We narrowed to 12,964 results for: BASE
-
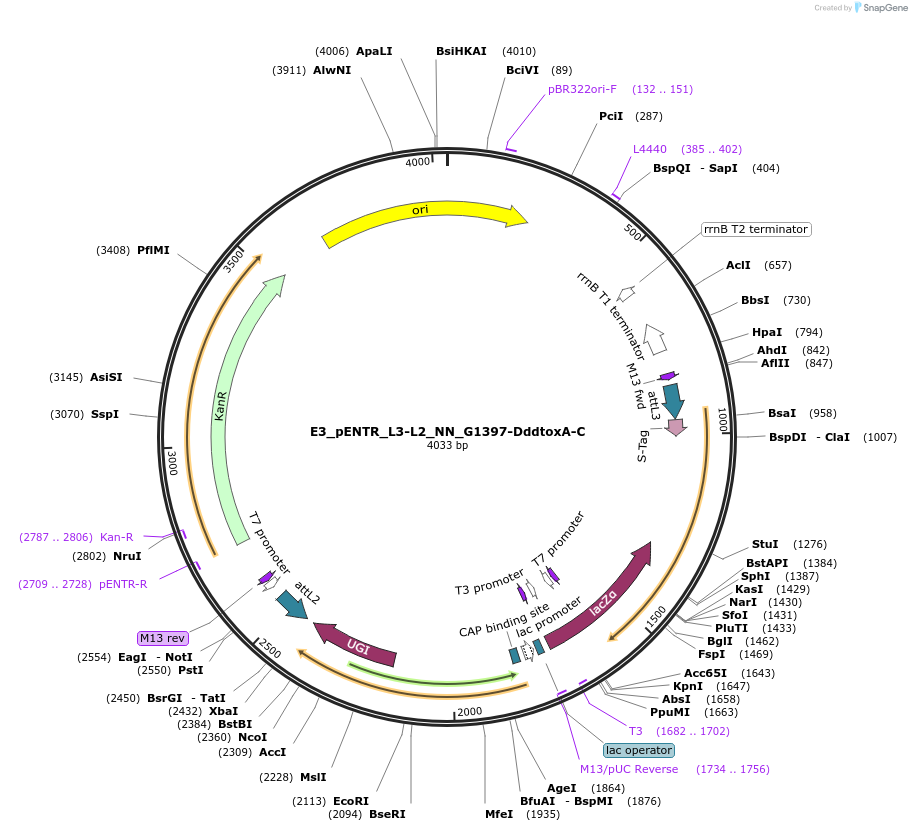
Plasmid#191595PurposeptpTALECD vector assembly, Entry clone 3 with attL2 and L3DepositorInsertC and N termini of platinum TALEN, 1/2 a cytidine deaminase domain of DddA of B. cenocepacia, and a uracil glycosylase inhibitor
UseOtherAvailable SinceFeb. 9, 2024AvailabilityAcademic Institutions and Nonprofits only -
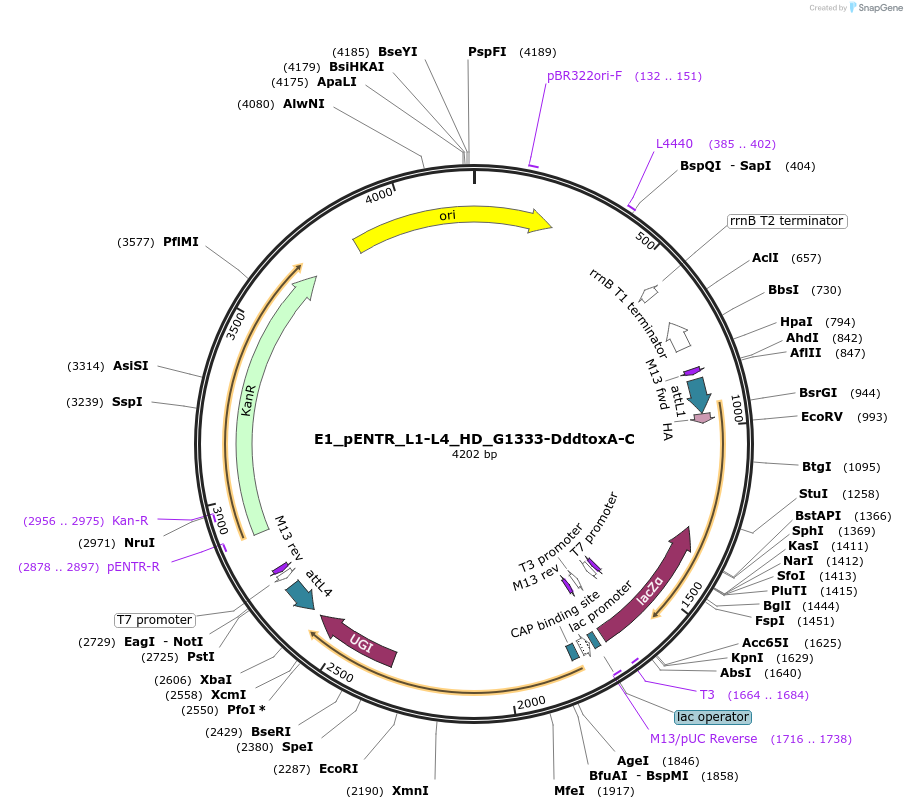
E1_pENTR_L1-L4_HD_G1333-DddtoxA-C
Plasmid#191579PurposeptpTALECD vector assembly, Entry clone 1 with attL1 and L4DepositorInsertC and N termini of platinum TALEN, 1/2 a cytidine deaminase domain of DddA of B. cenocepacia, and a uracil glycosylase inhibitor
UseOtherAvailable SinceFeb. 9, 2024AvailabilityAcademic Institutions and Nonprofits only -
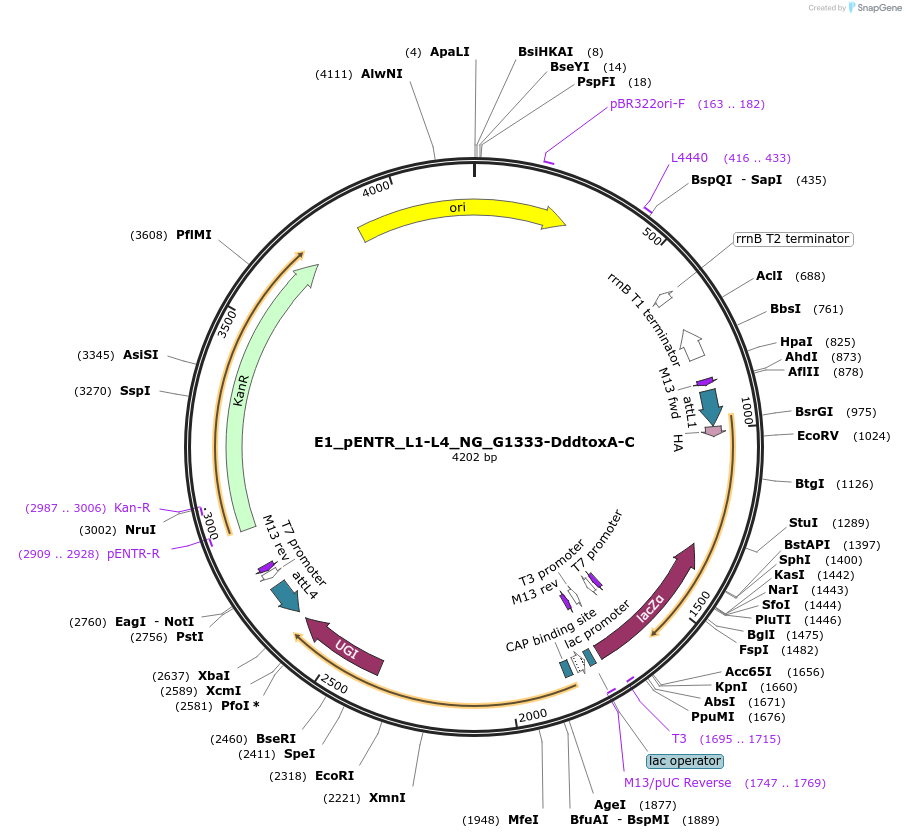
E1_pENTR_L1-L4_NG_G1333-DddtoxA-C
Plasmid#191580PurposeptpTALECD vector assembly, Entry clone 1 with attL1 and L4DepositorInsertC and N termini of platinum TALEN, 1/2 a cytidine deaminase domain of DddA of B. cenocepacia, and a uracil glycosylase inhibitor
UseOtherAvailable SinceFeb. 9, 2024AvailabilityAcademic Institutions and Nonprofits only -
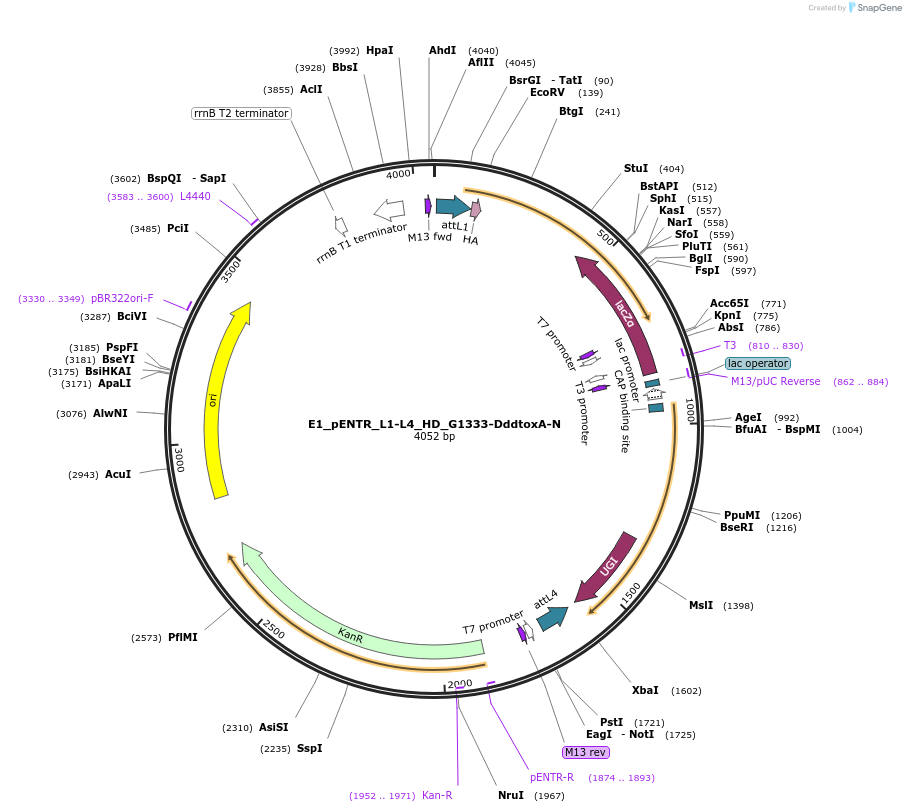
E1_pENTR_L1-L4_HD_G1333-DddtoxA-N
Plasmid#191581PurposeptpTALECD vector assembly, Entry clone 1 with attL1 and L4DepositorInsertC and N termini of platinum TALEN, 1/2 a cytidine deaminase domain of DddA of B. cenocepacia, and a uracil glycosylase inhibitor
