We narrowed to 4,526 results for: golden gate
-
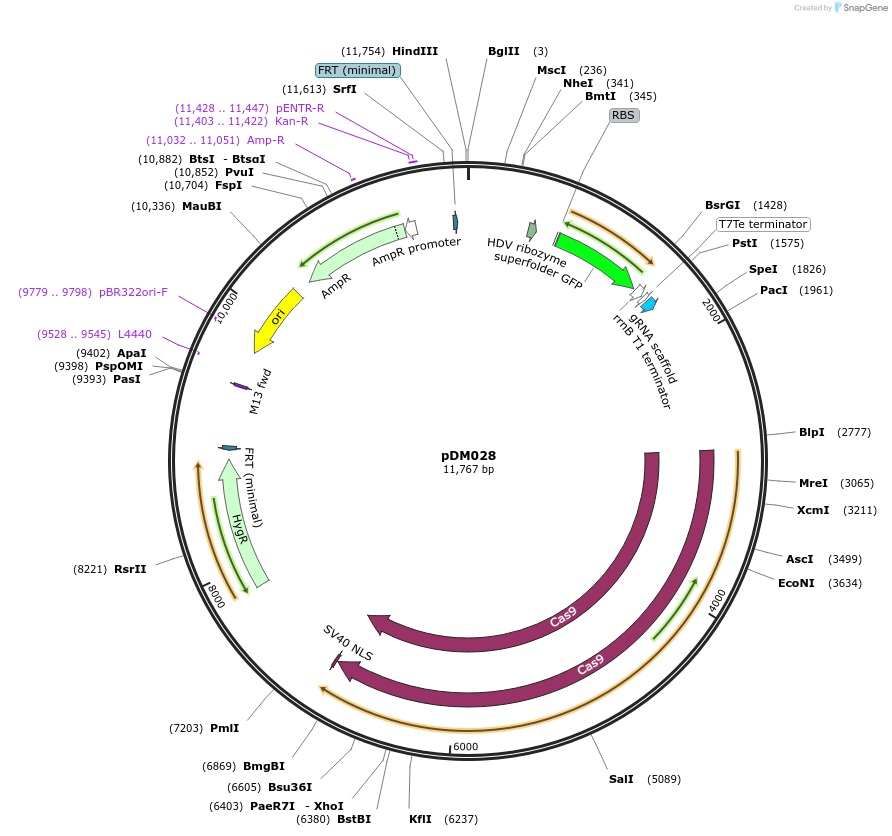
Plasmid#216809PurposeVector for Aspergillus CRISPR-Cas9 genetic engineering with Golden Gate cloning drop-out cassette for protospacer insertion and hph selectable marker.DepositorTypeEmpty backboneUseCRISPR; Fungal expressionExpressionBacterialAvailable SinceSept. 5, 2024AvailabilityAcademic Institutions and Nonprofits only
-
pDM068
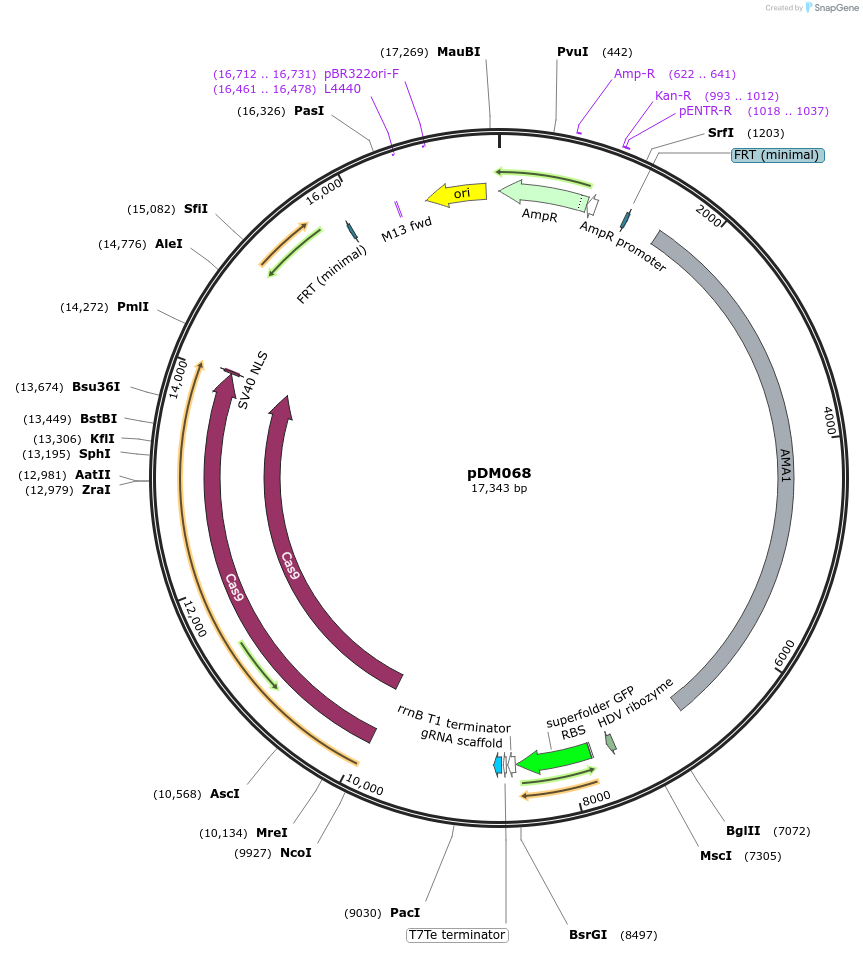
Plasmid#216811PurposeVector for Aspergillus CRISPR-Cas9 genetic engineering with Golden Gate cloning drop-out cassette for protospacer insertion and NAT selectable marker.DepositorTypeEmpty backboneUseCRISPR; Fungal expressionExpressionBacterialAvailable SinceSept. