We narrowed to 10,828 results for: 158
-
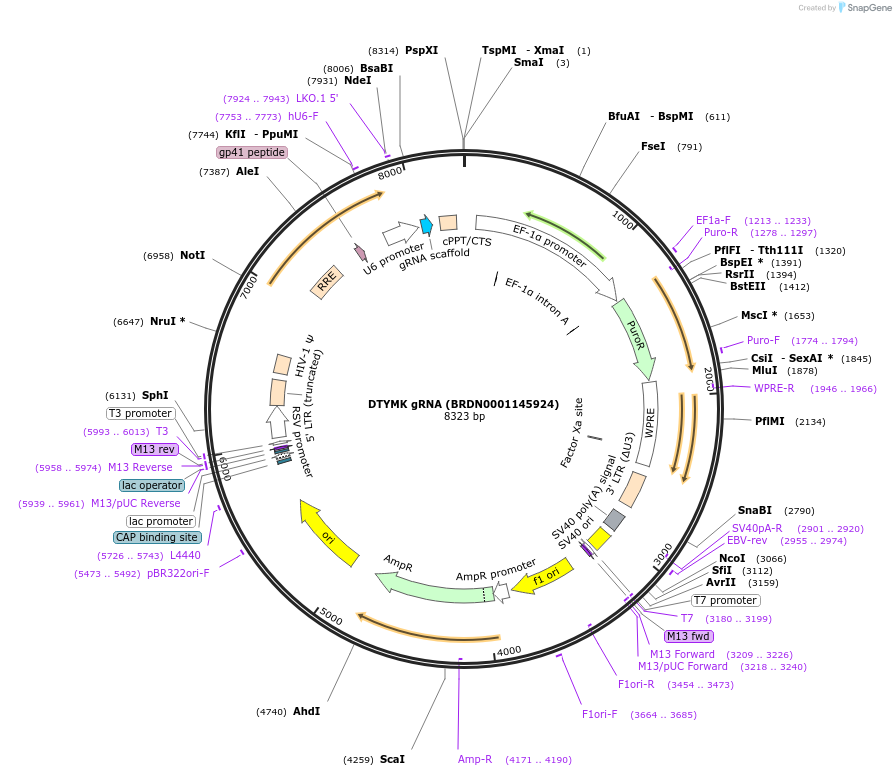
Plasmid#77513Purpose3rd generation lentiviral gRNA plasmid targeting human DTYMKDepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only
-
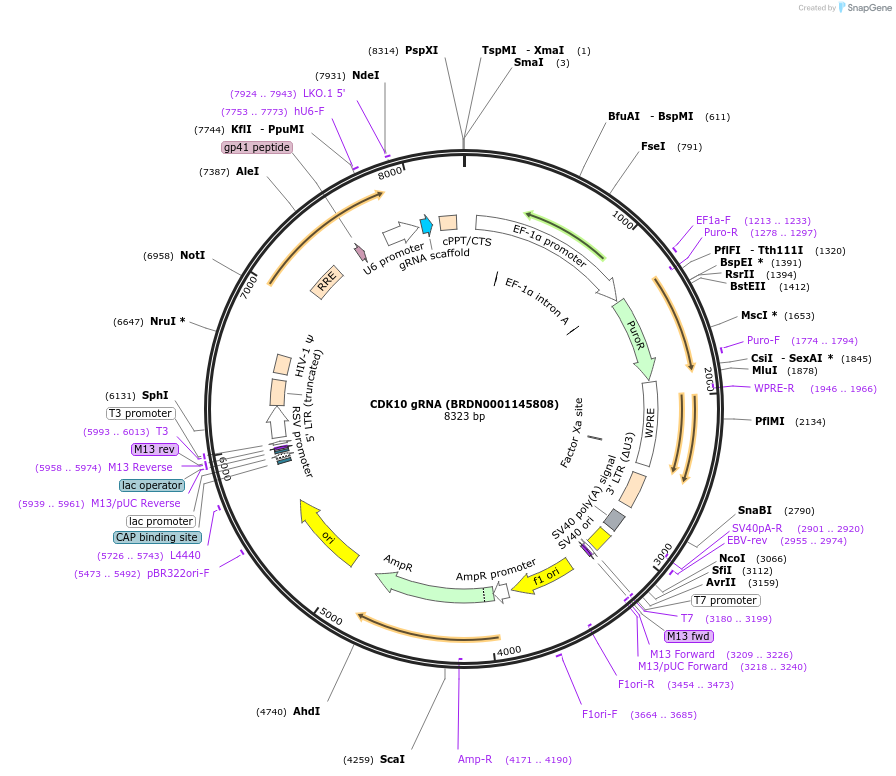
CDK10 gRNA (BRDN0001145808)
Plasmid#77578Purpose3rd generation lentiviral gRNA plasmid targeting human CDK10DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
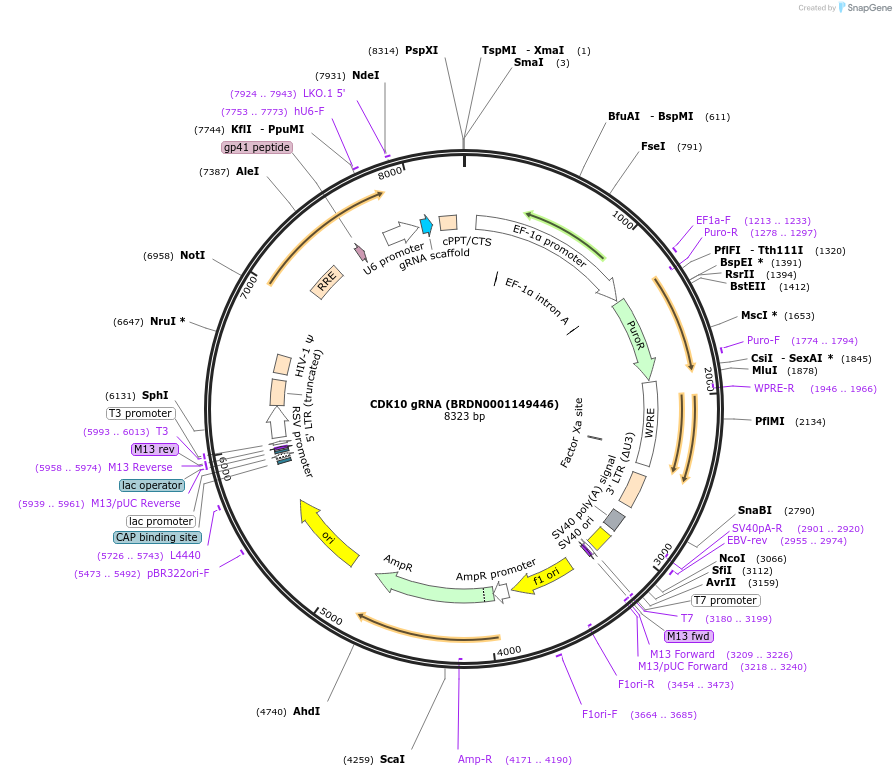
CDK10 gRNA (BRDN0001149446)
Plasmid#77579Purpose3rd generation lentiviral gRNA plasmid targeting human CDK10DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
MAP3K12 gRNA (BRDN0001144753)
Plasmid#76760Purpose3rd generation lentiviral gRNA plasmid targeting human MAP3K12DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
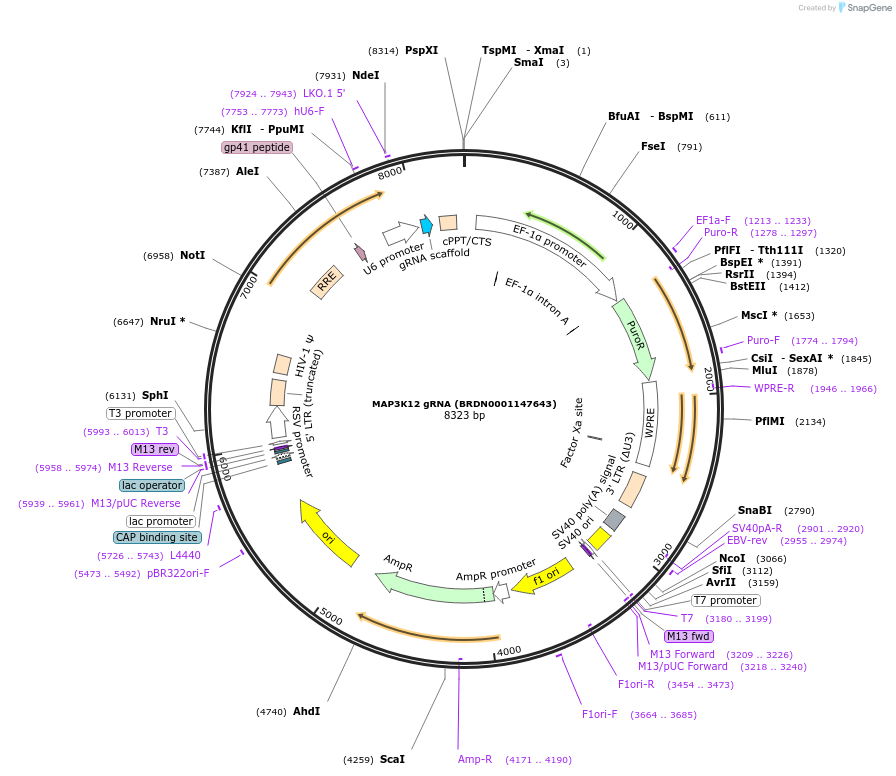