UseOtherAvailable SinceFeb. 9, 2024AvailabilityAcademic Institutions and Nonprofits only -
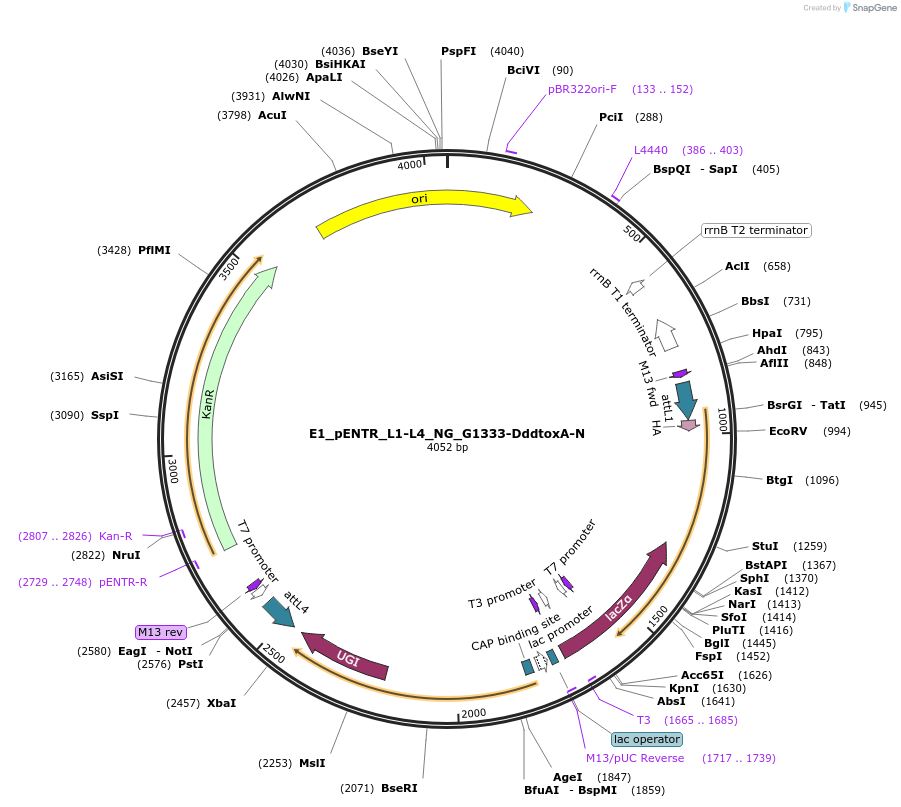
E1_pENTR_L1-L4_NG_G1333-DddtoxA-N
Plasmid#191582PurposeptpTALECD vector assembly, Entry clone 1 with attL1 and L4DepositorInsertC and N termini of platinum TALEN, 1/2 a cytidine deaminase domain of DddA of B. cenocepacia, and a uracil glycosylase inhibitor
UseOtherAvailable SinceFeb. 9, 2024AvailabilityAcademic Institutions and Nonprofits only -
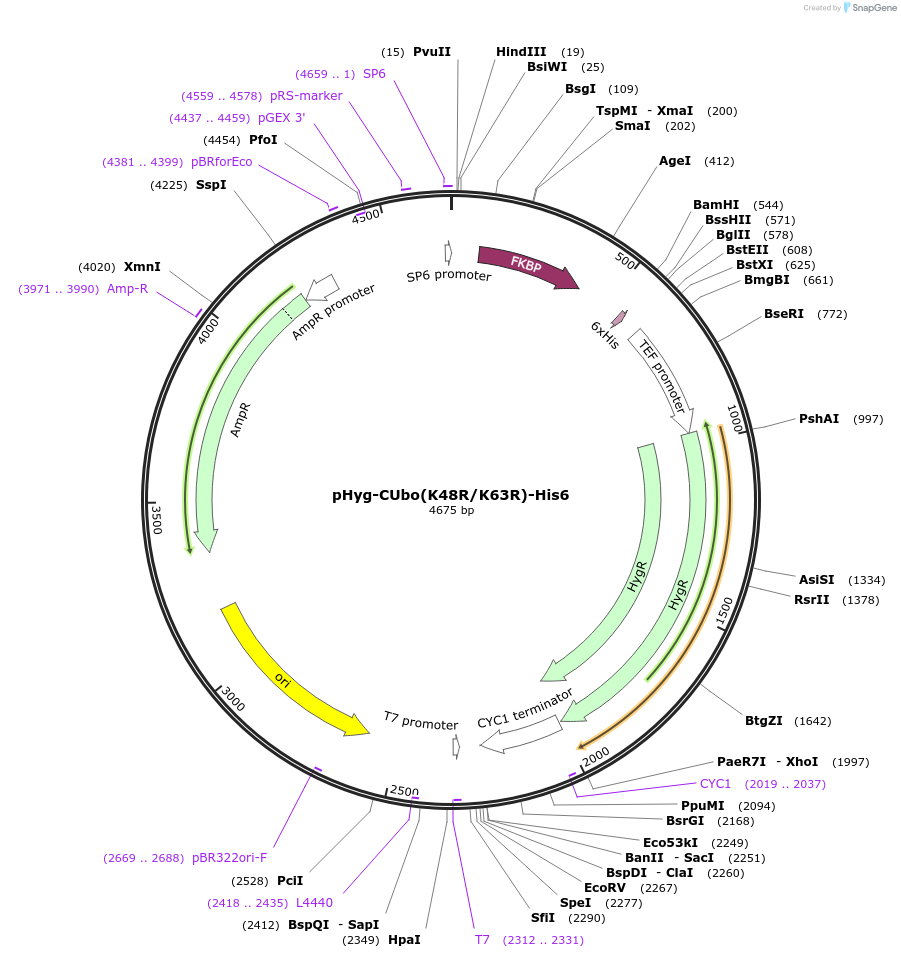
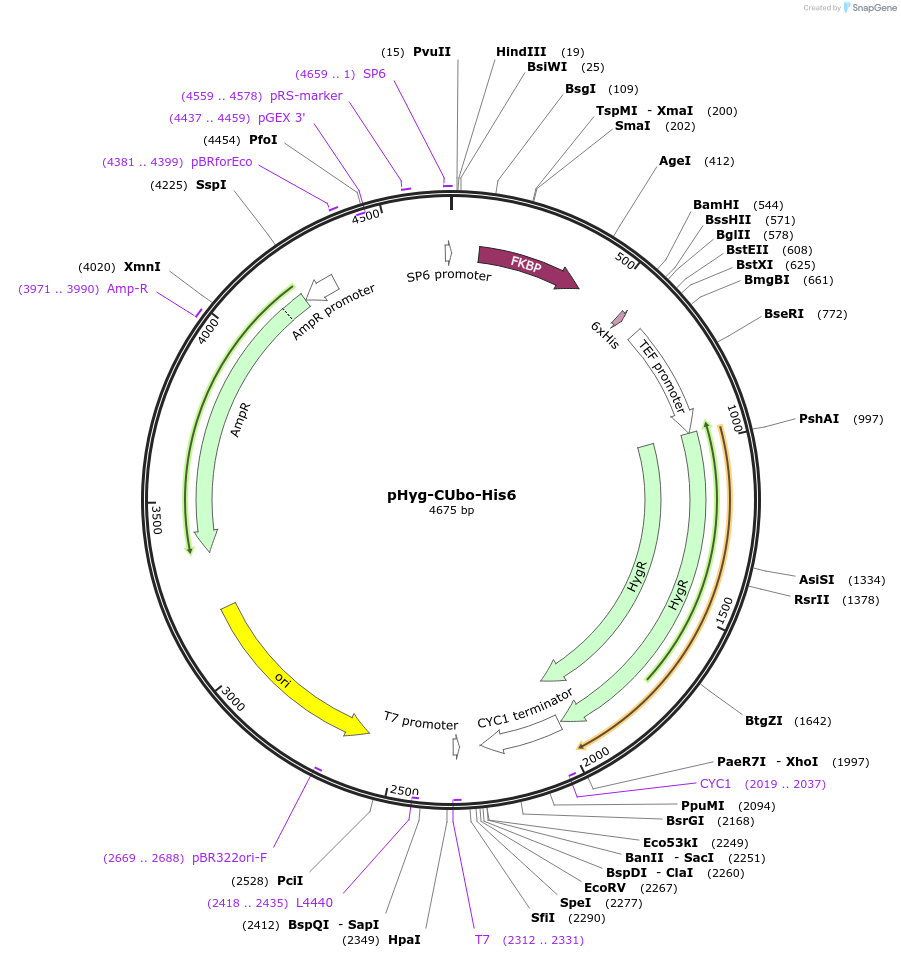
pHyg-CUbo(K48R/K63R)-His6
Plasmid#212780PurposeC-terminal PCR-based tagging with CUbo(K48R/K63R)-His6 in budding yeast with HygR selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceFeb. 8, 2024AvailabilityAcademic Institutions and Nonprofits only -
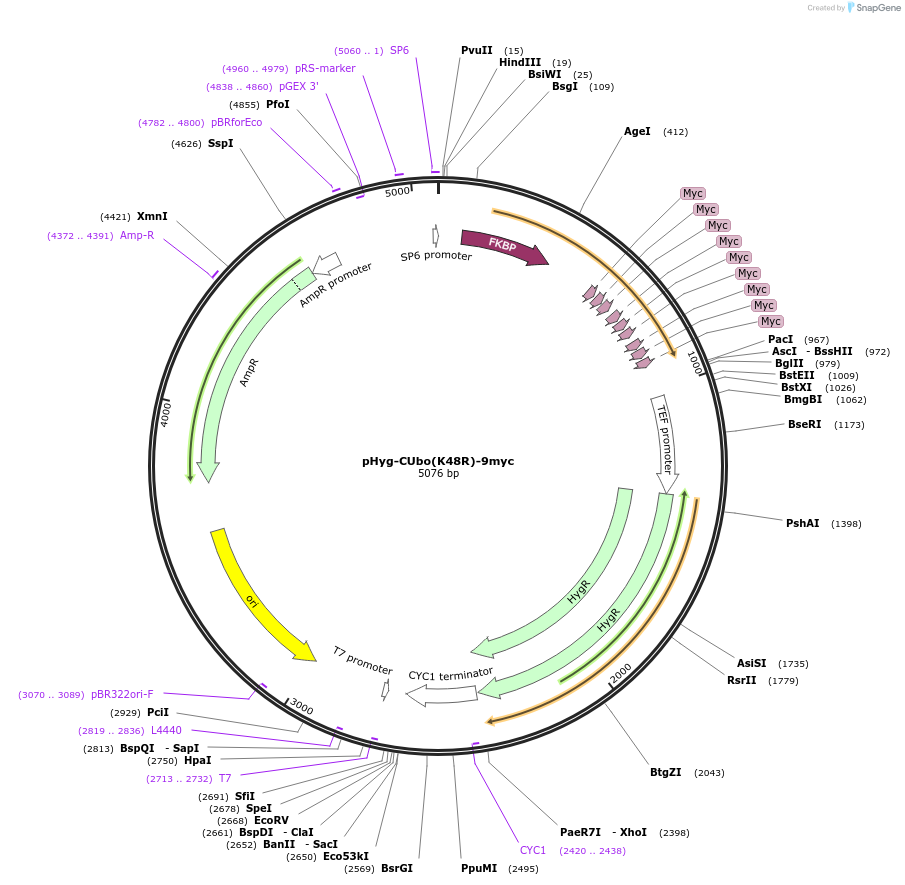
pHyg-CUbo(K48R)-9myc
Plasmid#212787PurposeC-terminal PCR-based tagging with CUbo(K48R)-9myc in budding yeast with HygR selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceFeb. 8, 2024AvailabilityAcademic Institutions and Nonprofits only -
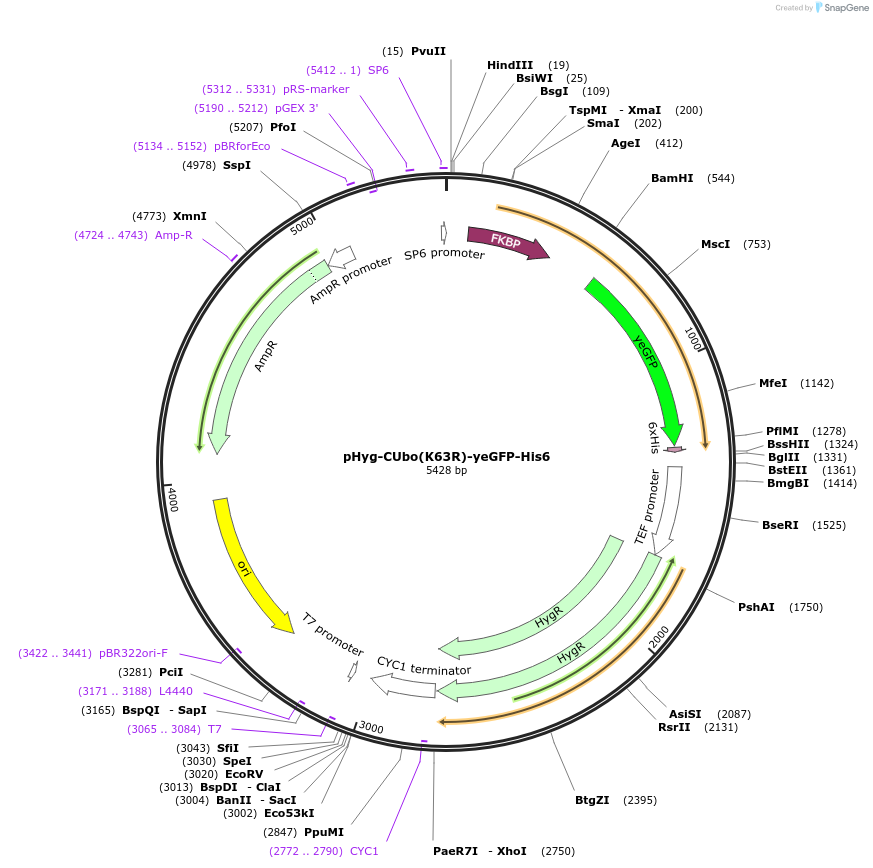
pHyg-CUbo(K63R)-yeGFP
Plasmid#212768PurposeC-terminal PCR-based tagging with CUbo(K63R)-yeGFP in budding yeast with HygR selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceFeb. 8, 2024AvailabilityAcademic Institutions and Nonprofits only -
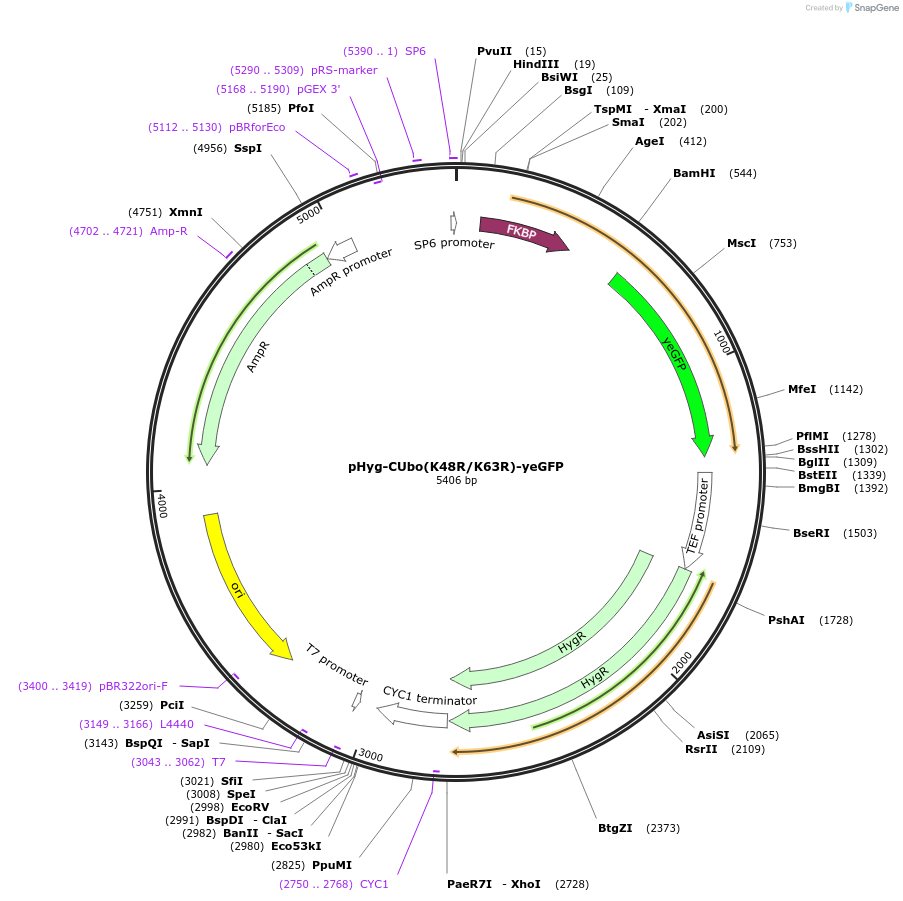
pHyg-CUbo(K48R/K63R)-yeGFP