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
pDM026
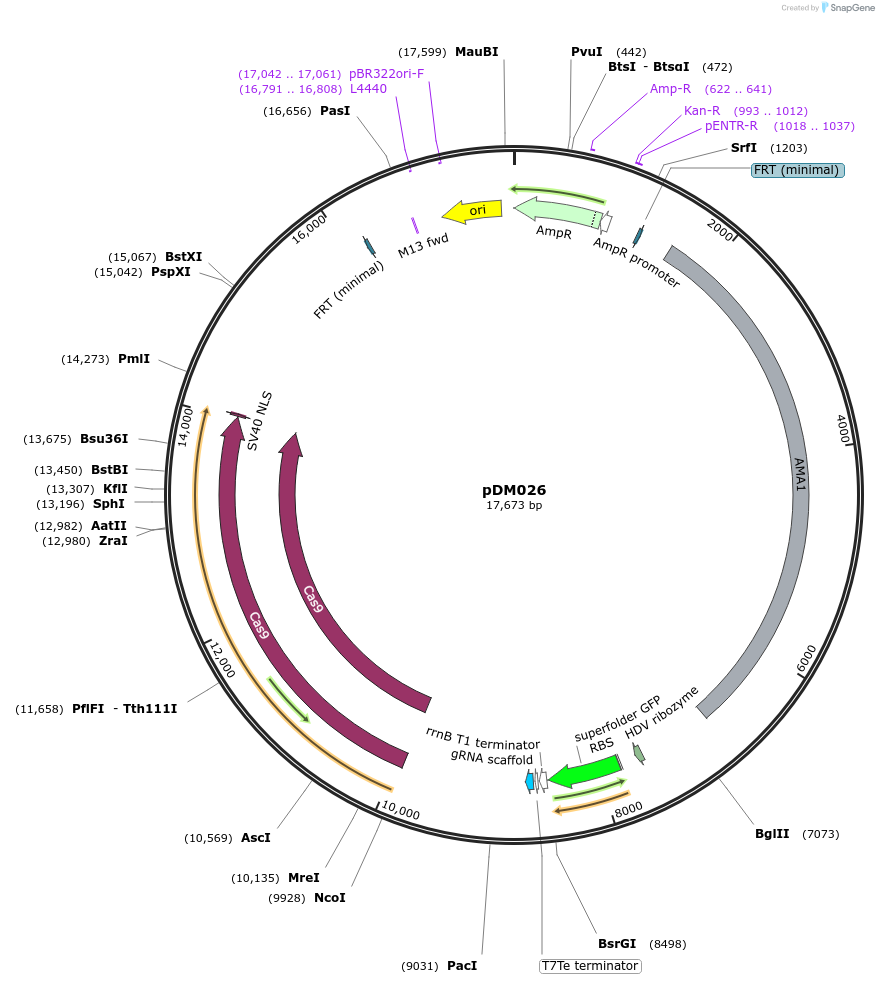
Plasmid#216808PurposeVector for Aspergillus CRISPR-Cas9 genetic engineering with Golden Gate cloning drop-out cassette for protospacer insertion and pyrG selectable marker.DepositorTypeEmpty backboneUseCRISPR; Fungal expressionExpressionBacterialAvailable SinceSept. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
pZA8
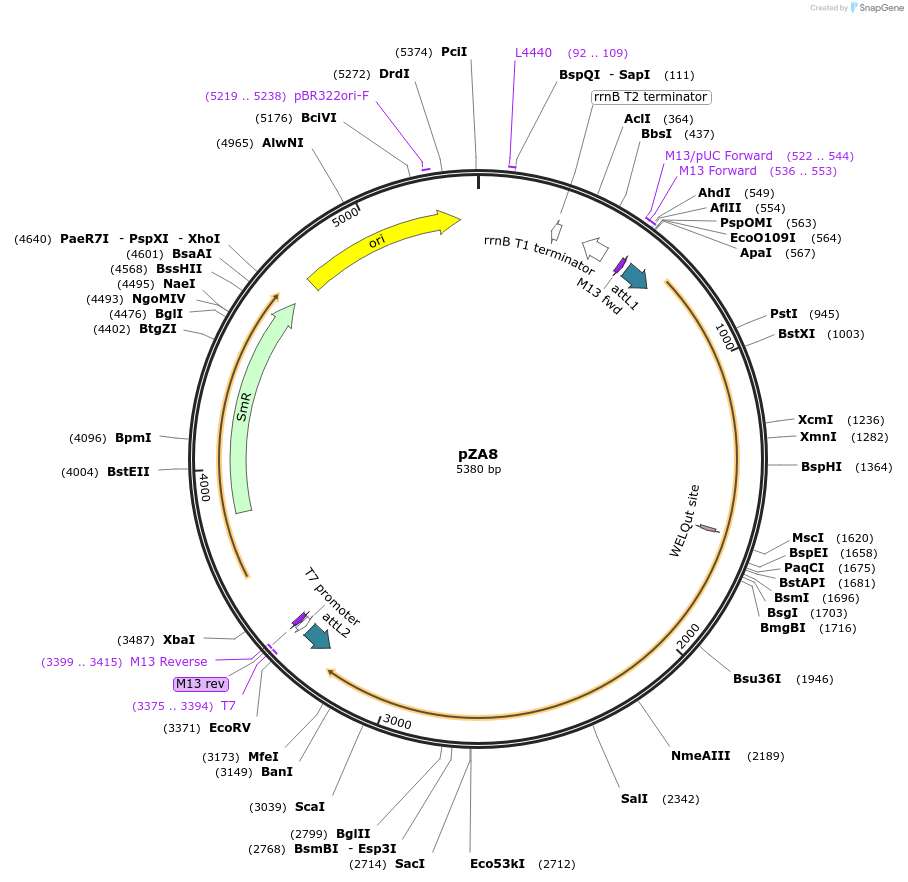
Plasmid#158486PurposeEntry clone for Golden Gate assembly. Golden gate compatible N. benthamiana ZAR1 L17E/D481V with STOP codonDepositorInsertZAR1
