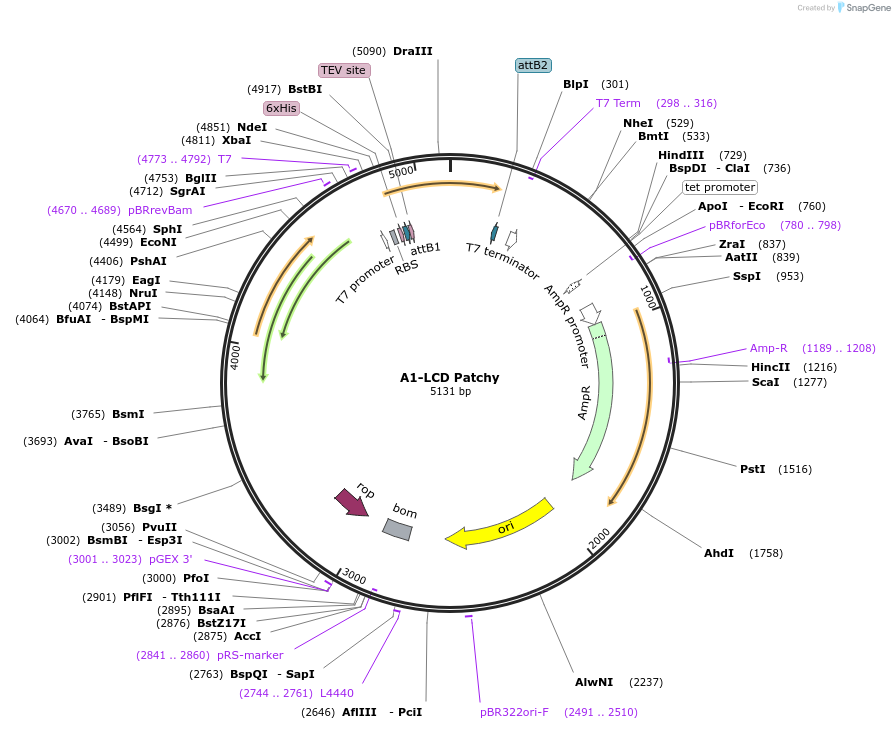
A1-LCD Patchy
Plasmid#169719PurposeBacterial Expression of human hnRNPA1-A Low complexity domain including amino acids 186-320, aromatic amino acids.DepositorInserthnRNPA1 (HNRNPA1 Human)
Tags6xHisExpressionBacterialMutationhnRNPA1-A Low complexity domain including amino a…PromoterT7Available SinceJune 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
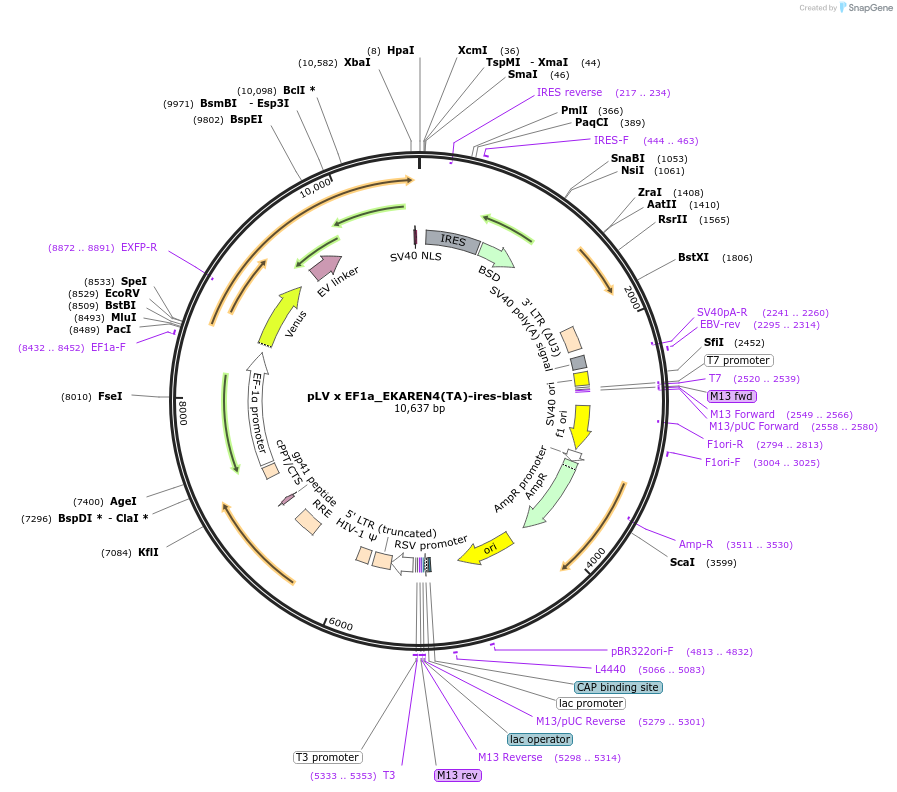
pLV x EF1a_EKAREN4(TA)-ires-blast
Plasmid#167831PurposeControl sensor for EKAREN4DepositorInsertEKAREN4(TA)
UseLentiviralTagsnls localization motifExpressionMammalianMutationK424P, K426W, T420APromoterEF1aAvailable SinceApril 20, 2021AvailabilityAcademic Institutions and Nonprofits only -