Plasmid#212769PurposeC-terminal PCR-based tagging with CUbo(K48R/K63R)-yeGFP in budding yeast with HygR selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceFeb. 8, 2024AvailabilityAcademic Institutions and Nonprofits only -
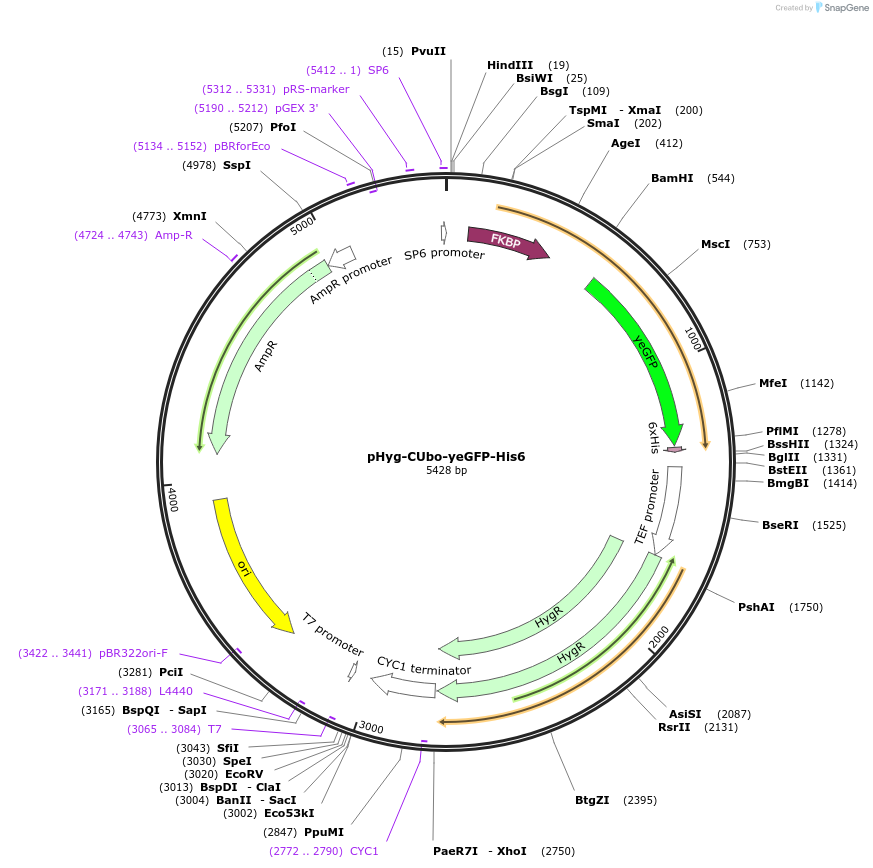
pHyg-CUbo-yeGFP
Plasmid#212766PurposeC-terminal PCR-based tagging with CUbo-yeGFP in budding yeast with HygR selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceFeb. 8, 2024AvailabilityAcademic Institutions and Nonprofits only -
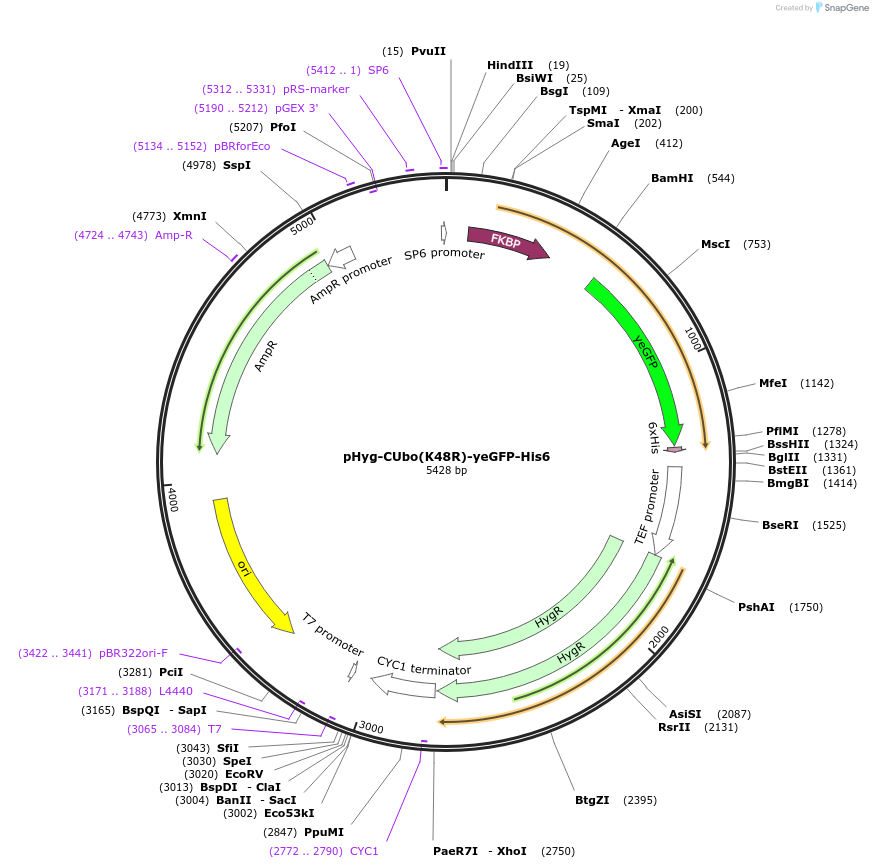
pHyg-CUbo(K48R)-yeGFP
Plasmid#212767PurposeC-terminal PCR-based tagging with CUbo(K48R)-yeGFP in budding yeast with HygR selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 30, 2024AvailabilityAcademic Institutions and Nonprofits only -
pCaUra3-CUbo-yeGFP
Plasmid#212771PurposeC-terminal PCR-based tagging with CUbo-yeGFP in budding yeast with CaUra3 selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 30, 2024AvailabilityAcademic Institutions and Nonprofits only -
pCaUra3-CUbo(K48R)-yeGFP
Plasmid#212772PurposeC-terminal PCR-based tagging with CUbo(K48R)-yeGFP in budding yeast with CaUra3 selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 30, 2024AvailabilityAcademic Institutions and Nonprofits only -
pCaUra3-CUbo(K63R)-yeGFP
Plasmid#212773PurposeC-terminal PCR-based tagging with CUbo(K63R)-yeGFP in budding yeast with CaUra3 selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 30, 2024AvailabilityAcademic Institutions and Nonprofits only -
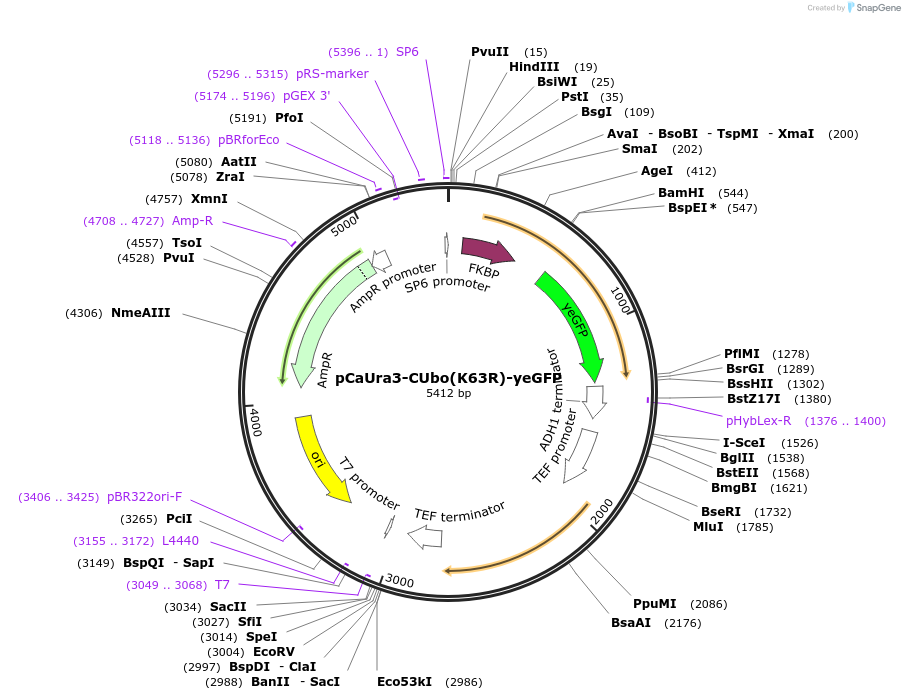
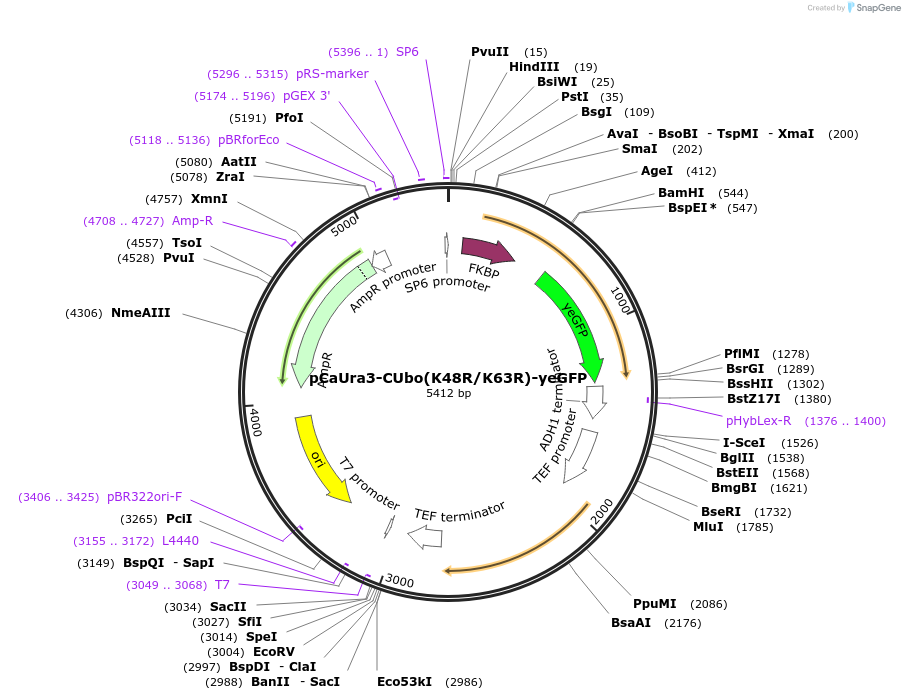