MAP3K12 gRNA (BRDN0001147643)
Plasmid#76761Purpose3rd generation lentiviral gRNA plasmid targeting human MAP3K12DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
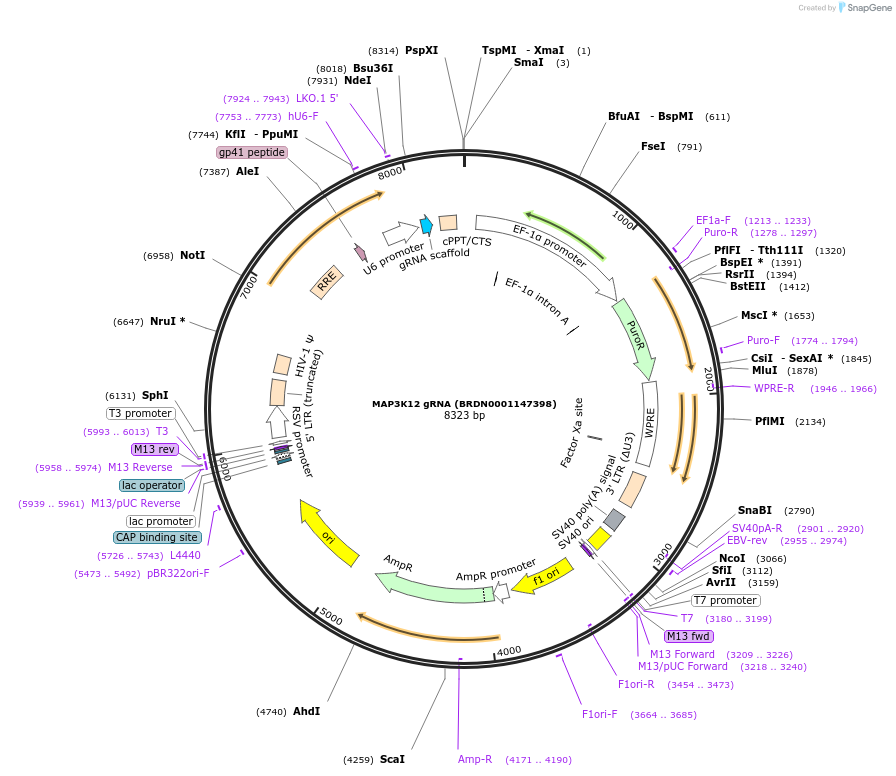
MAP3K12 gRNA (BRDN0001147398)
Plasmid#76762Purpose3rd generation lentiviral gRNA plasmid targeting human MAP3K12DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
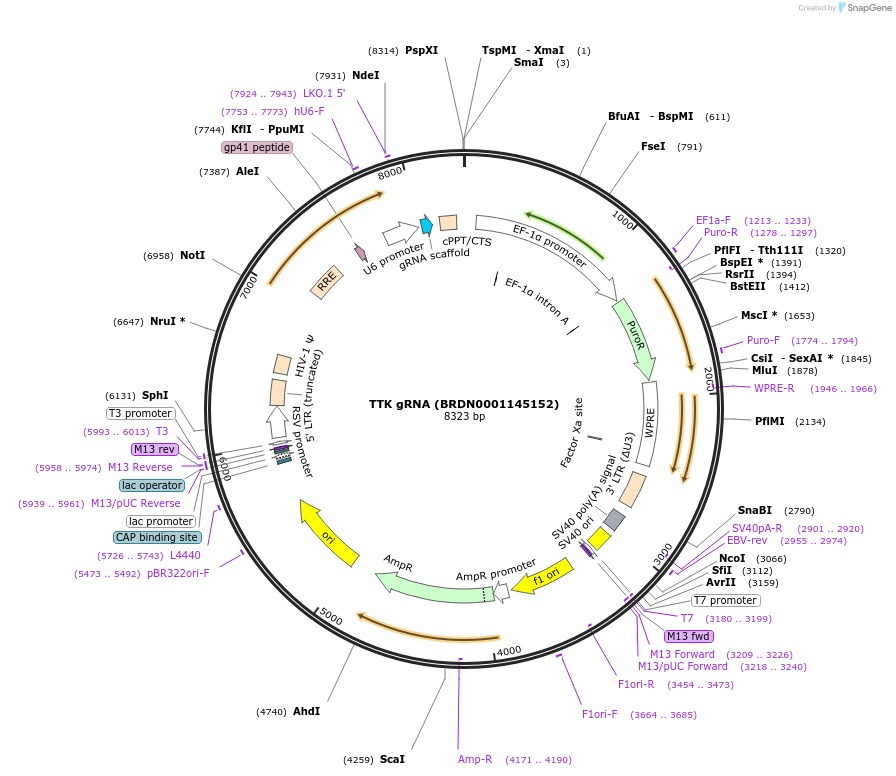
TTK gRNA (BRDN0001145152)
Plasmid#75579Purpose3rd generation lentiviral gRNA plasmid targeting human TTKDepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
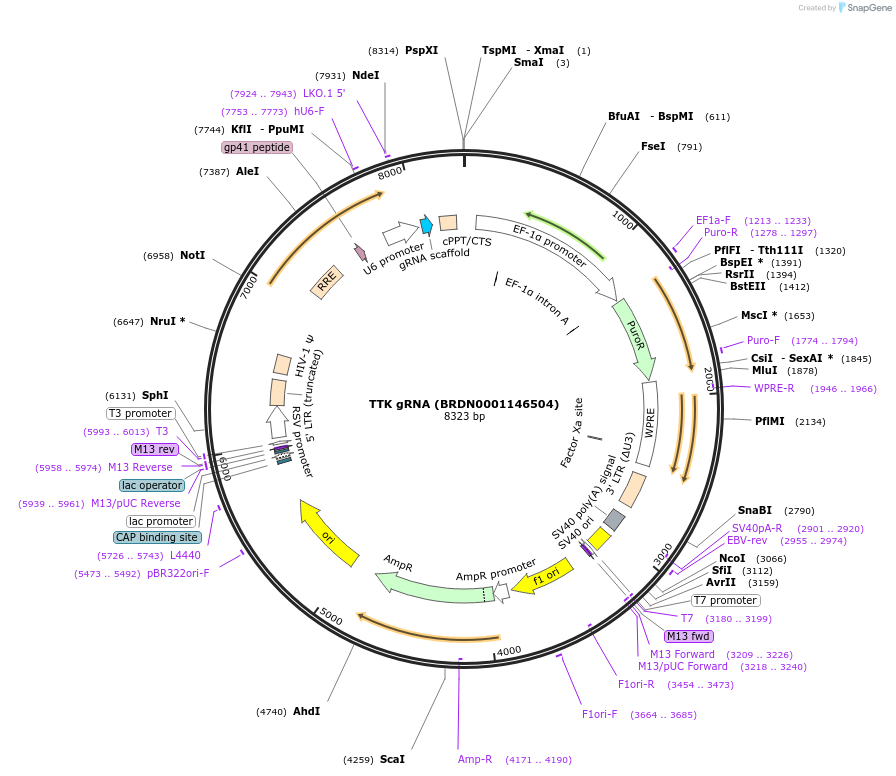
TTK gRNA (BRDN0001146504)
Plasmid#75580Purpose3rd generation lentiviral gRNA plasmid targeting human TTKDepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
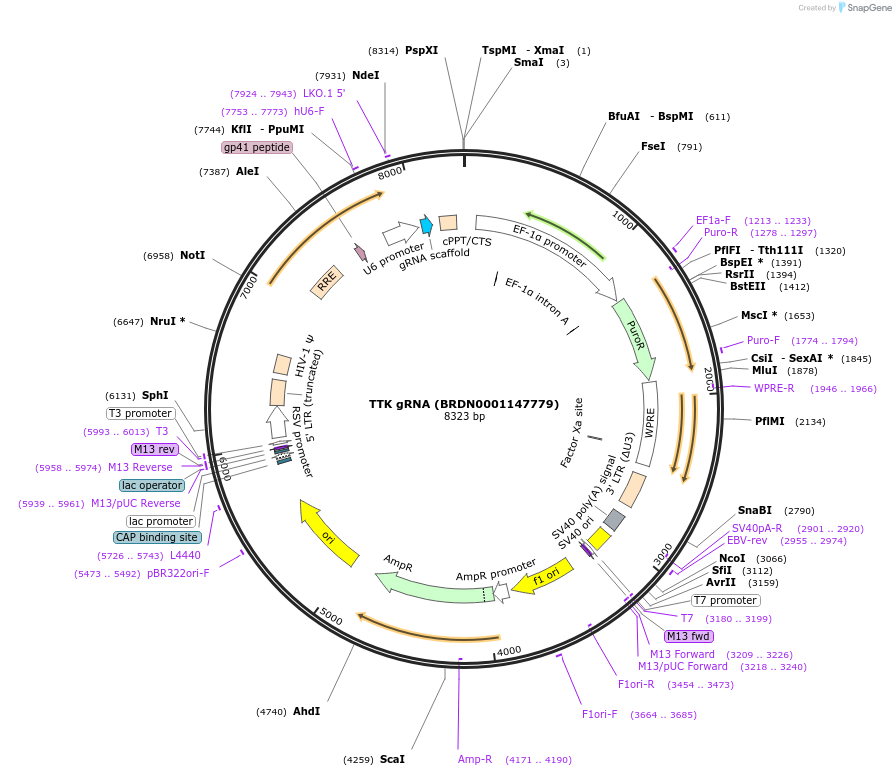