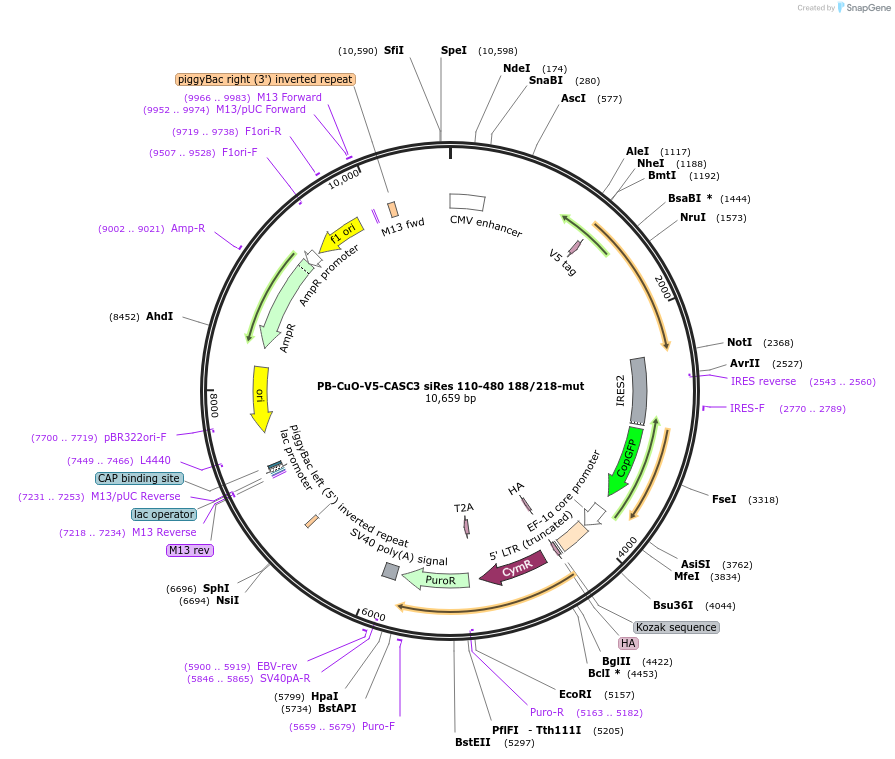
PB-CuO-V5-CASC3 siRes 110-480 188/218-mut
Plasmid#158544PurposeFor PiggyBac-mediated integration of V5-tagged siRNA-resistant EJC-binding deficient CASC3 110-480DepositorInsertCASC3; BTZ; MLN51 (CASC3 Human)
TagsV5ExpressionMammalianMutationAmino acids 110-480, C-terminal deletion, F188D, …PromoterCMVAvailable SinceOct. 21, 2020AvailabilityAcademic Institutions and Nonprofits only -
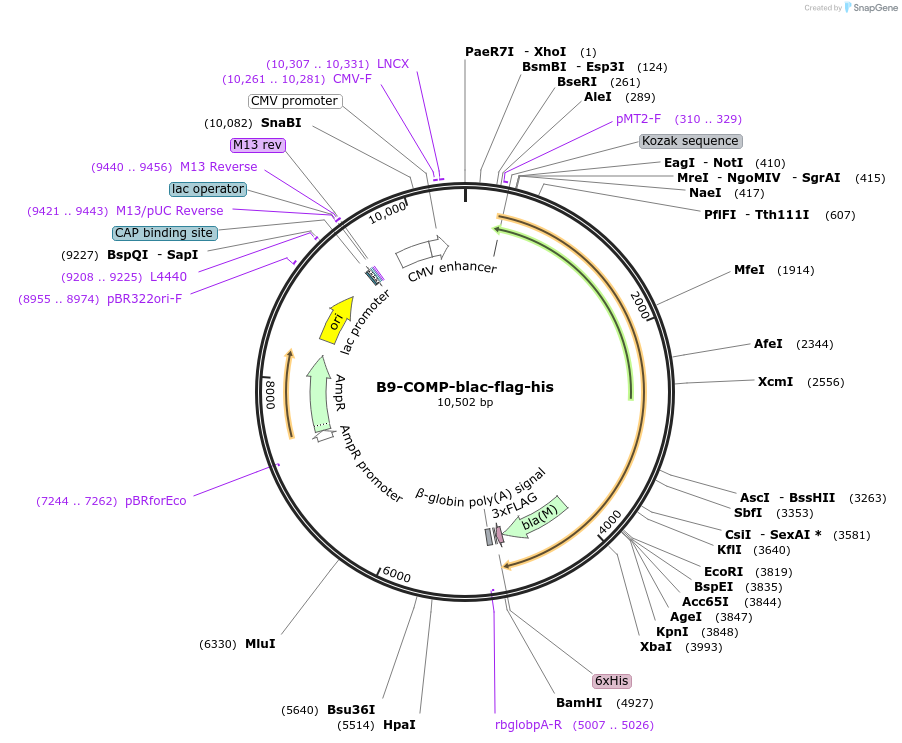
B9-COMP-blac-flag-his
Plasmid#111020PurposeExpresses pentamerised full-length protein ectodomain with no N-linked glycans in mammalian cells. Contains a C-terminal rat Cd4 d3+4 tag, COMP pentamerisation domain, flag- and 6xHis tags.DepositorInsert6-cysteine protein (B9) (PF3D7_0317100 )