pCaUra3-CUbo(K48R/K63R)-yeGFP
Plasmid#212774PurposeC-terminal PCR-based tagging with CUbo(K48R/K63R)-yeGFP in budding yeast with CaUra3 selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 30, 2024AvailabilityAcademic Institutions and Nonprofits only -
pHyg-CUbo-His6
Plasmid#212777PurposeC-terminal PCR-based tagging with CUbo-His6 in budding yeast with HygR selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 30, 2024AvailabilityAcademic Institutions and Nonprofits only -
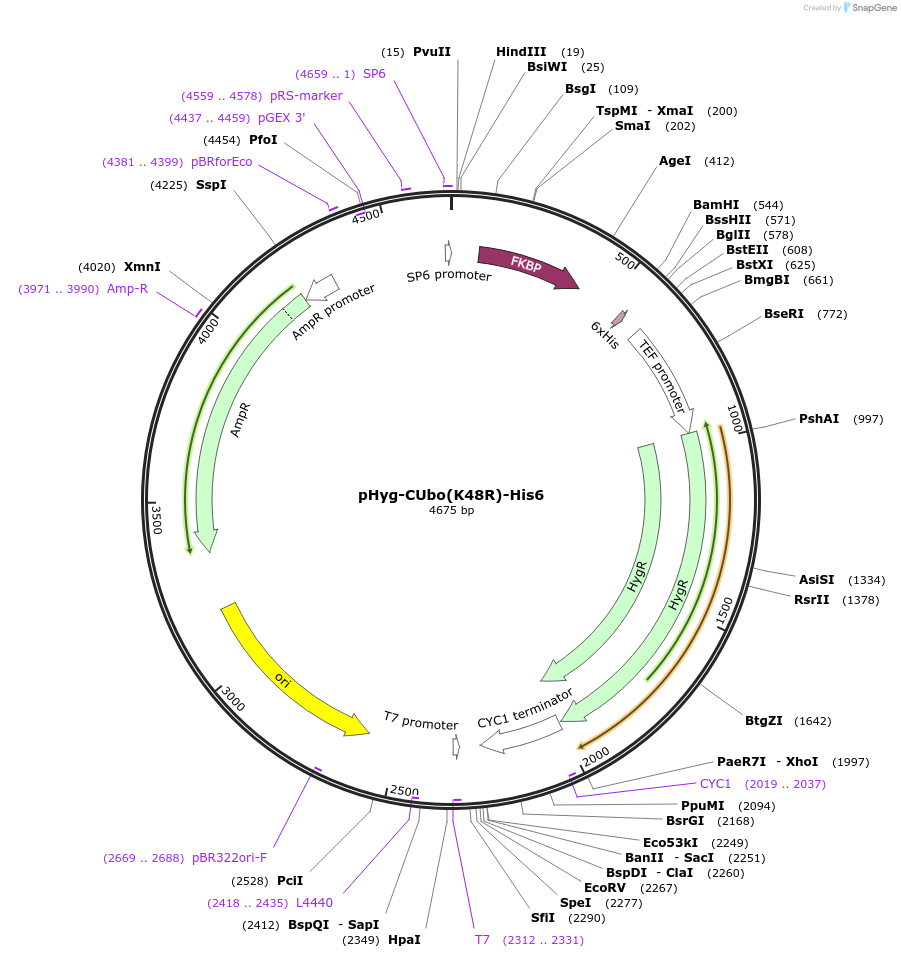
pHyg-CUbo(K48R)-His6
Plasmid#212778PurposeC-terminal PCR-based tagging with CUbo(K48R)-His6 in budding yeast with HygR selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 30, 2024AvailabilityAcademic Institutions and Nonprofits only -
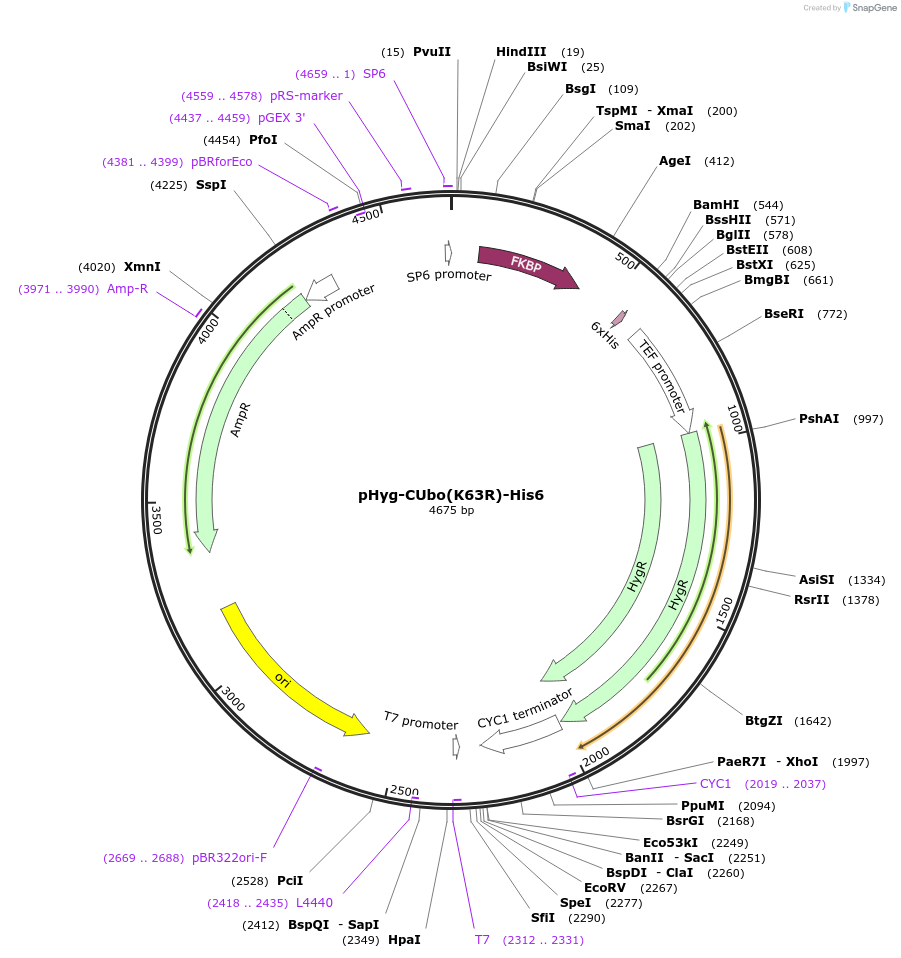
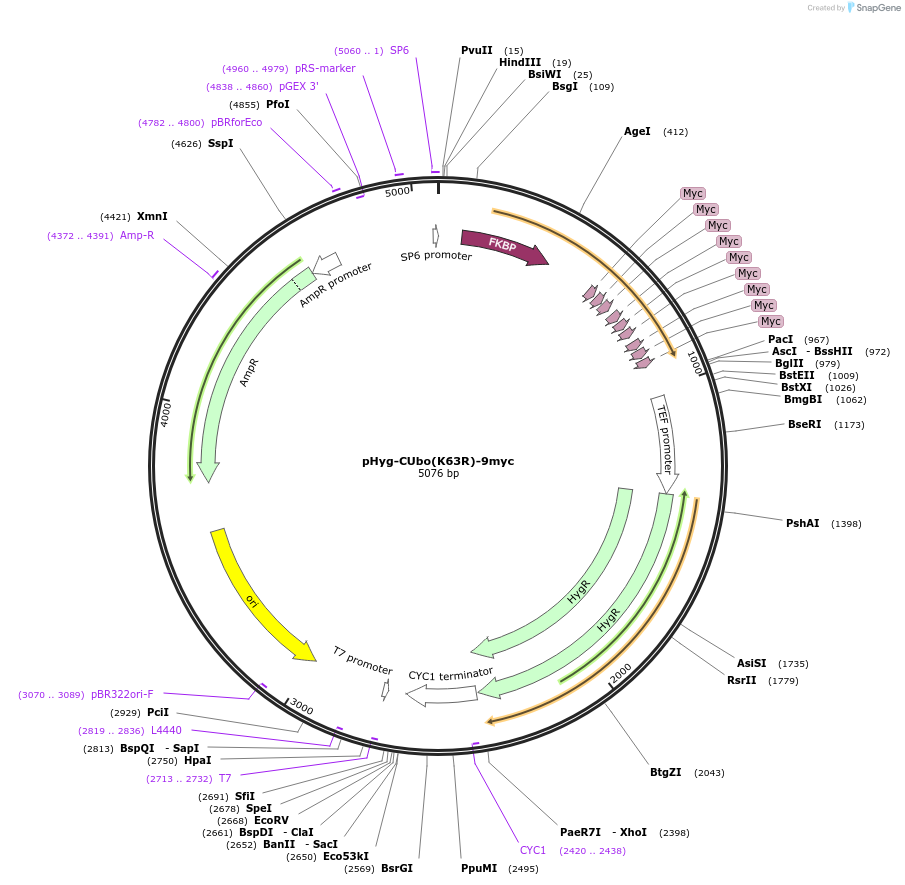
pHyg-CUbo(K63R)-His6
Plasmid#212779PurposeC-terminal PCR-based tagging with CUbo(K63R)-His6 in budding yeast with HygR selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 30, 2024AvailabilityAcademic Institutions and Nonprofits only -
pHyg-CUbo-yeGFP-His6
Plasmid#212782PurposeC-terminal PCR-based tagging with CUbo-yeGFP-His6 in budding yeast with HygR selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 30, 2024AvailabilityAcademic Institutions and Nonprofits only -
pHyg-CUbo(K48R)-yeGFP-His6
Plasmid#212783PurposeC-terminal PCR-based tagging with CUbo(K48R)-yeGFP-His6 in budding yeast with HygR selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 30, 2024AvailabilityAcademic Institutions and Nonprofits only -