TTK gRNA (BRDN0001147779)
Plasmid#75581Purpose3rd generation lentiviral gRNA plasmid targeting human TTKDepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
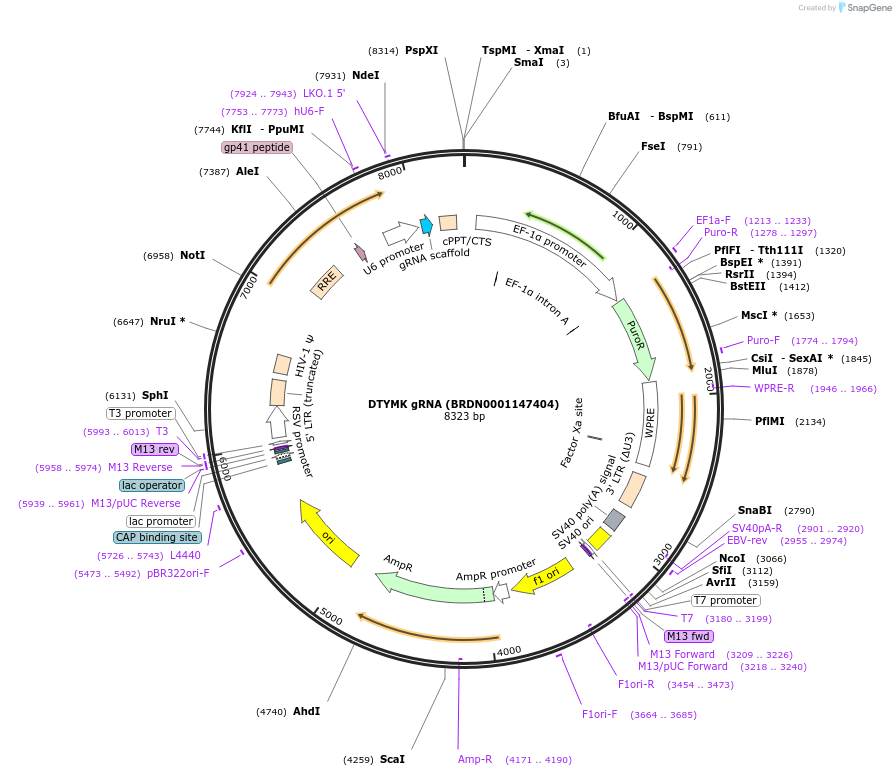
DTYMK gRNA (BRDN0001147404)
Plasmid#77511Purpose3rd generation lentiviral gRNA plasmid targeting human DTYMKDepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
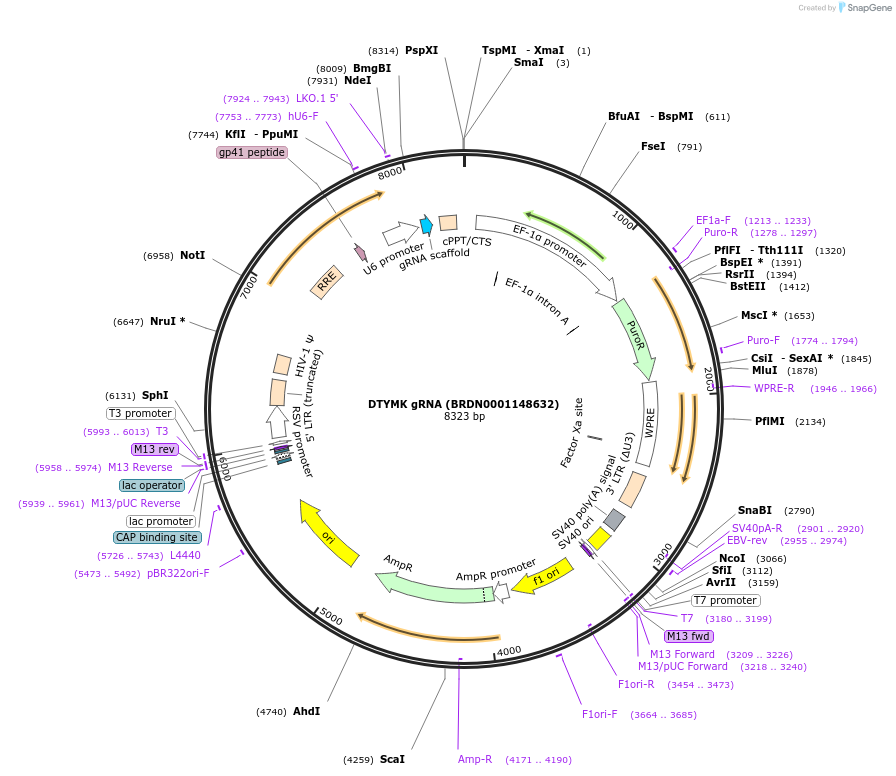
DTYMK gRNA (BRDN0001148632)
Plasmid#77512Purpose3rd generation lentiviral gRNA plasmid targeting human DTYMKDepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
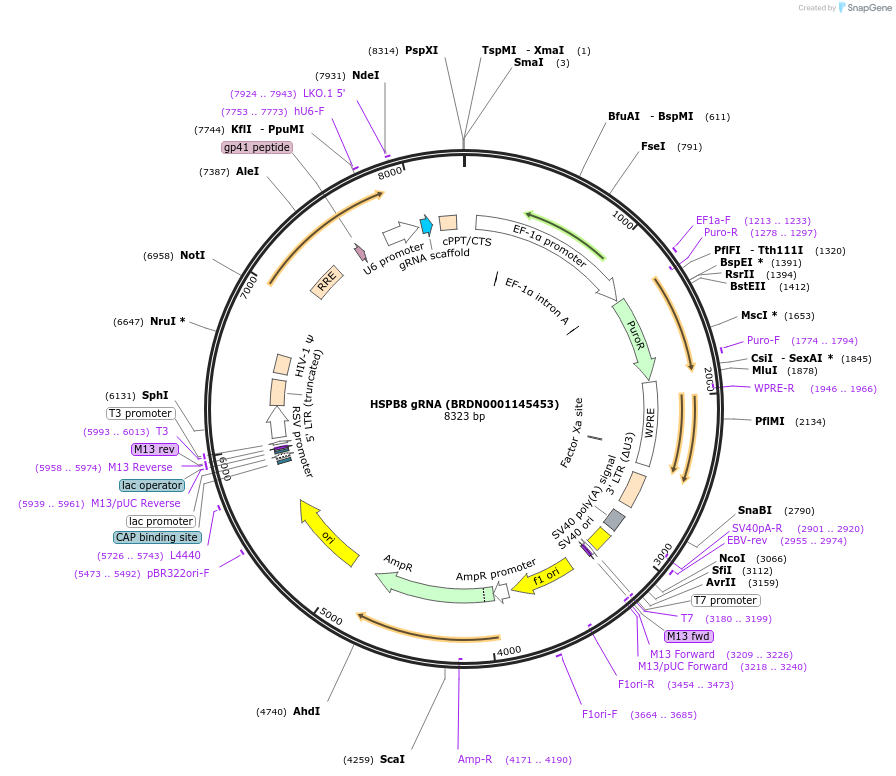
HSPB8 gRNA (BRDN0001145453)
Plasmid#75640Purpose3rd generation lentiviral gRNA plasmid targeting human HSPB8DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
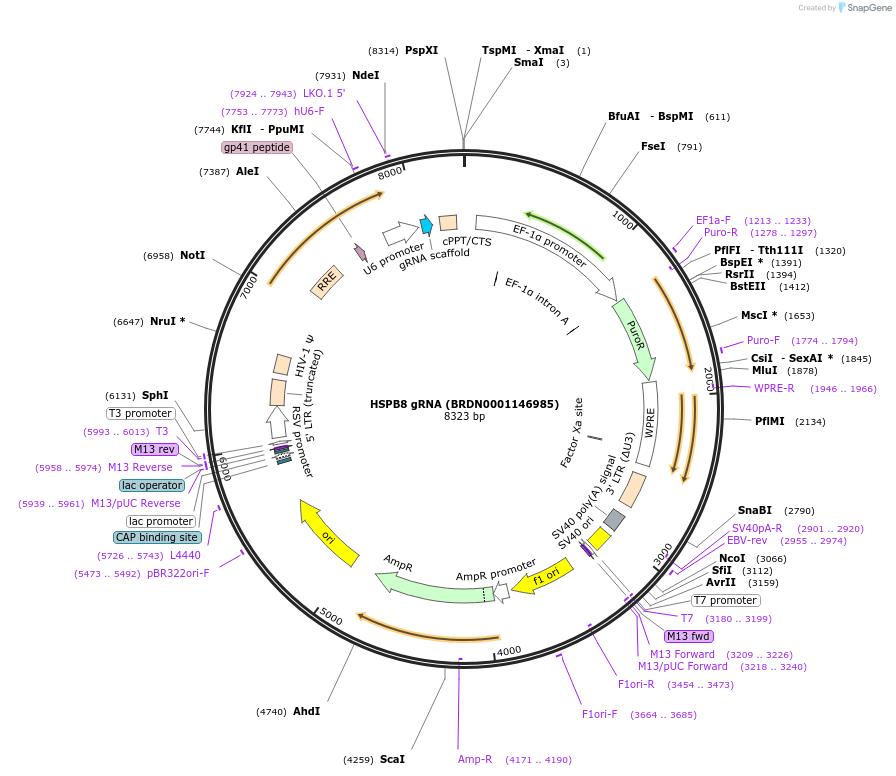