UseGolden gate entry vectorMutationL17E, D481V, STOP codonAvailable SinceSept. 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
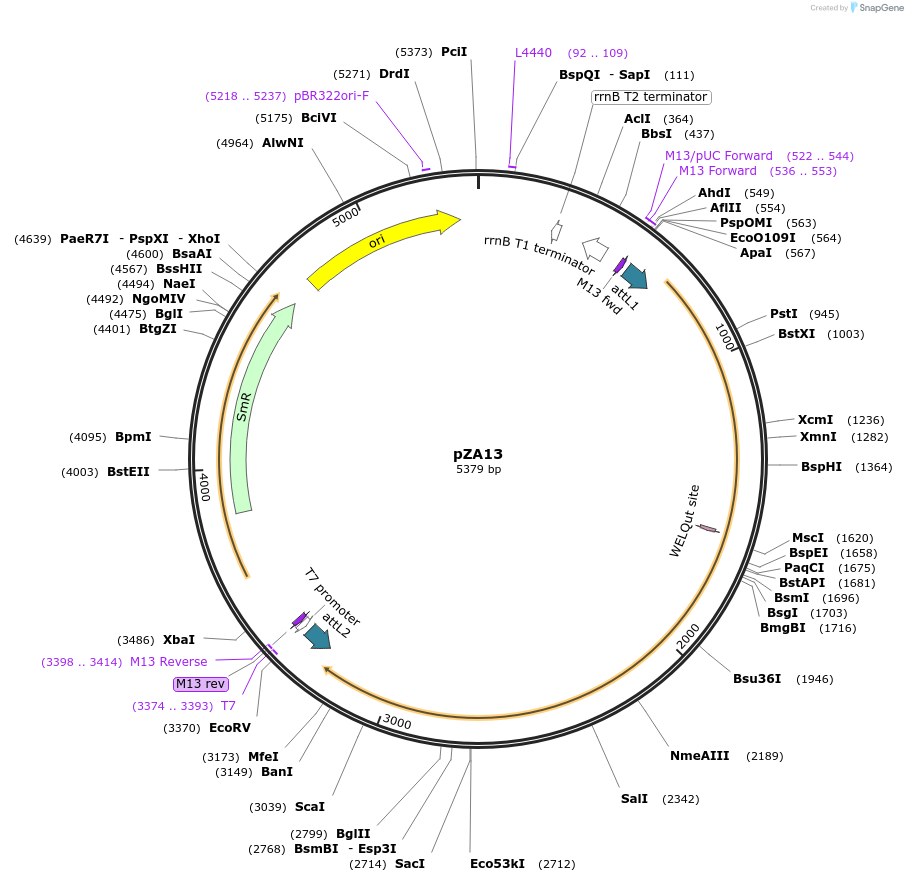
pZA13
Plasmid#158491PurposeEntry clone for Golden Gate assembly. Golden gate compatible N. benthamiana ZAR1 without STOP codonDepositorInsertZAR1
UseGolden gate entry vectorMutationno STOP codonAvailable SinceSept. 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
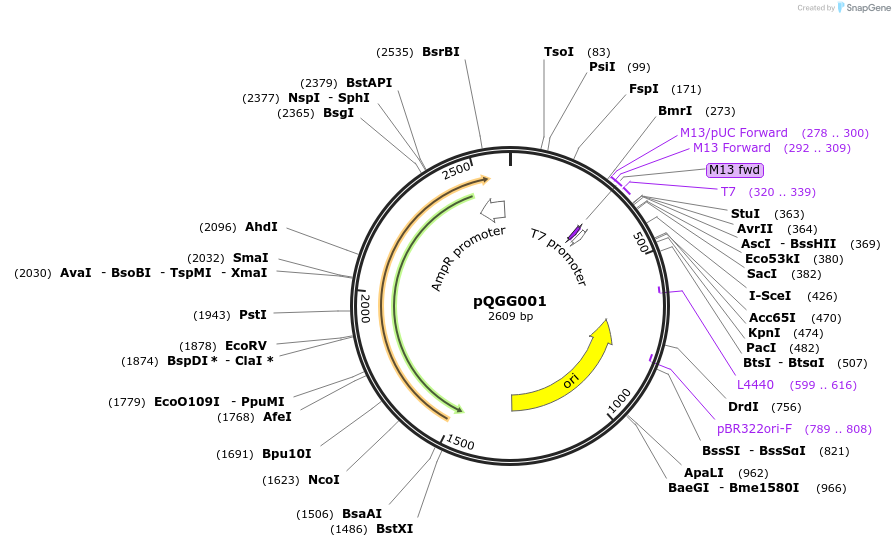
pQGG001
Plasmid#231313PurposeVector part for BEVA; BEVA2.0 Position 4 I-SceI recognition/cut site, SpR.DepositorInsertBEVA2.0 Position 4 I-SceI recognition/cut site, SpR.