pHyg-CUbo(K63R)-yeGFP-His6
Plasmid#212784PurposeC-terminal PCR-based tagging with CUbo(K63R)-yeGFP-His6 in budding yeast with HygR selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 30, 2024AvailabilityAcademic Institutions and Nonprofits only -
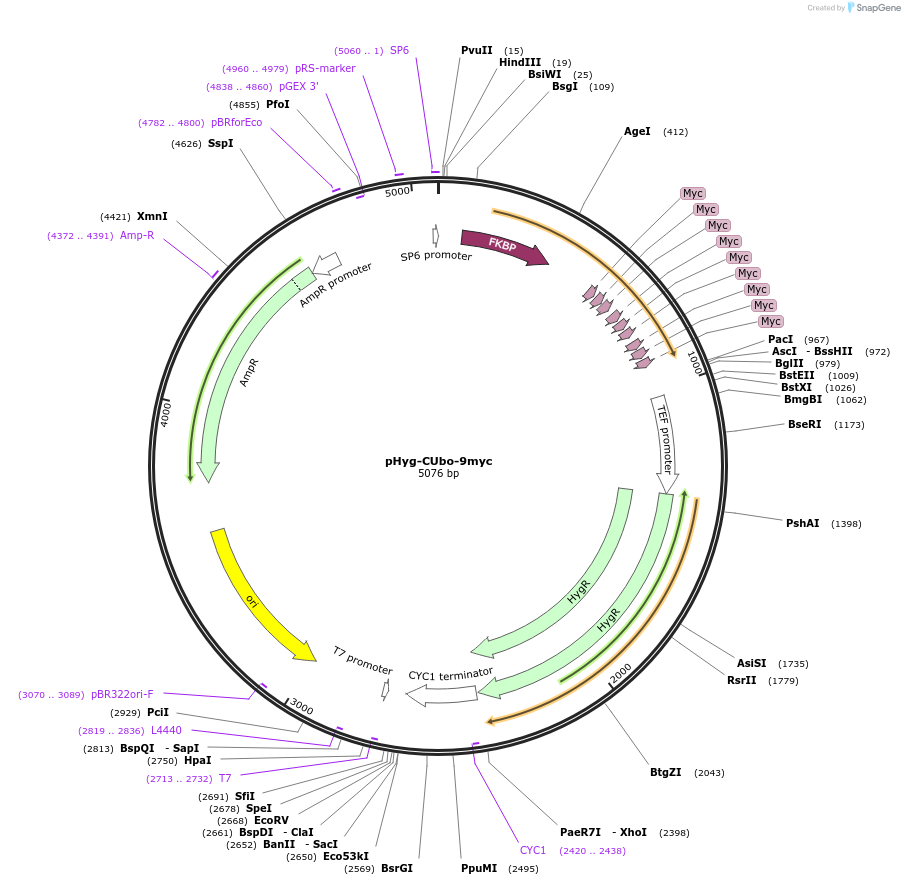
pHyg-CUbo-9myc
Plasmid#212786PurposeC-terminal PCR-based tagging with CUbo-9myc in budding yeast with HygR selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 30, 2024AvailabilityAcademic Institutions and Nonprofits only -
pHyg-CUbo(K63R)-9myc
Plasmid#212788PurposeC-terminal PCR-based tagging with CUbo(K63R)-9myc in budding yeast with HygR selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
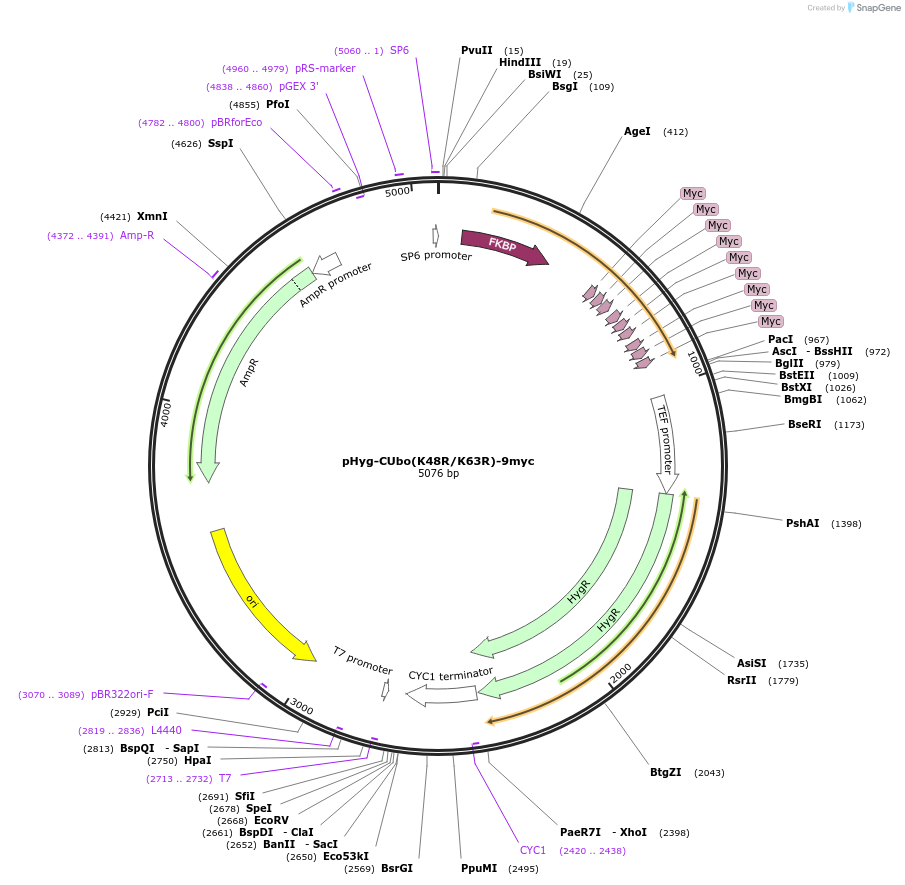
pHyg-CUbo(K48R/K63R)-9myc
Plasmid#212789PurposeC-terminal PCR-based tagging with CUbo(K48R/K63R)-9myc in budding yeast with HygR selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
pHyg-CUbo-AID*-GFP
Plasmid#212770PurposeC-terminal PCR-based tagging with CUbo-AID*-yeGFP in budding yeast with HygR selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
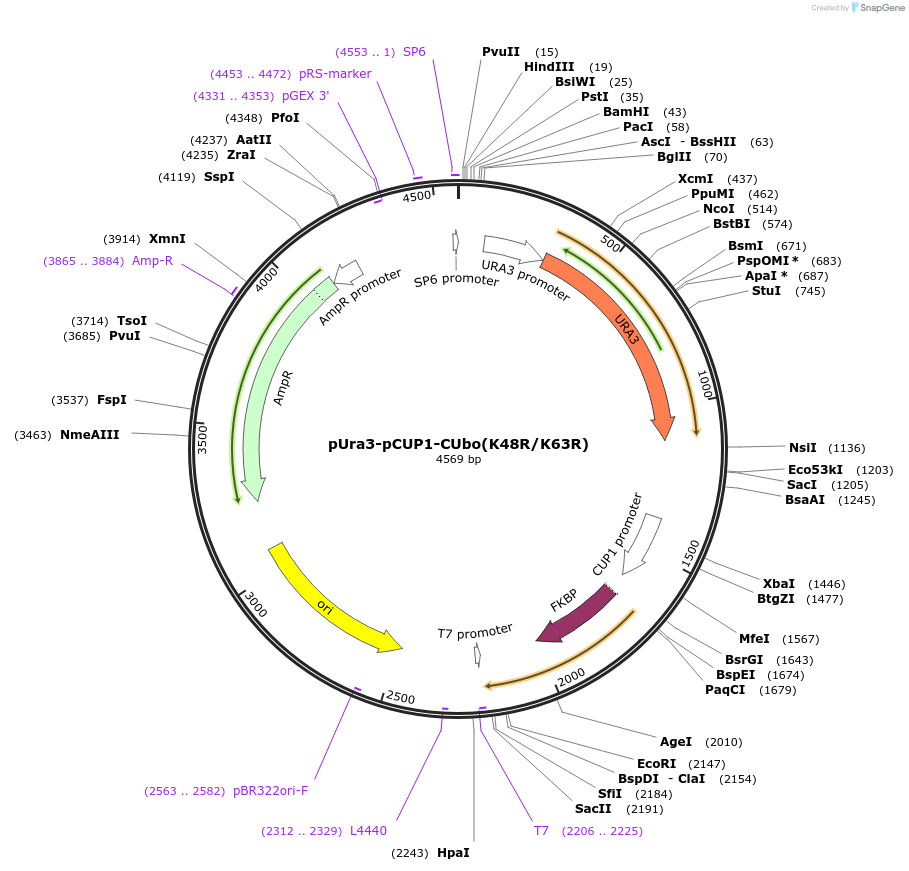
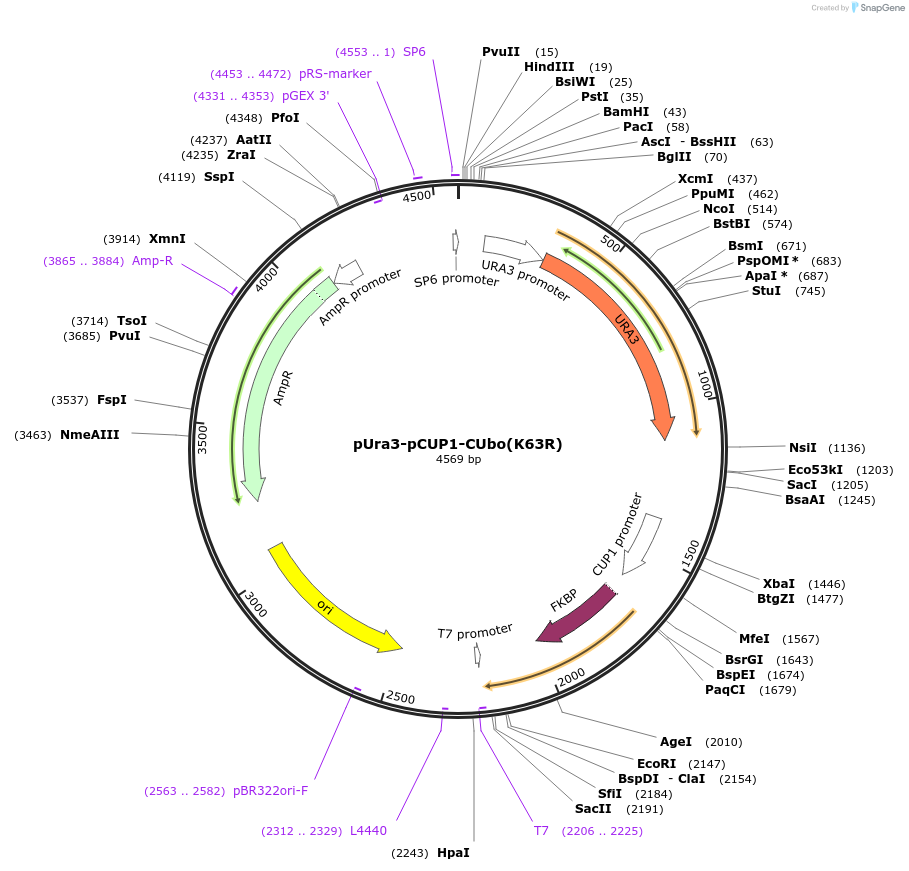
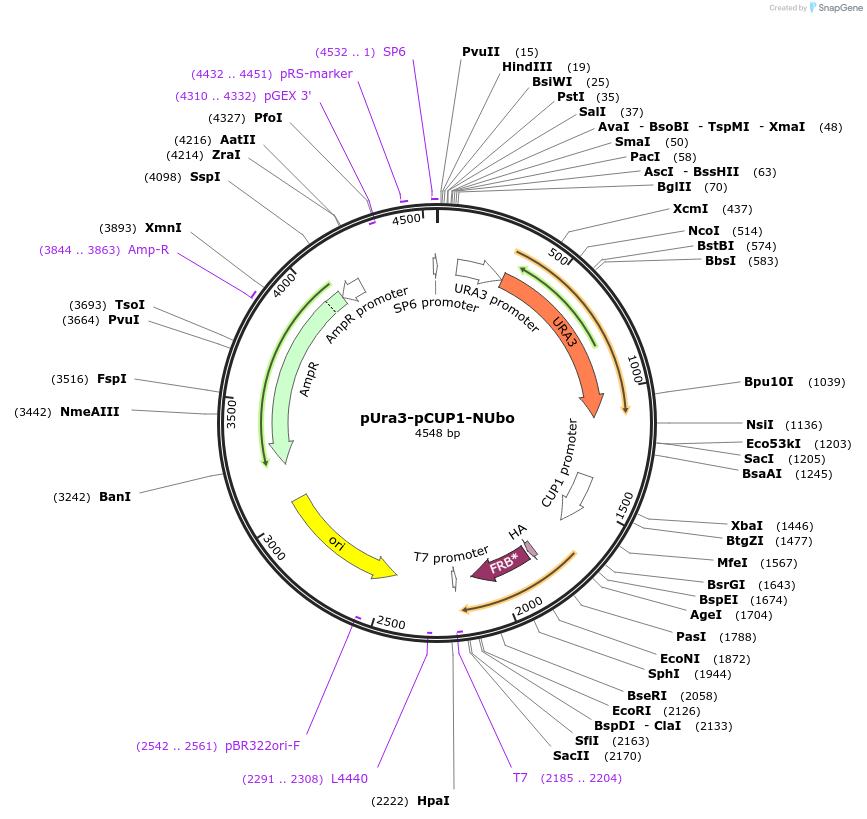
pUra3-pCUP1-CUbo(K48R/K63R)
Plasmid#212765PurposeN-terminal PCR-based tagging with pCUP1-CUbo(K48R/K63R) in budding yeast with URA3 selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
pUra3-pCUP1-CUbo(K63R)