HSPB8 gRNA (BRDN0001146985)
Plasmid#75641Purpose3rd generation lentiviral gRNA plasmid targeting human HSPB8DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
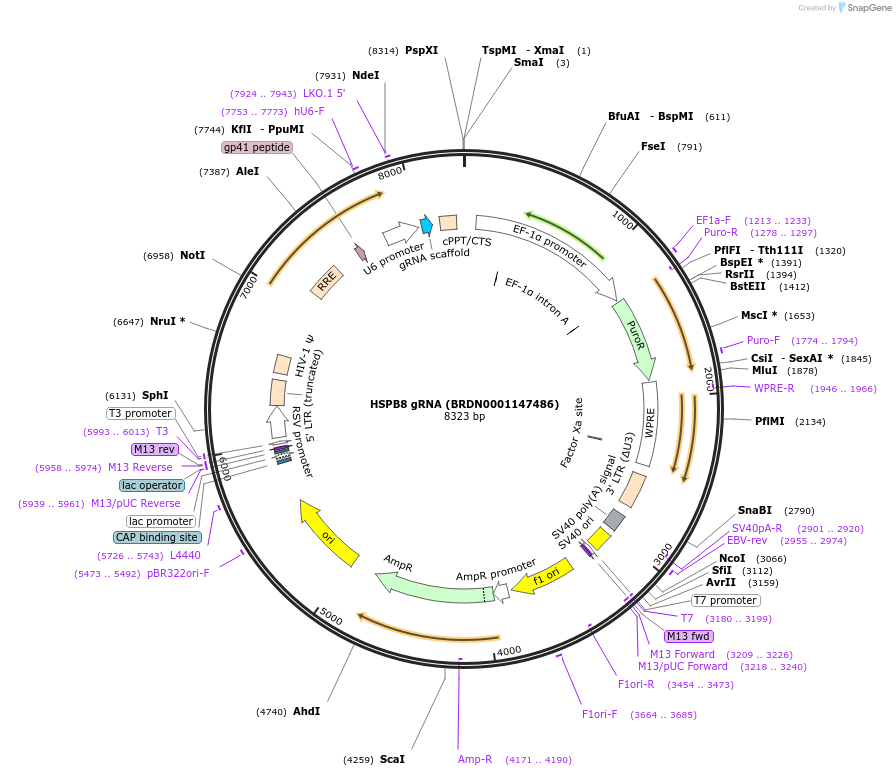
HSPB8 gRNA (BRDN0001147486)
Plasmid#75642Purpose3rd generation lentiviral gRNA plasmid targeting human HSPB8DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
pCMV-FLAG-CHD5-Y619E-FRAG
Plasmid#68871PurposeMammalian expression of CHD5 histone binding domain (331-672aa) with Y619 mutationDepositorAvailable SinceNov. 20, 2015AvailabilityAcademic Institutions and Nonprofits only -
NC12 pCDNA3.1 VENUS WTX delta ETGE
Plasmid#36967DepositorInsertWTX (delta ETGE motif) (AMER1 Human)
TagsVenusExpressionMammalianMutationlacks amino acids 288 –291PromoterCMVAvailable SinceJuly 24, 2012AvailabilityAcademic Institutions and Nonprofits only -