ExpressionBacterialAvailable SinceOct. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
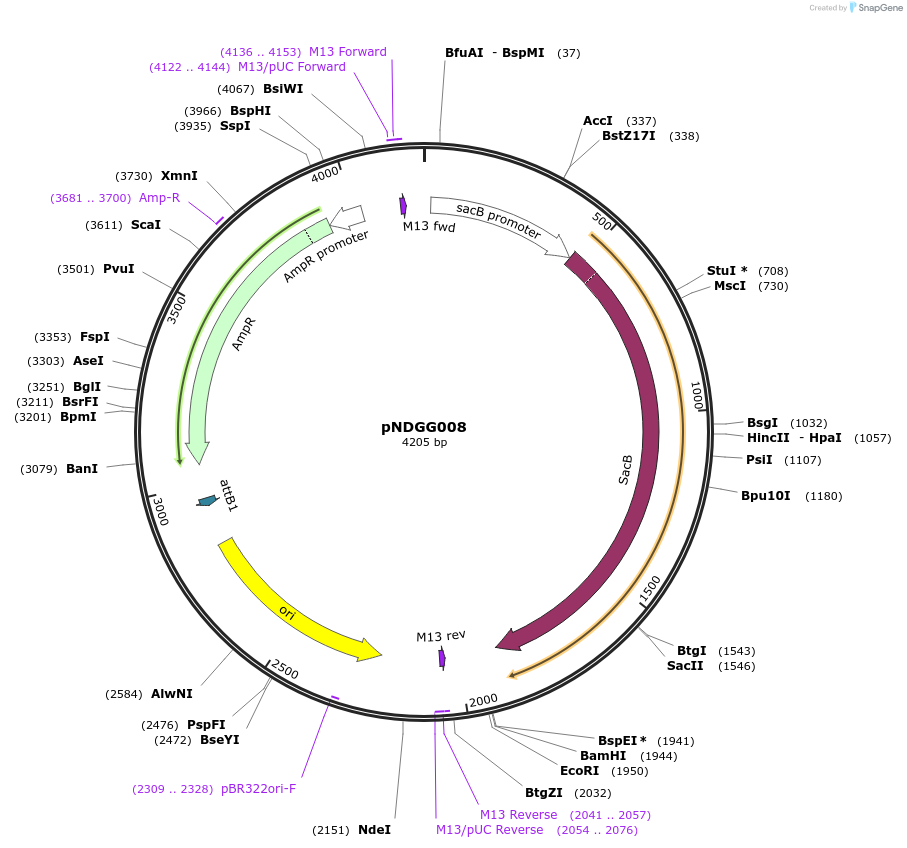
pNDGG008
Plasmid#231312PurposeVector part for BEVA; BEVA2.0 Position 4 sacB sucrose counterselection module, AmpR.DepositorInsertBEVA2.0 Position 4 sacB sucrose counterselection module, AmpR.