TagsExogenous signal peptide of mouse origin and rat …ExpressionMammalianMutationAll NXT/S sites mutated to NXA, where X is any am…PromoterCMVAvailable SinceOct. 10, 2018AvailabilityAcademic Institutions and Nonprofits only -
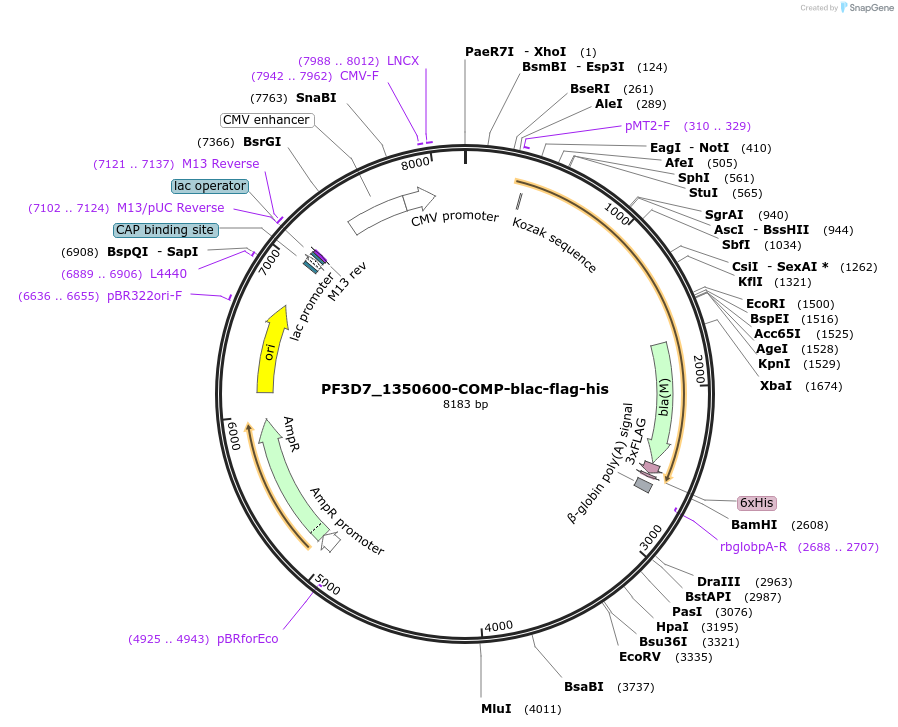
PF3D7_1350600-COMP-blac-flag-his
Plasmid#110968PurposeExpresses pentamerised full-length protein ectodomain with no N-linked glycans in mammalian cells. Contains a C-terminal rat Cd4 d3+4 tag, COMP pentamerisation domain, flag- and 6xHis tags.DepositorInsertPF3D7_1350600 (PF3D7_1350600 )