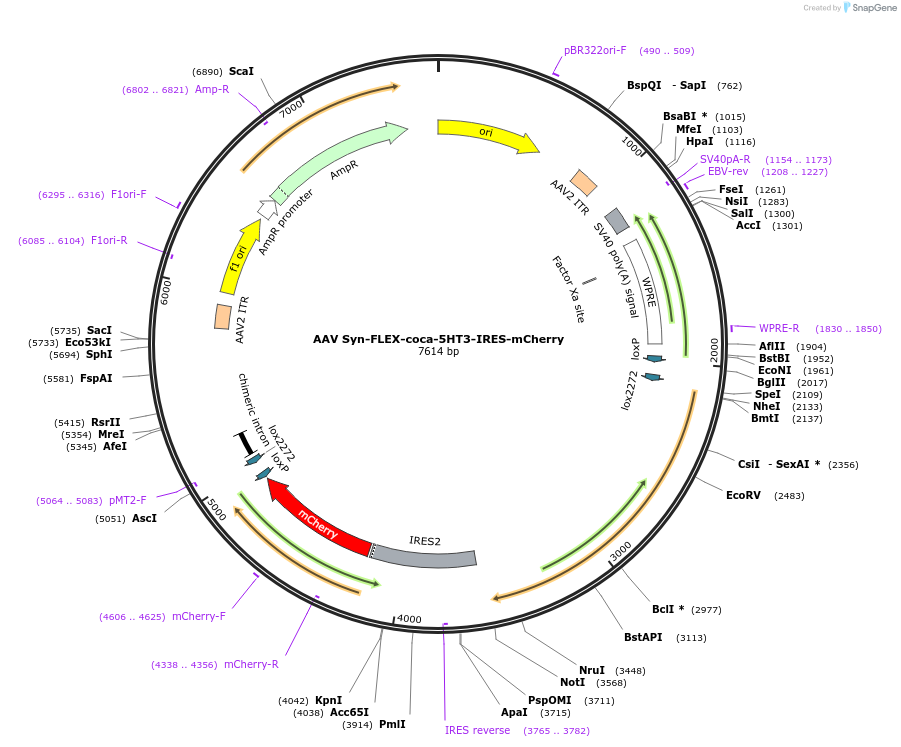
AAV Syn-FLEX-coca-5HT3-IRES-mCherry
Plasmid#242195PurposeCre-dependent AAV vector for cocaine-gated chemogenetic cation channelDepositorInsertSyn-FLEX-coca-5HT3-IRES-mCherry (Htr3a Mouse)
UseAAVExpressionMammalianMutationL141G G175K Y210F Y217FPromoterSynapsinAvailable SinceOct. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
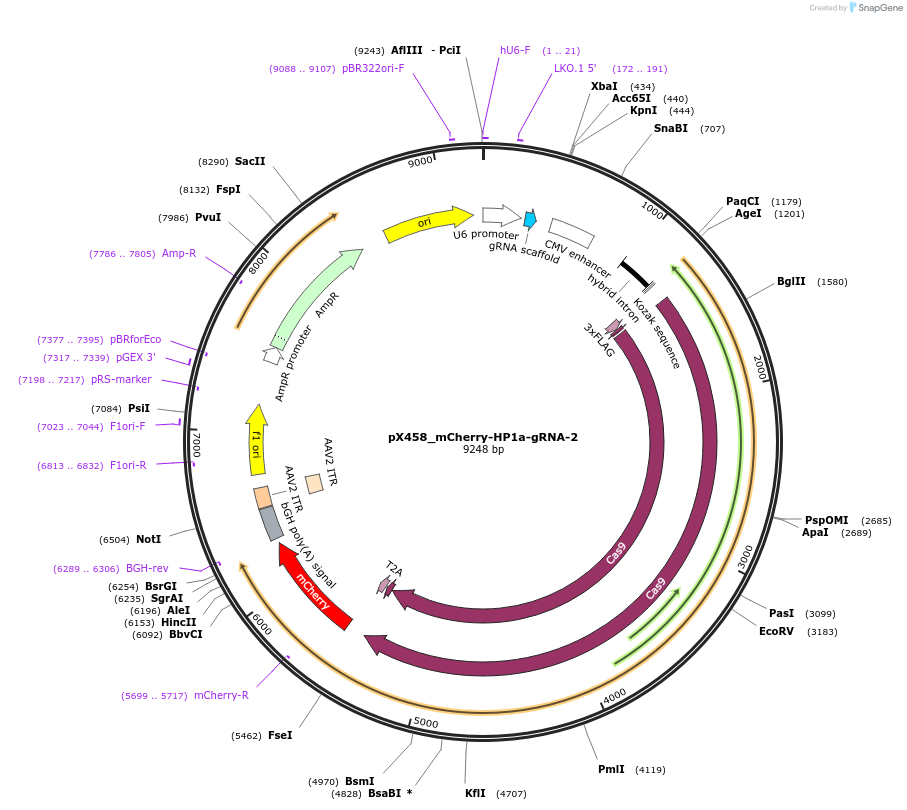
pX458_mCherry-HP1a-gRNA-2
Plasmid#237635PurposeCas9 from S. pyogenes with 2A-mCherry, gRNA to knock-in msfGFP to HP1a locusDepositorAvailable SinceMay 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
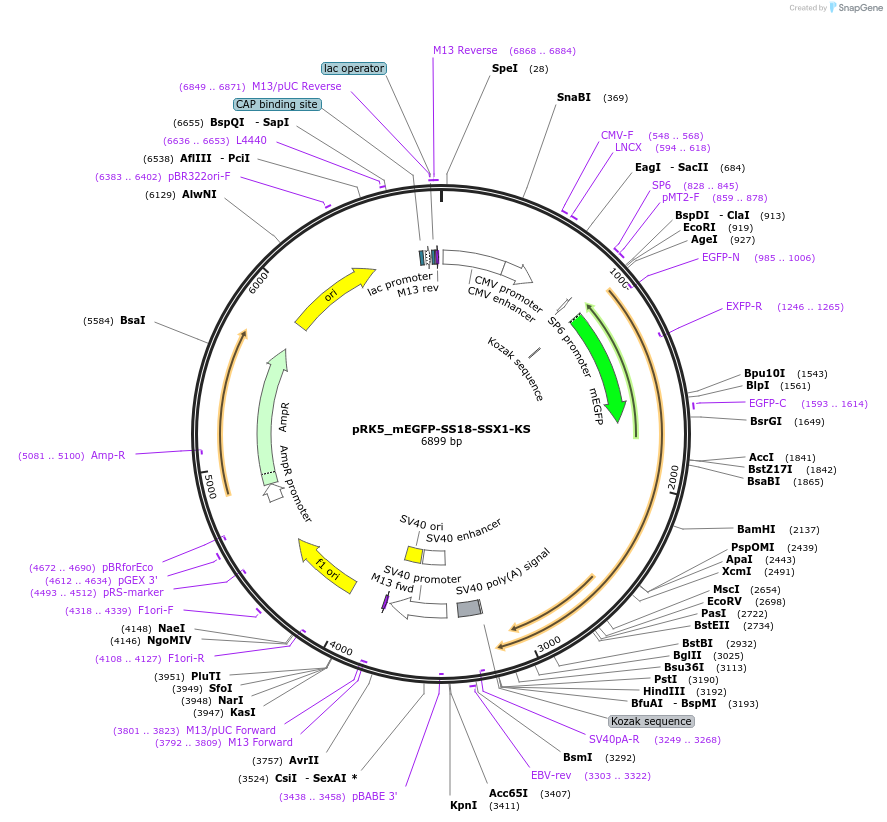
pRK5_mEGFP-SS18-SSX1-KS
Plasmid#237679PurposeFor overexpression of mEGFP-SS18-SSX1-KSDepositorAvailable SinceMay 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