ExpressionBacterialAvailable SinceFeb. 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
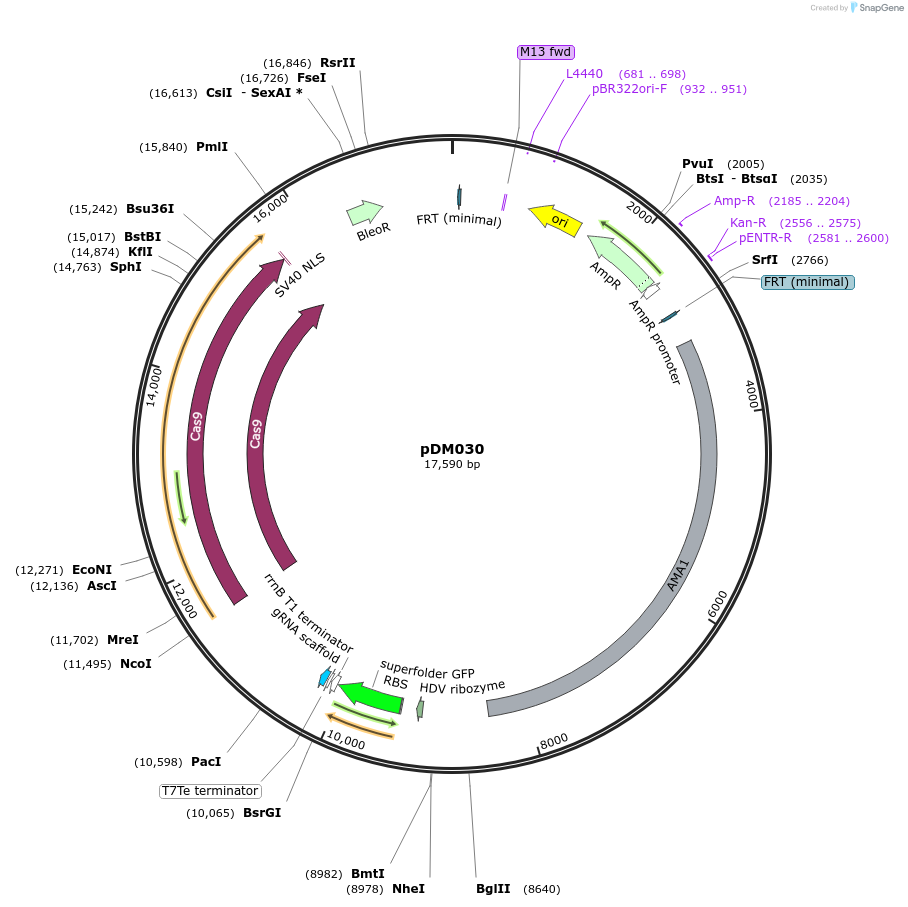
pDM030
Plasmid#216810PurposeVector for Aspergillus CRISPR-Cas9 genetic engineering with Golden Gate cloning drop-out cassette for protospacer insertion and ble selectable marker.DepositorTypeEmpty backboneUseCRISPR; Fungal expressionExpressionBacterialAvailable SinceSept. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
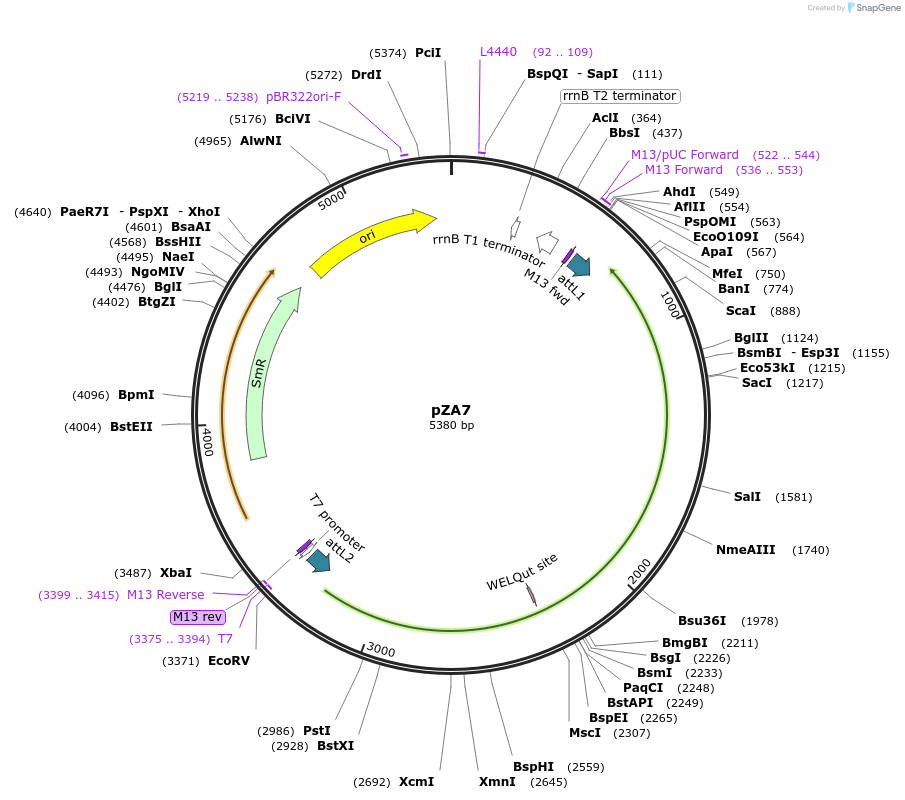
pZA7
Plasmid#158485PurposeEntry clone for Golden Gate assembly. Golden gate compatible N. benthamiana ZAR1 L17E with STOP codonDepositorInsertZAR1