TagsExogenous signal peptide of mouse origin and rat …ExpressionMammalianMutationAll NXT/S sites mutated to NXA, where X is any am…PromoterCMVAvailable SinceOct. 3, 2018AvailabilityAcademic Institutions and Nonprofits only -
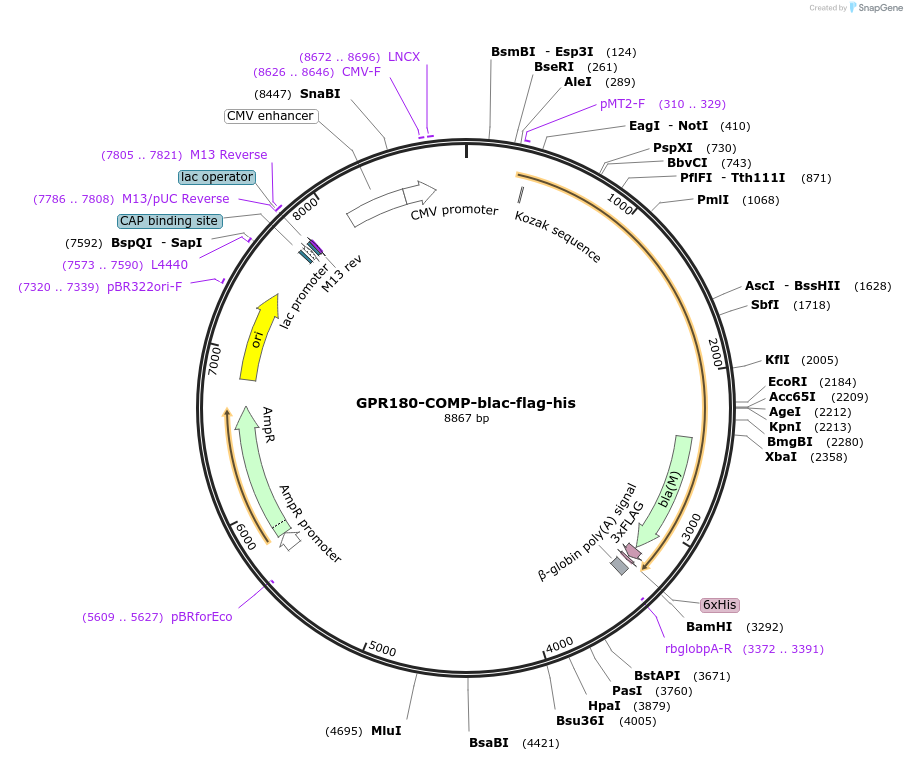
GPR180-COMP-blac-flag-his
Plasmid#110955PurposeExpresses pentamerised full-length protein ectodomain with no N-linked glycans in mammalian cells. Contains a C-terminal rat Cd4 d3+4 tag, COMP pentamerisation domain, flag- and 6xHis tags.DepositorInsertPF3D7_1213500 (PF3D7_1213500 )
TagsExogenous signal peptide of mouse origin and rat …ExpressionMammalianMutationAll NXT/S sites mutated to NXA, where X is any am…PromoterCMVAvailable SinceSept. 28, 2018AvailabilityAcademic Institutions and Nonprofits only