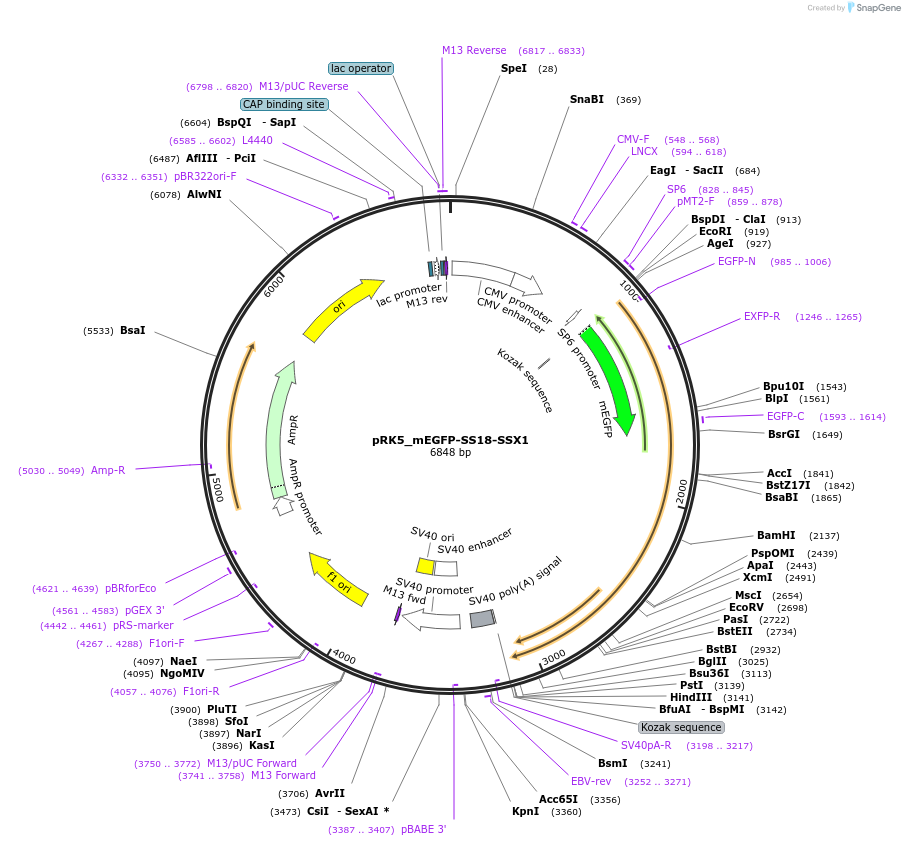
pRK5_mEGFP-SS18-SSX1
Plasmid#237678PurposeFor overexpression of mEGFP-SS18-SSX1DepositorAvailable SinceMay 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
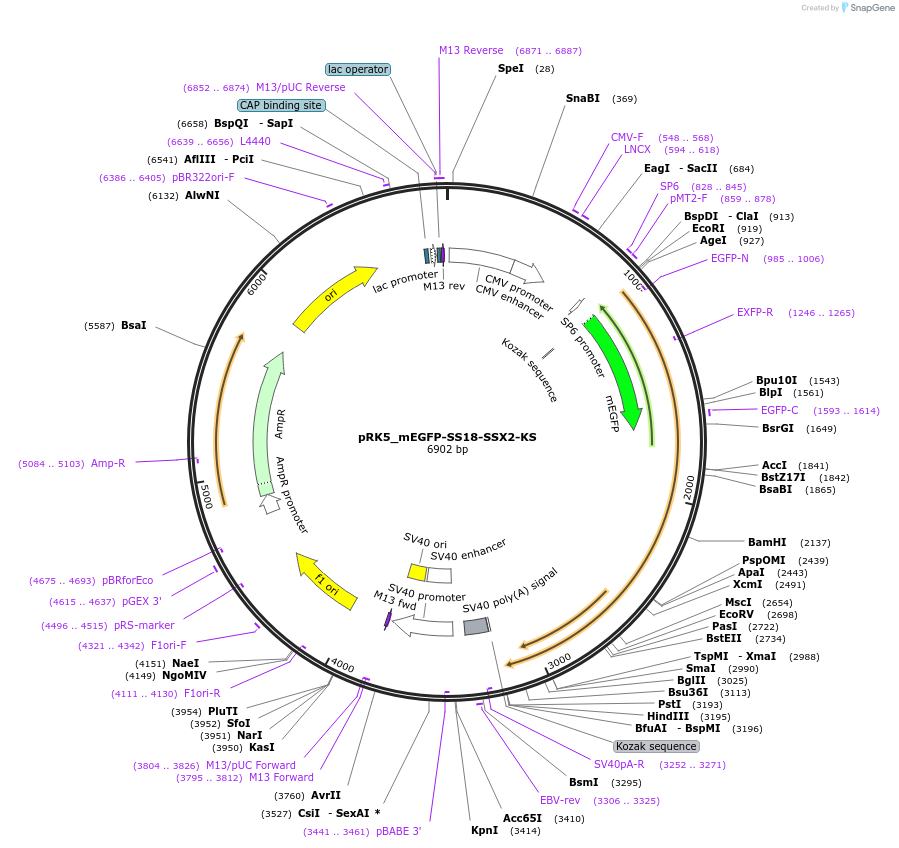
pRK5_mEGFP-SS18-SSX2-KS
Plasmid#237680PurposeFor overexpression of mEGFP-SS18-SSX2-KSDepositorAvailable SinceMay 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
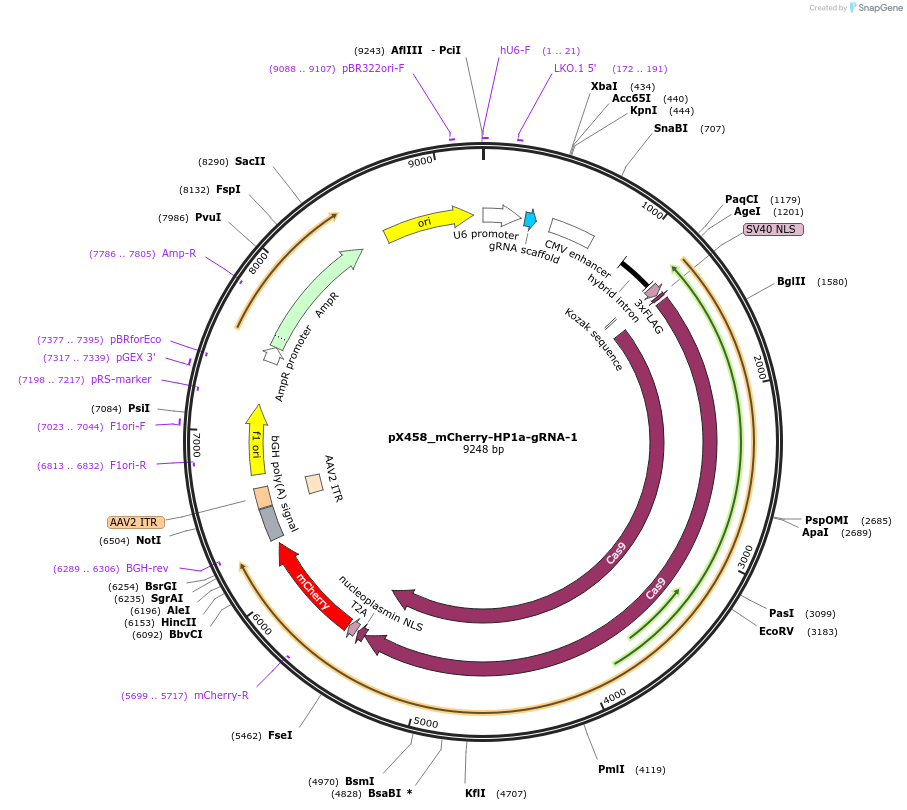
pX458_mCherry-HP1a-gRNA-1
Plasmid#237634PurposeCas9 from S. pyogenes with 2A-mCherry, gRNA to knock-in msfGFP to HP1a locusDepositorAvailable SinceMay 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
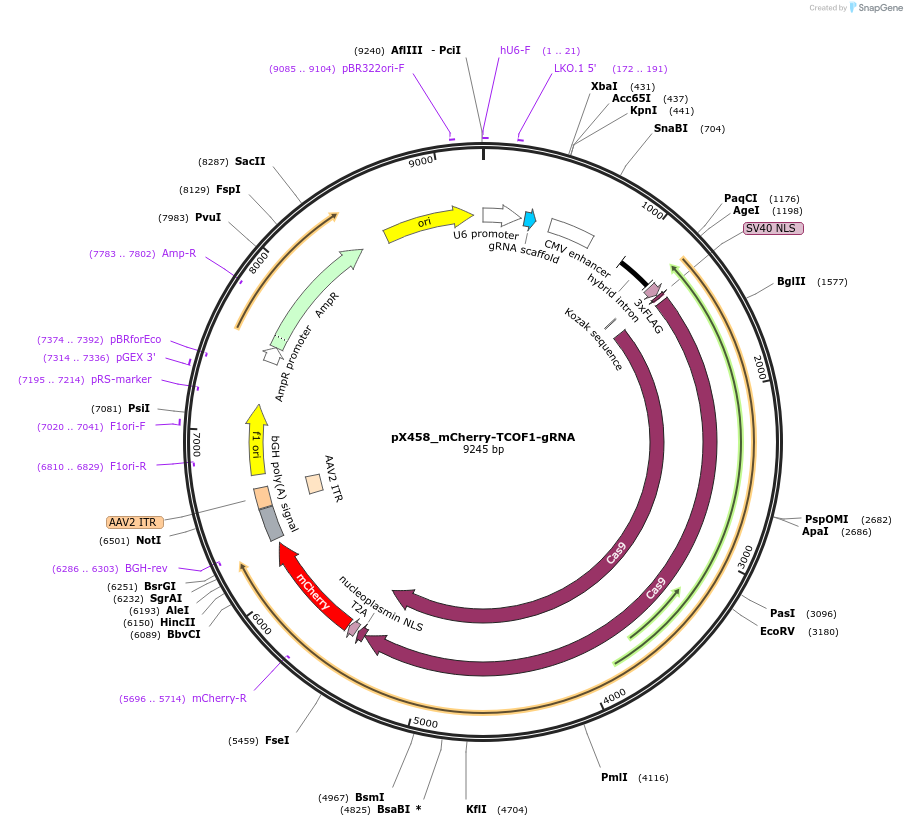
pX458_mCherry-TCOF1-gRNA
Plasmid#237633PurposeCas9 from S. pyogenes with 2A-mCherry, gRNA to knock-in msfGFP to TCOF1 locusDepositorAvailable SinceMay 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