UseGolden gate entry vectorMutationL17E, STOP codonAvailable SinceSept. 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
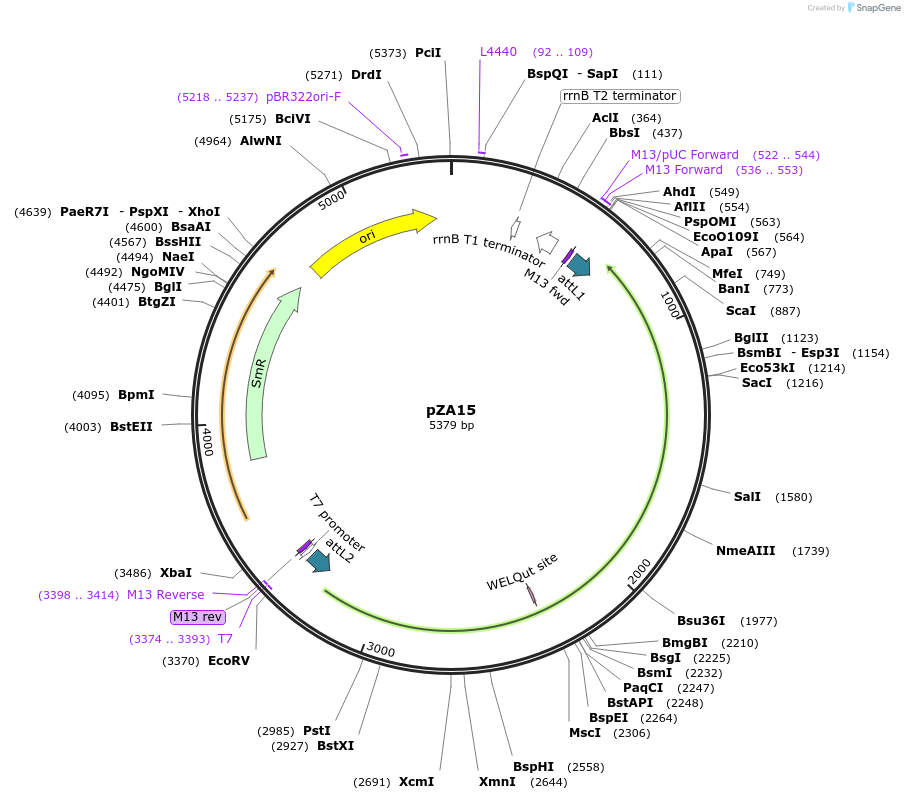
pZA15
Plasmid#158493PurposeEntry clone for Golden Gate assembly. Golden gate compatible N. benthamiana ZAR1 L17E without STOP codonDepositorInsertZAR1
UseGolden gate entry vectorMutationL17E, no STOP codonAvailable SinceSept. 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
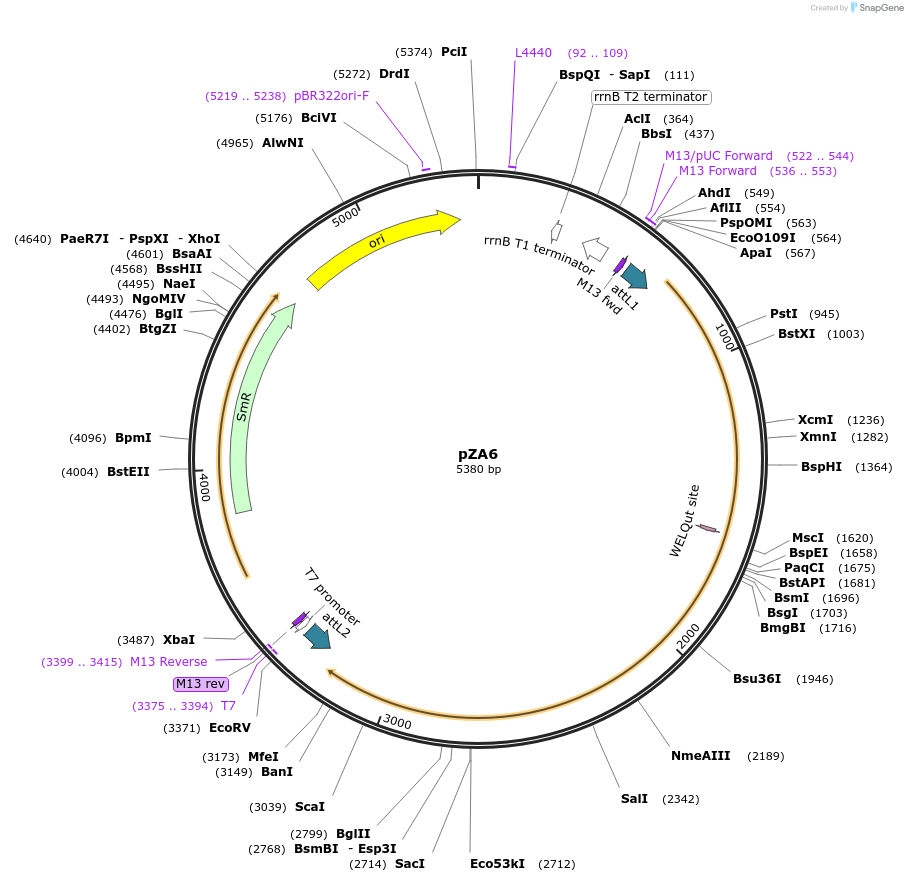
pZA6
Plasmid#158484PurposeEntry clone for Golden Gate assembly. Golden gate compatible N. benthamiana ZAR1 D481V with STOP codonDepositorInsertZAR1
UseGolden gate entry vectorMutationD481V, STOP codonAvailable SinceAug. 27, 2020AvailabilityAcademic Institutions and Nonprofits only -