Plasmid#212764PurposeN-terminal PCR-based tagging with pCUP1-CUbo(K63R) in budding yeast with URA3 selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
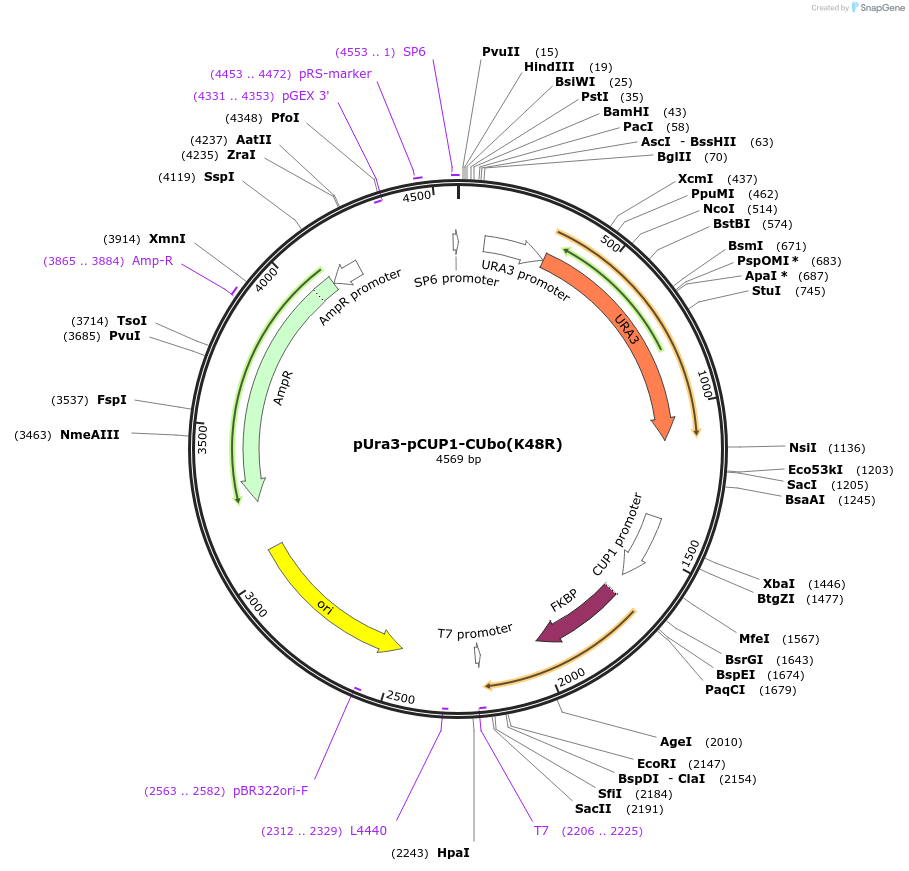
pUra3-pCUP1-CUbo(K48R)
Plasmid#212763PurposeN-terminal PCR-based tagging with pCUP1-CUbo(K48R) in budding yeast with URA3 selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
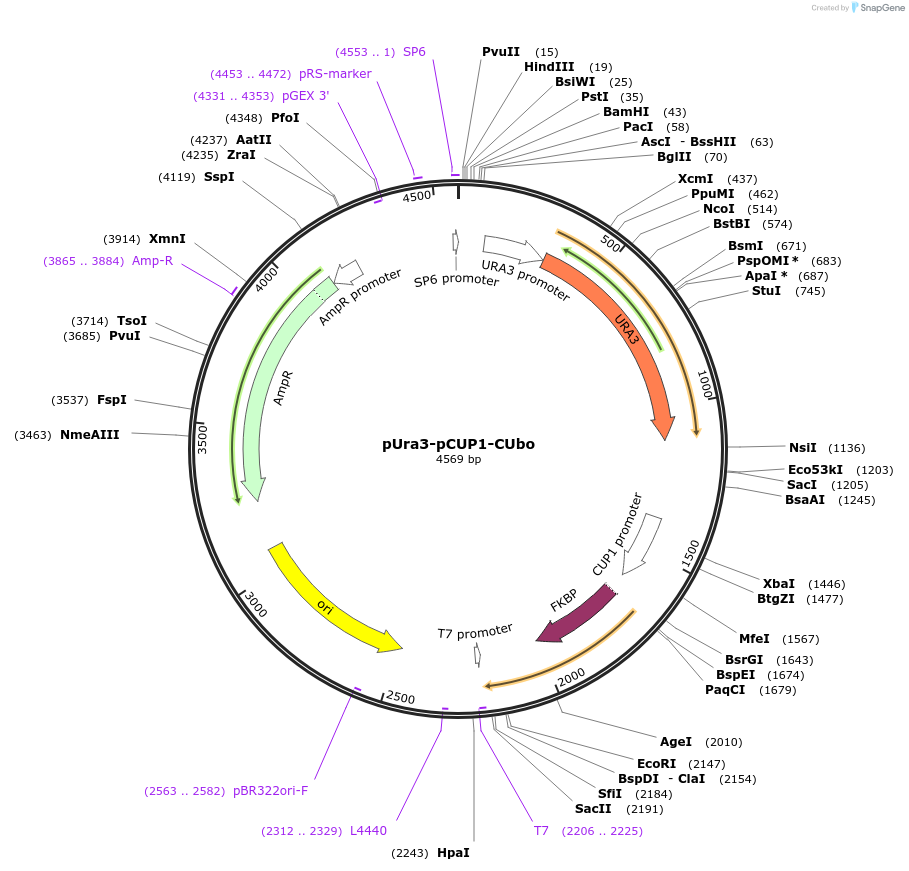
pUra3-pCUP1-CUbo
Plasmid#212762PurposeN-terminal PCR-based tagging with pCUP1-CUbo in budding yeast with URA3 selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
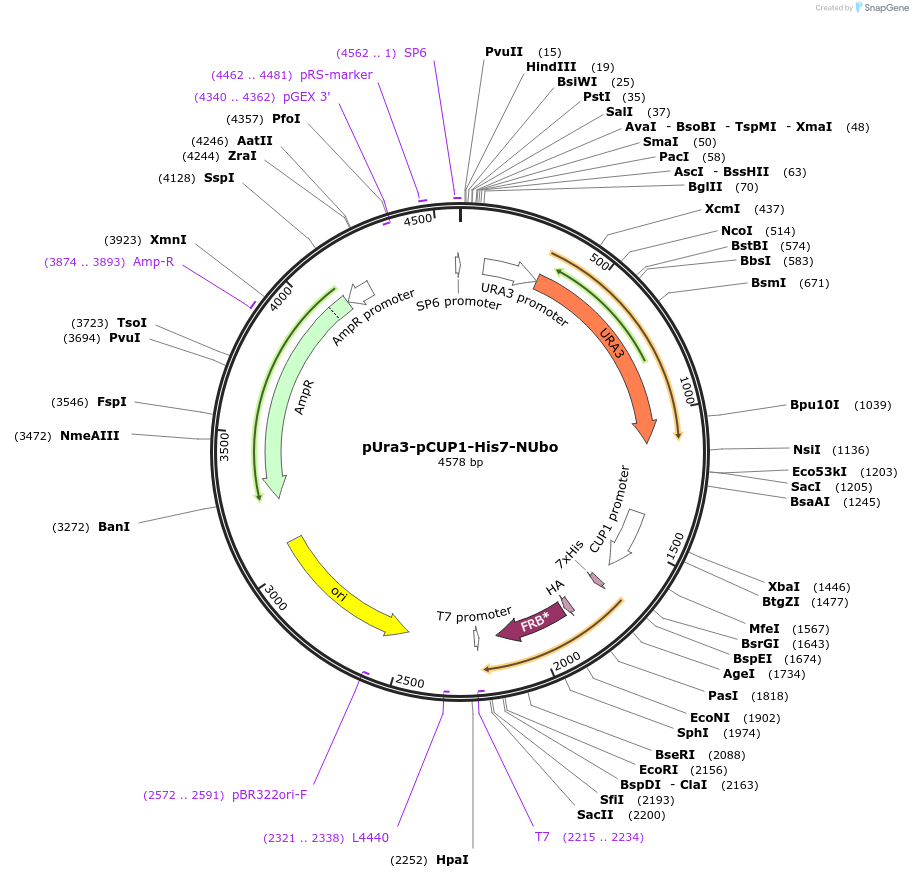
pUra3-pCUP1-His7-NUbo
Plasmid#212761PurposeN-terminal PCR-based tagging with pCUP1-His7-NUbo in budding yeast with URA3 selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
pUra3-pCUP1-NUbo
Plasmid#212760PurposeN-terminal PCR-based tagging with pCUP1-NUbo in budding yeast with URA3 selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
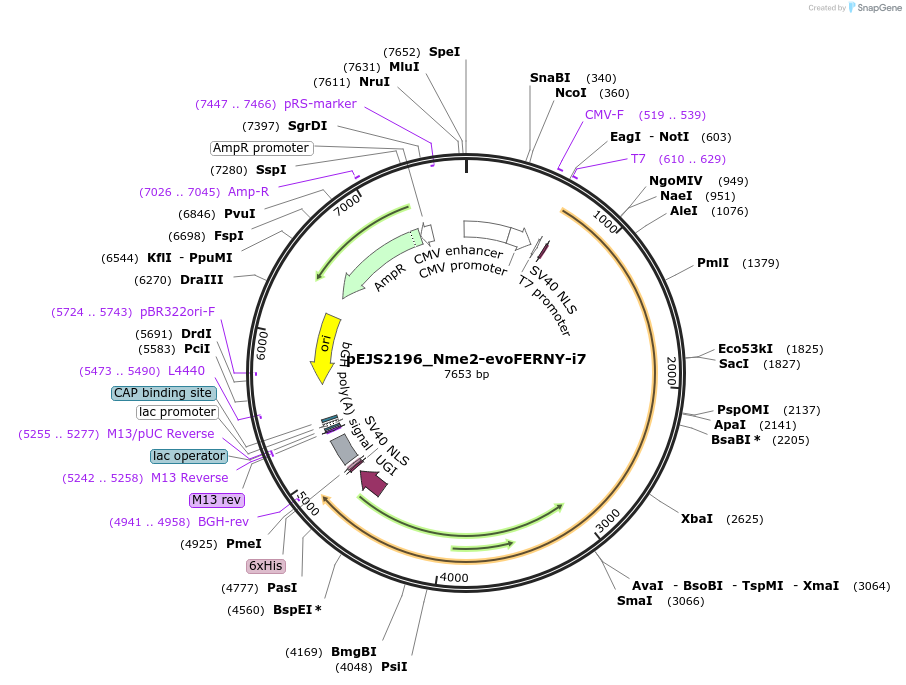
pEJS2196_Nme2-evoFERNY-i7
Plasmid#201529PurposeMammalian expression of domain inlaid Nme2-evoFERNY-i7DepositorInsertNme2-evoFERNY-i7
ExpressionMammalianPromoterCMVAvailable SinceJan. 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
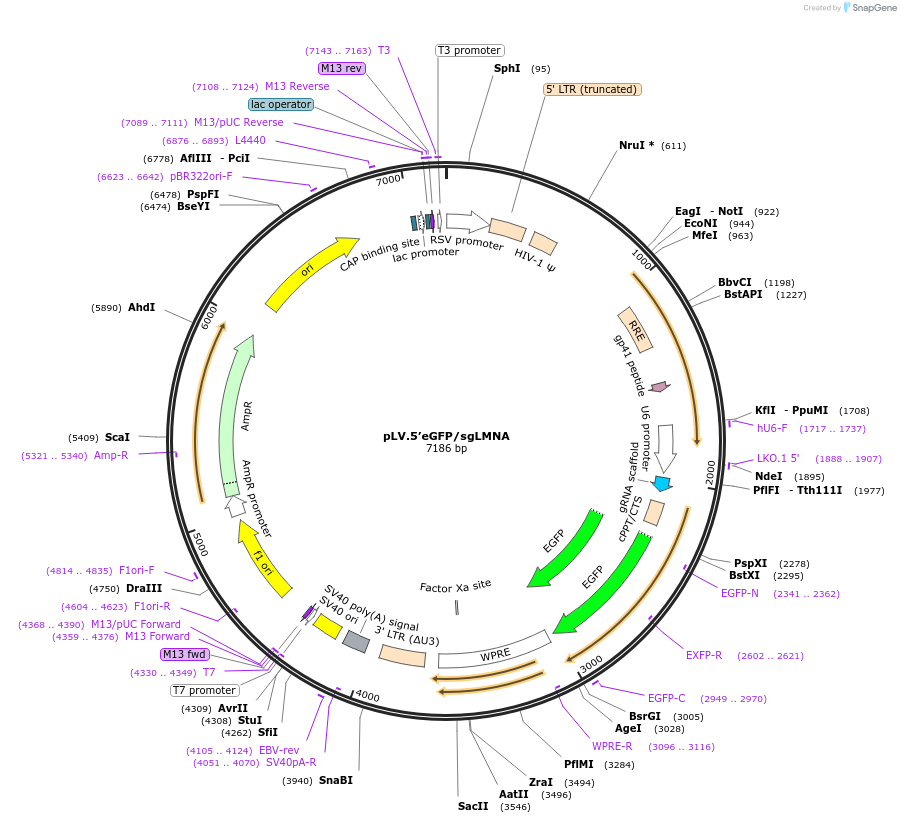
pLV.5’eGFP/sgLMNA
Plasmid#170546PurposeExpresses a sgRNA for 5' tagging to LMNA and contains a cassette with eGFP without homology armsDepositorInsertsgRNA targeting 5'- end of LMNA