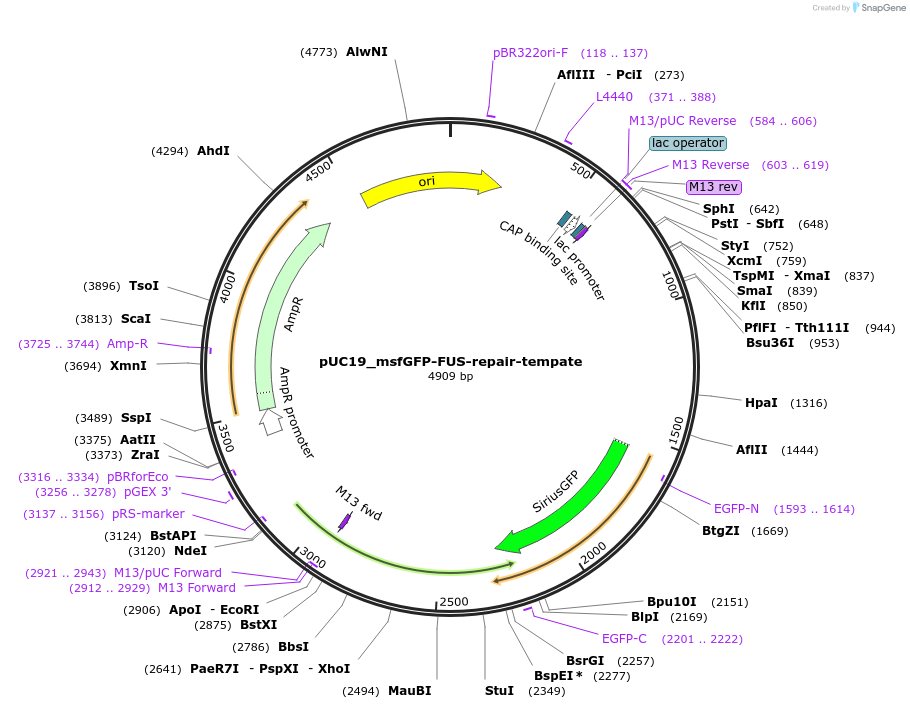
pUC19_msfGFP-FUS-repair-tempate
Plasmid#237684PurposeFor knock-in msfGFP to FUS locusDepositorAvailable SinceMay 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
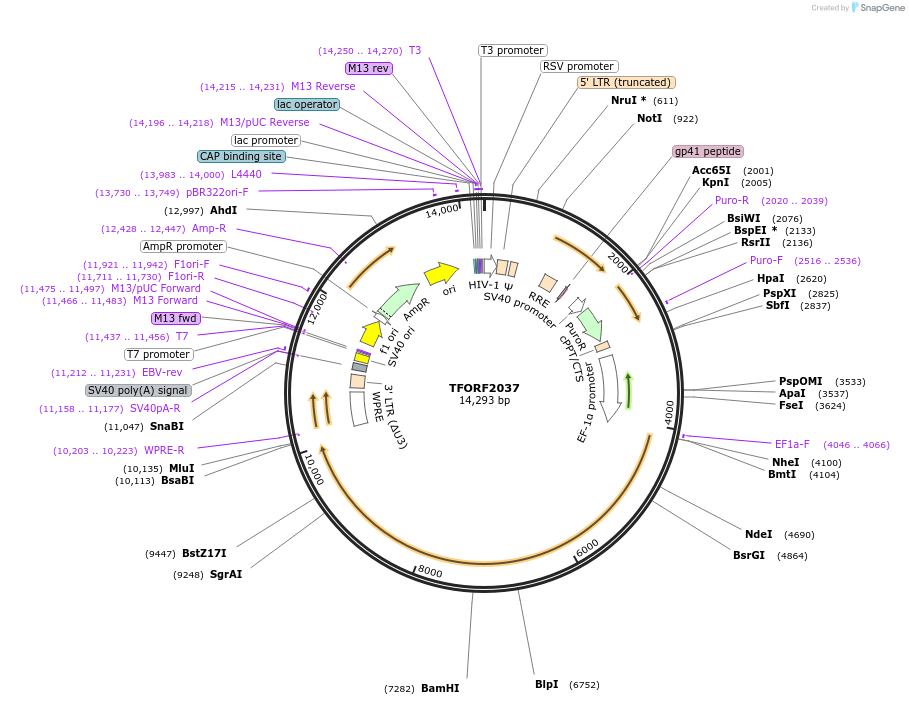
TFORF2037
Plasmid#144378PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceMay 1, 2025AvailabilityAcademic Institutions and Nonprofits only -
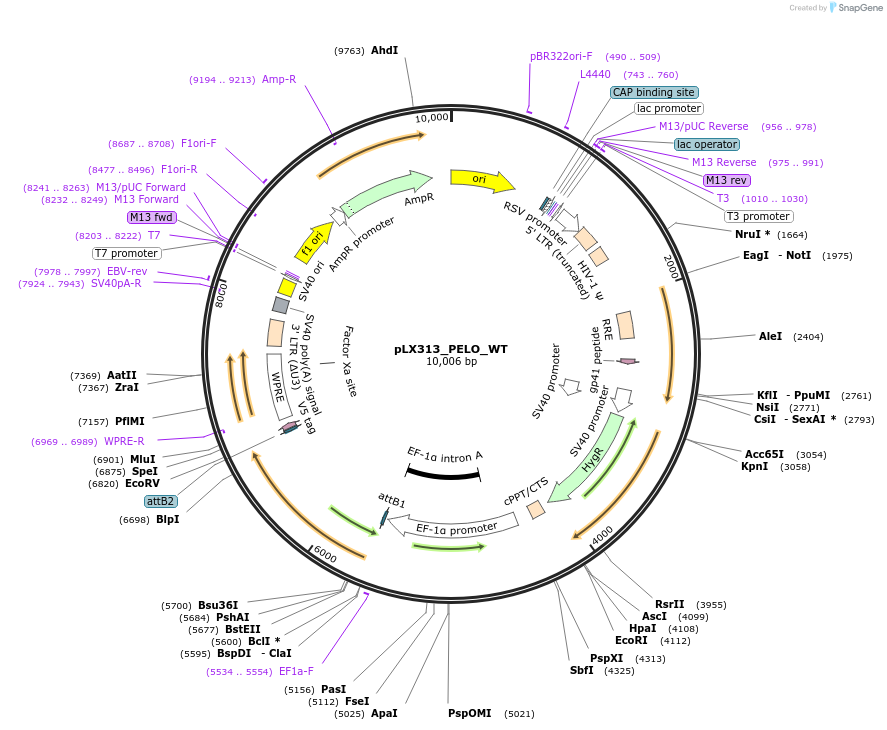
pLX313_PELO_WT
Plasmid#228938PurposeConstitutive expression of wild-type human PELODepositorAvailable SinceApril 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
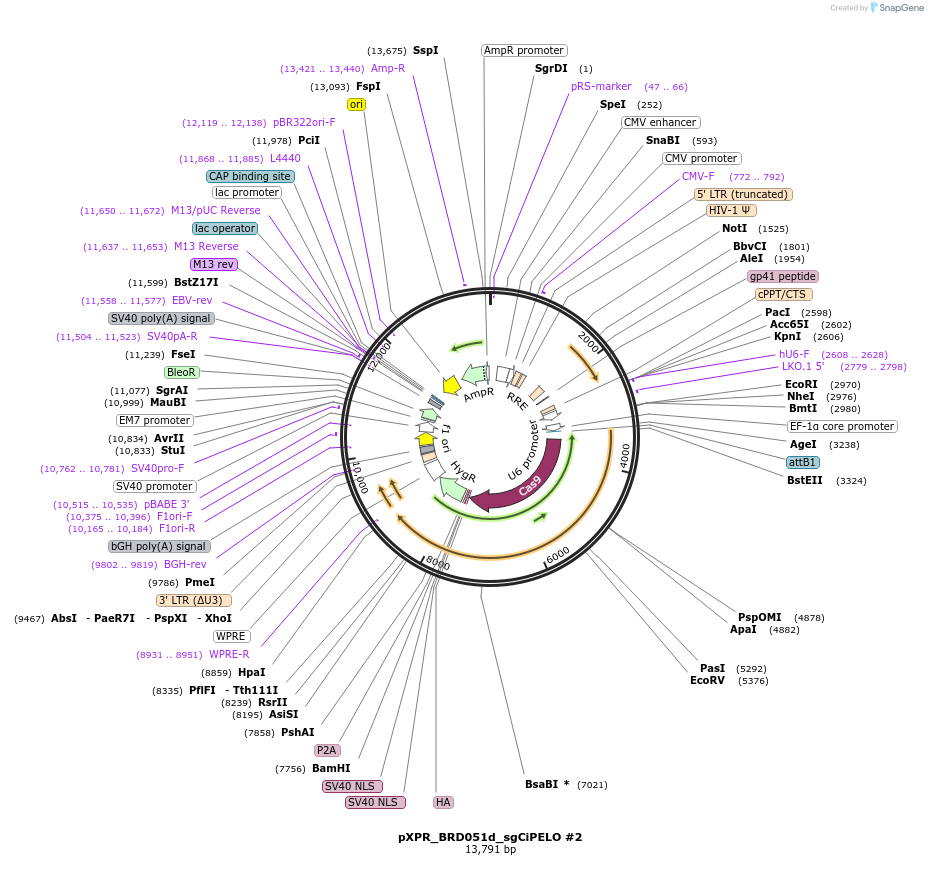
pXPR_BRD051d_sgCiPELO #2
Plasmid#229018PurposeExpression of CRISPRi PELO sgRNA #2DepositorAvailable SinceMarch 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