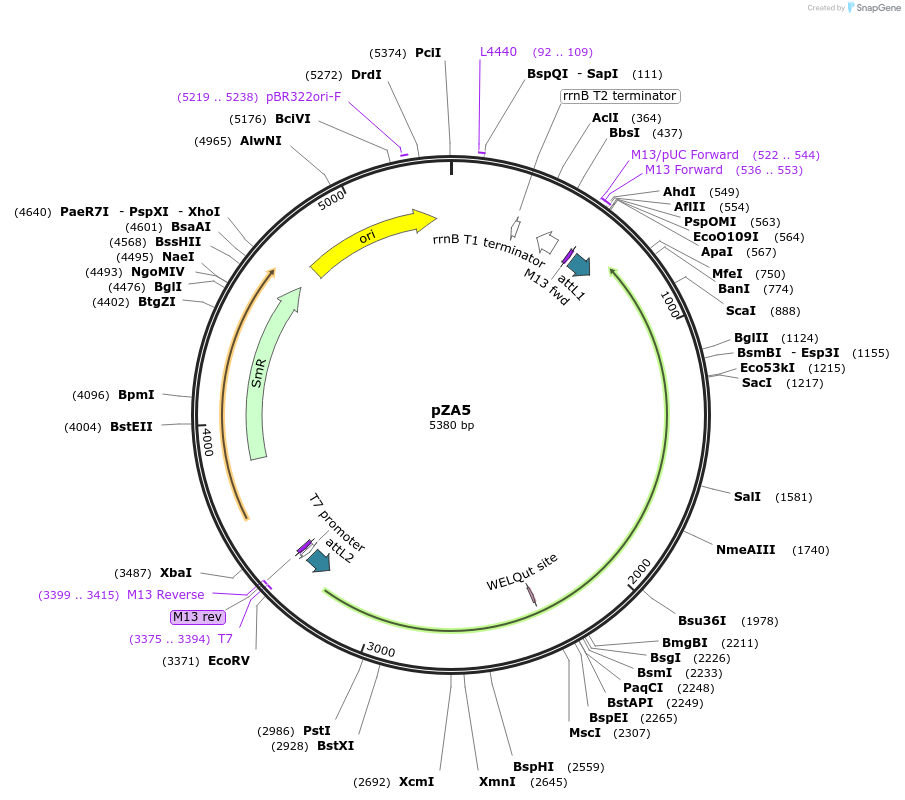
pZA5
Plasmid#158483PurposeEntry clone for Golden Gate assembly. Golden gate compatible N. benthamiana ZAR1 with STOP codonDepositorInsertZAR1
UseGolden gate entry vectorMutationSTOP codonAvailable SinceAug. 27, 2020AvailabilityAcademic Institutions and Nonprofits only -
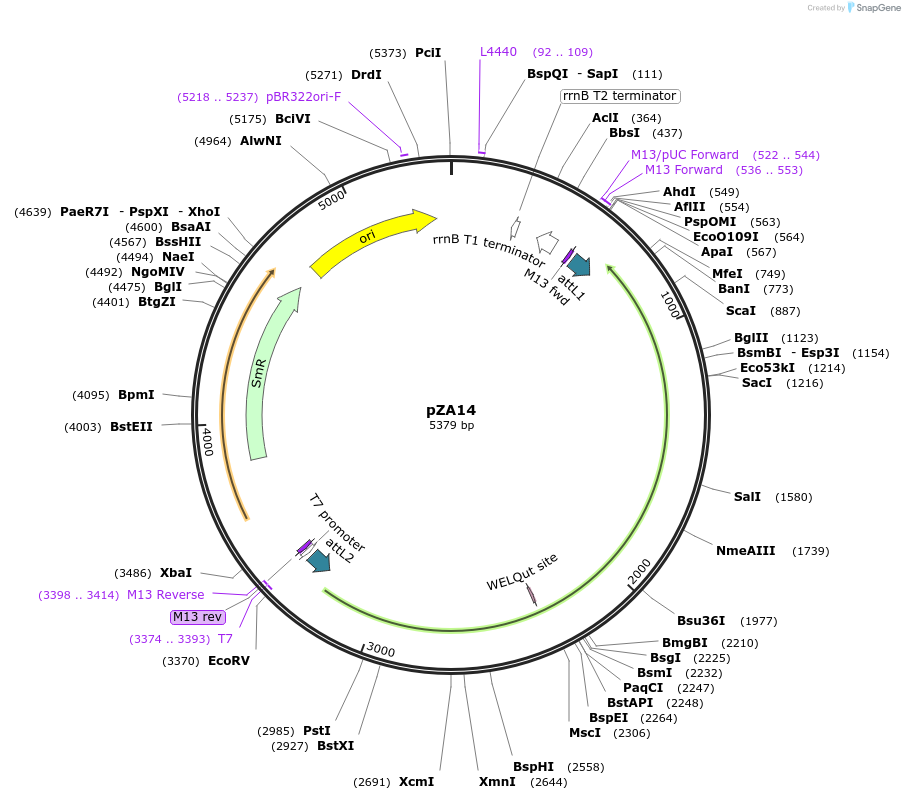
pZA14
Plasmid#158492PurposeEntry clone for Golden Gate assembly. Golden gate compatible N. benthamiana ZAR1 D481V without STOP codonDepositorInsertZAR1
UseGolden gate entry vectorMutationD481V, no STOP codonAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
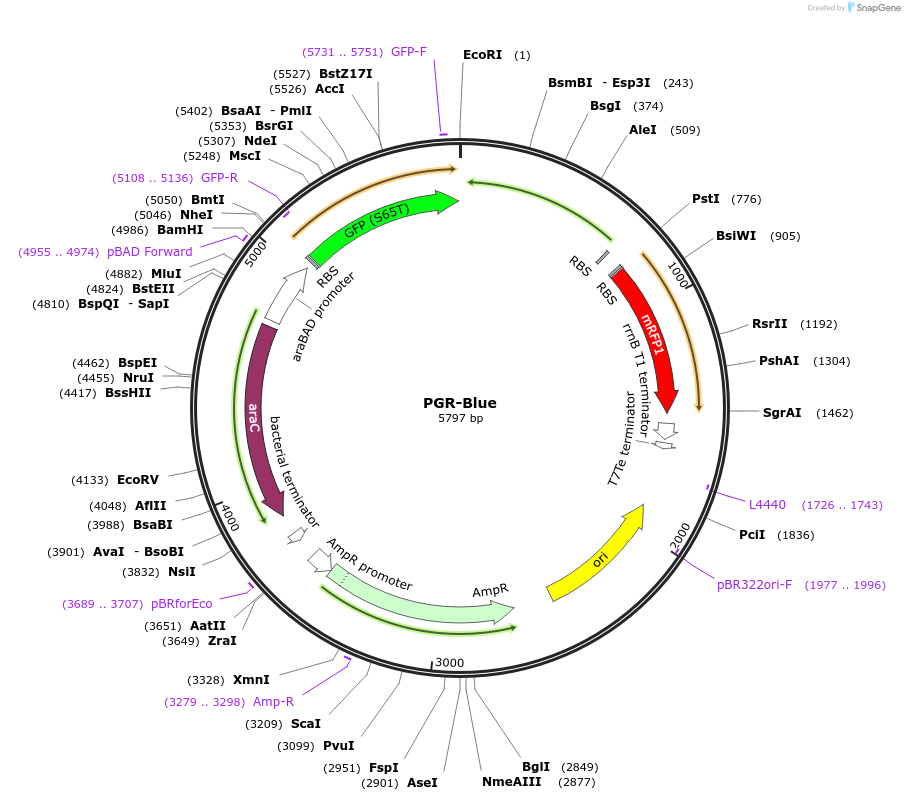
PGR-Blue