UseCRISPR and LentiviralAvailable SinceJan. 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
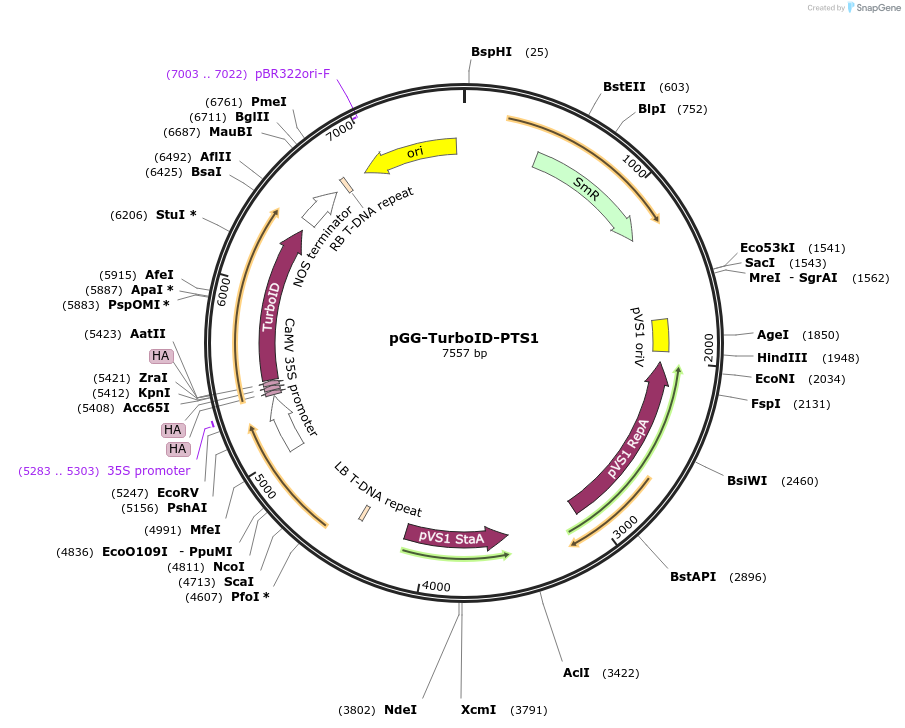
pGG-TurboID-PTS1
Plasmid#209398PurposeTransiently expressing TurboID fused with a peroxisomal targeting signal type I (PTS1) in plantaDepositorInsert3XHA-TurboID-PTS1
ExpressionPlantMutationTurboID gene sequence C249APromoterCauliflower mosaic virus (CaMV) 35S promoterAvailable SinceJan. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
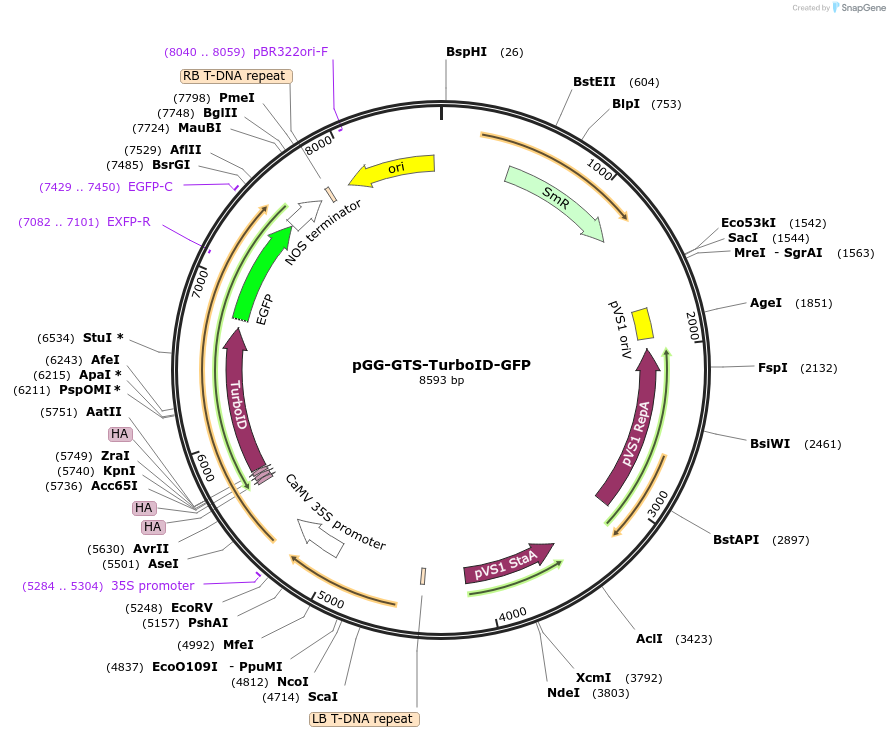
pGG-GTS-TurboID-GFP
Plasmid#209405PurposeTransiently expressing and visualizing the subcellular localization of TurboID fused with a Golgi targeting sequence (GTS) in plantaDepositorInsertGTS-3XHA-TurboID
TagsGFPExpressionPlantMutationTurboID gene sequence C249APromoterCauliflower mosaic virus (CaMV) 35S promoterAvailable SinceJan. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
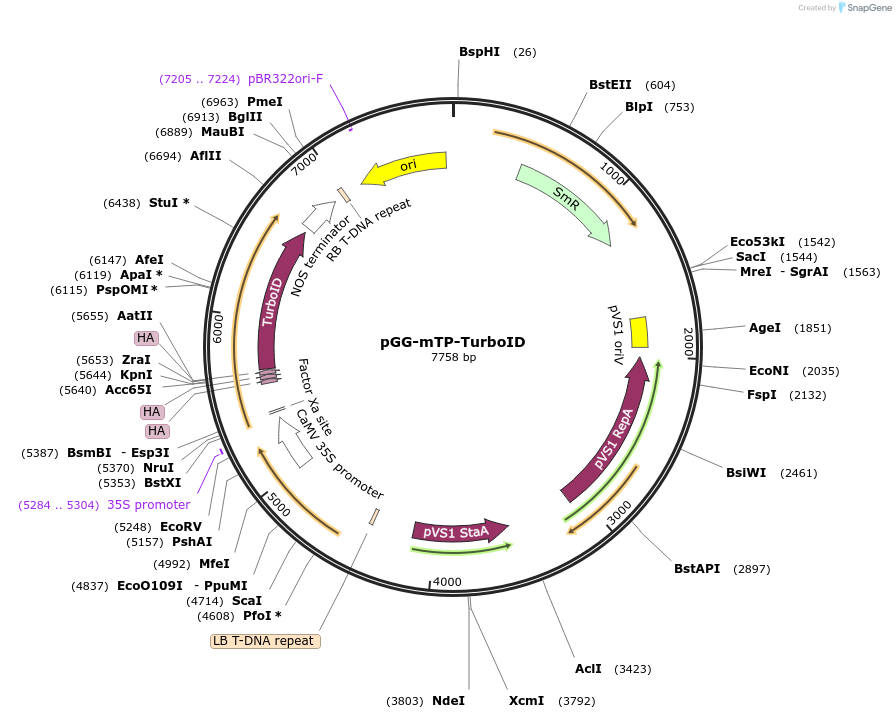
pGG-mTP-TurboID
Plasmid#209396PurposeTransiently expressing TurboID fused with a Mitochondrial transit peptide (mTP) in plantaDepositorInsertmTP-3XHA-TurboID
ExpressionPlantMutationTurboID gene sequence C249APromoterCauliflower mosaic virus (CaMV) 35S promoterAvailable SinceJan. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
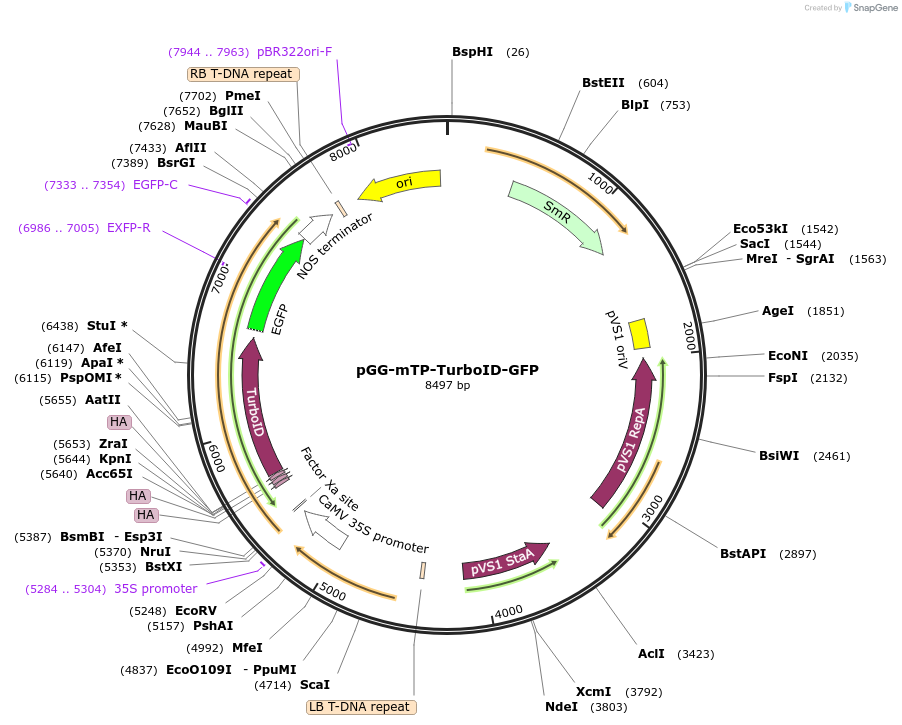
pGG-mTP-TurboID-GFP
Plasmid#209404PurposeTransiently expressing and visualizing the subcellular localization of TurboID fused with a Mitochondrial transit peptide (mTP) in plantaDepositorInsertmTP-3XHA-TurboID