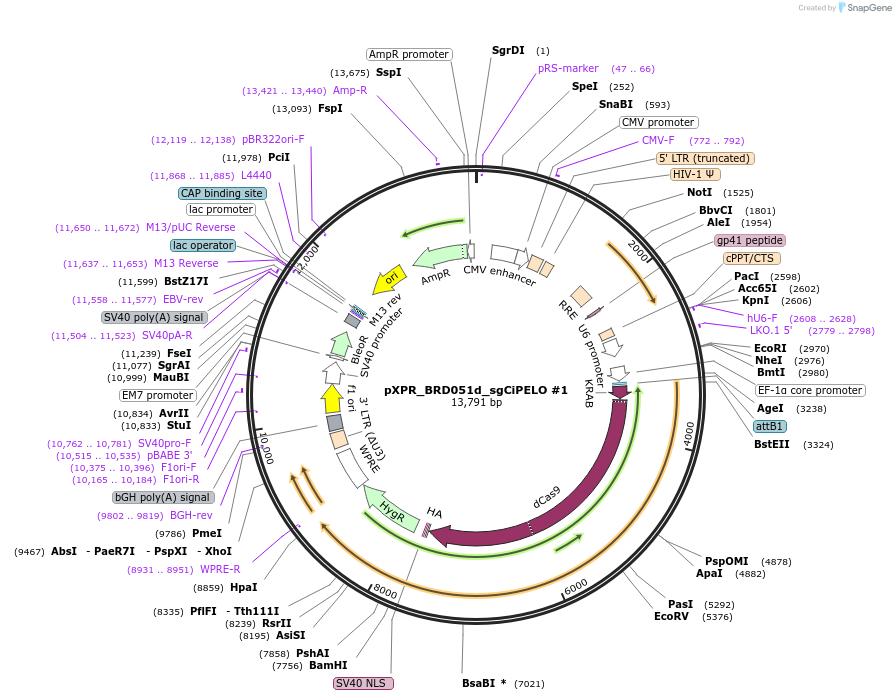
pXPR_BRD051d_sgCiPELO #1
Plasmid#229017PurposeExpression of CRISPRi PELO sgRNA #1DepositorAvailable SinceMarch 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
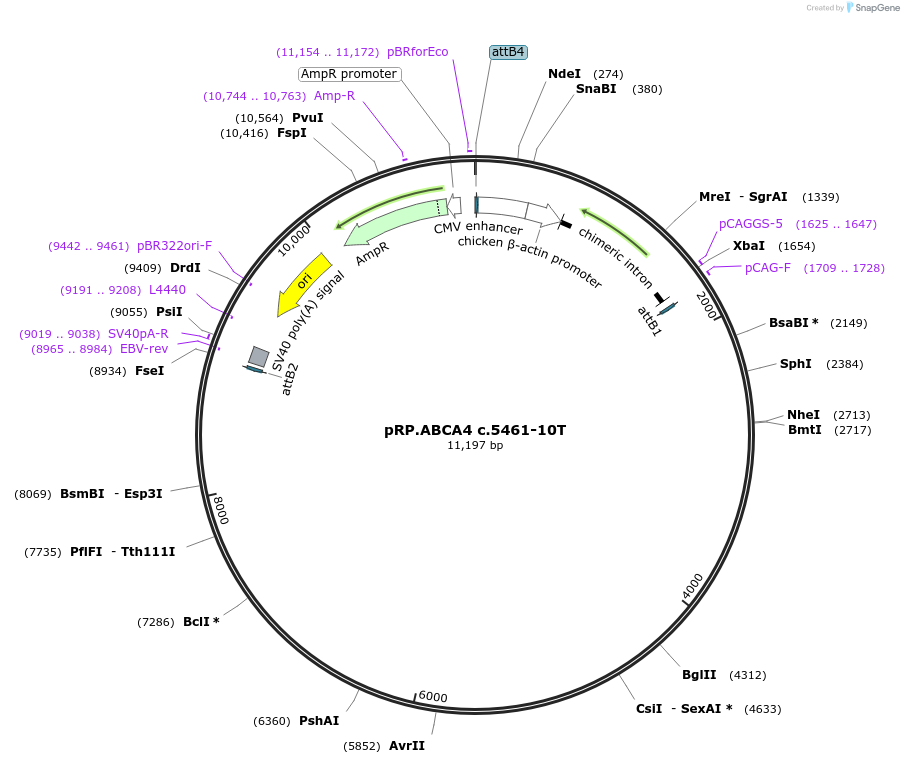
pRP.ABCA4 c.5461-10T
Plasmid#226553PurposeMinigene encoding the ABCA4 c.5461-10T mutationDepositorAvailable SinceJan. 13, 2025AvailabilityAcademic Institutions and Nonprofits only -
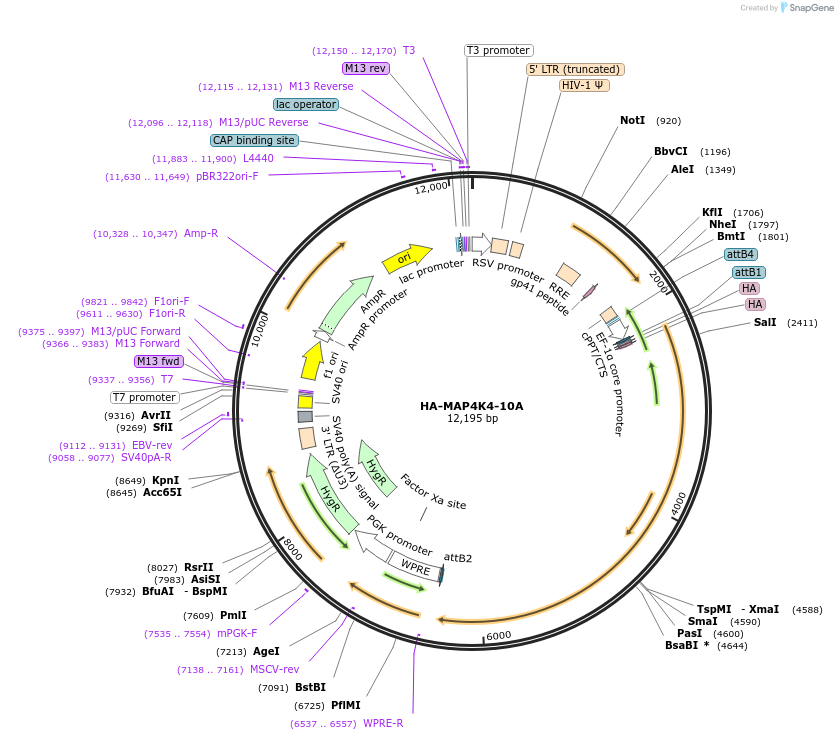
HA-MAP4K4-10A
Plasmid#229403PurposeLentiviral or overpexression of cDNADepositorInsertMAP4K4 (MAP4K4 Human)
UseLentiviralTagsHAExpressionMammalianMutationMARK2 phosphosites (S523,S536,S547,S643,T684,S726…PromoterEFSAvailable SinceDec. 12, 2024AvailabilityAcademic Institutions and Nonprofits only