TagsGFPExpressionPlantMutationTurboID gene sequence C249APromoterCauliflower mosaic virus (CaMV) 35S promoterAvailable SinceJan. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
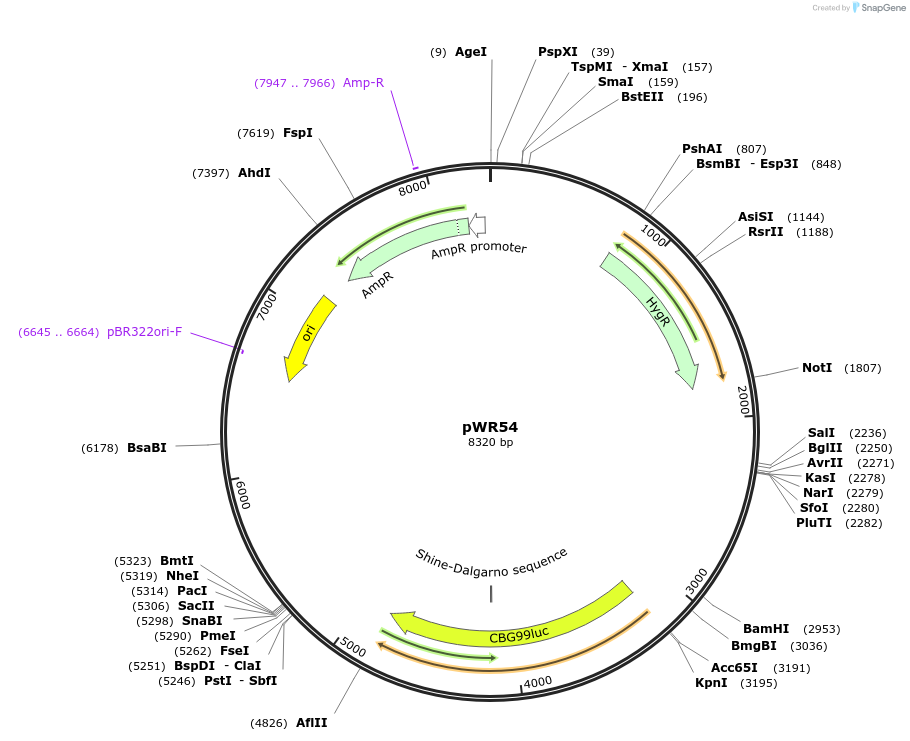
pWR54
Plasmid#203330Purposeepisomal bioluminescent reporter plasmid for S. stipitisDepositorInsertsTEF1 promoter (S. stipitis) (TEF1 S. stipitis (yeast))
coHPH
ACT1 terminator (S. stipitis)
CCW12 promoter (S. stipitis)
coCBG
ADH2 terminator (S. stipitis)
ARS from S. stipitis chromosome 1
ExpressionYeastAvailable SinceDec. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
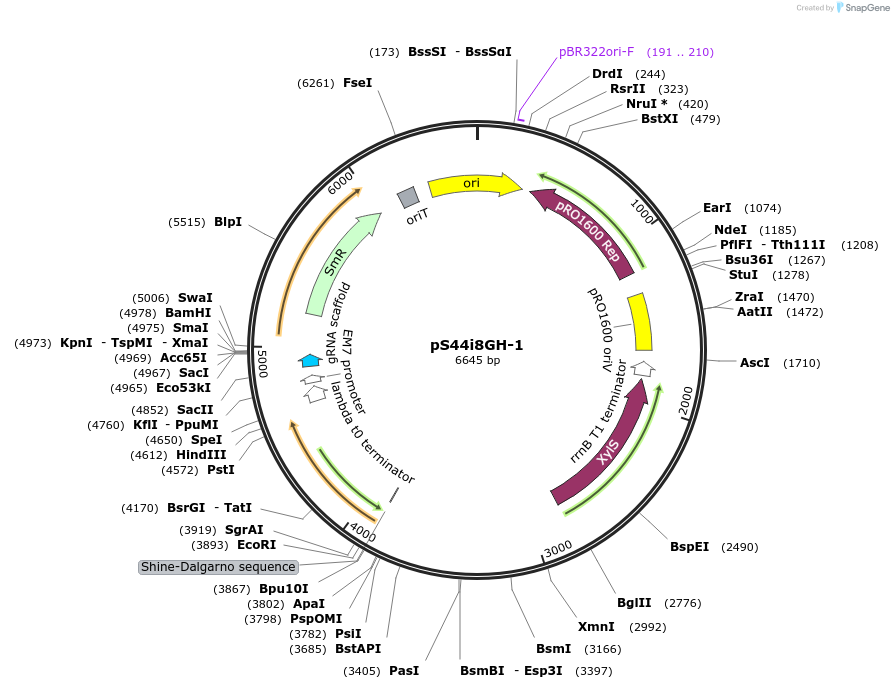
pS44i8GH-1
Plasmid#207525PurposeConditionally-replicating in Pseudomonas vector, high-copy-number; oriV(pRO1600/ColE1), xylS (cured of BsaI-sites), Pm→repA, P14b with a translational BCD2 coupler from BG35 (53)→msfGFP; SmR/SpRDepositorInsertoriV(pRO1600/ColE1), xylS, Pm→repA, P14b with a translational BCD2 coupler from BG35 (53)→msfGFP;
UseCRISPR; Pseudomonas vectorAvailable SinceDec. 12, 2023AvailabilityAcademic Institutions and Nonprofits only -
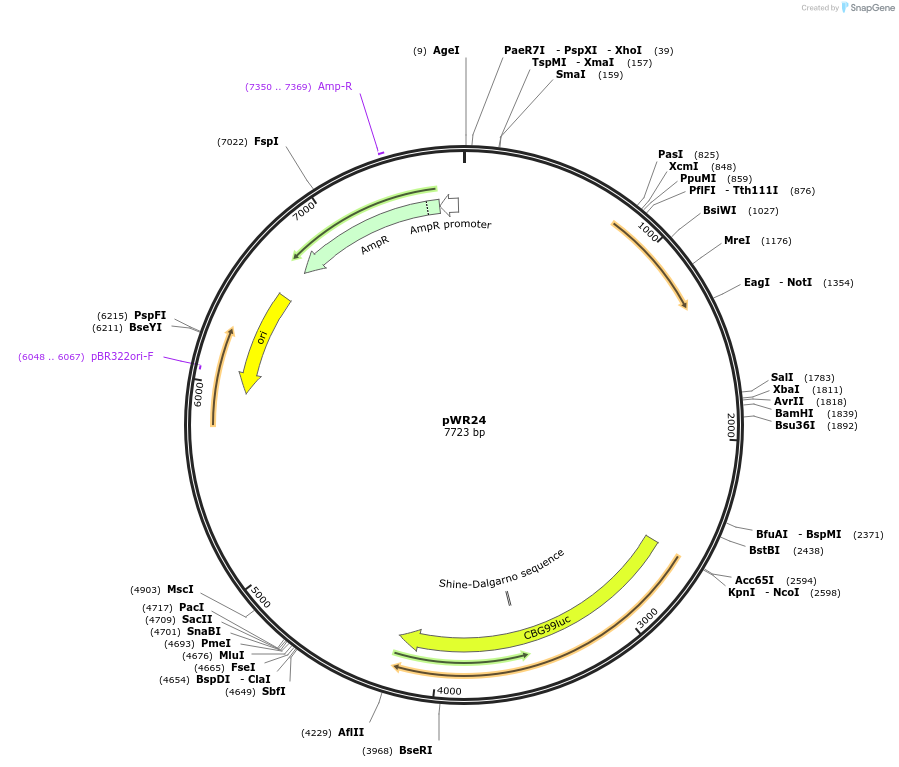
pWR24
Plasmid#202771Purposeepisomal bioluminescent reporter plasmid for S. stipitisDepositorInsertsTEF1 promoter (S. stipitis) (TEF1 S. stipitis (yeast))
coNAT
ACT1 terminator (S. stipitis)
TDH3 promoter (S. stipitis)
coCBG
ADH2 terminator (S. stipitis)
ARS from S. stipitis chromosome 1
ExpressionYeastAvailable SinceOct. 4, 2023AvailabilityAcademic Institutions and Nonprofits only