Plasmid#68374PurposeTerminator quantification vector utilizing Golden Gate Assembly and amilCP. Positive colonies are green and false positives are blue. Terminators are placed between GFP and RFP - induced by arabinose.DepositorTypeEmpty backboneUseSynthetic Biology; Golden gate assemblyExpressionBacterialPromoterpBADAvailable SinceOct. 29, 2015AvailabilityAcademic Institutions and Nonprofits only -
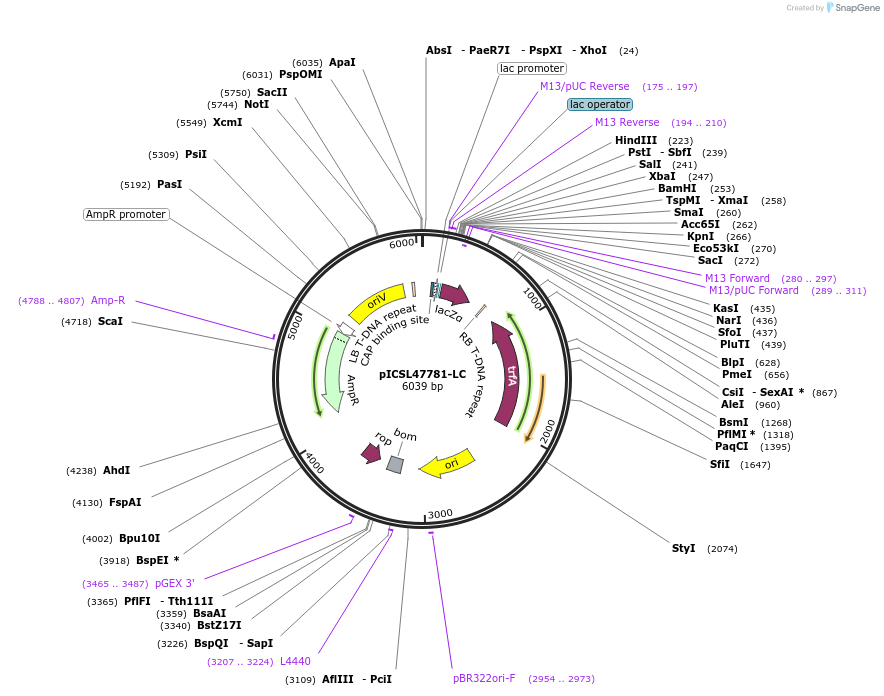
pICSL47781-LC
Plasmid#245717PurposeLevel 1 Position 6 FORWARD acceptor Low - copy (with pBR322 origin of replication) GGAG - GCTTDepositorTypeEmpty backboneUseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceJan. 28, 2026AvailabilityAcademic Institutions and Nonprofits only -
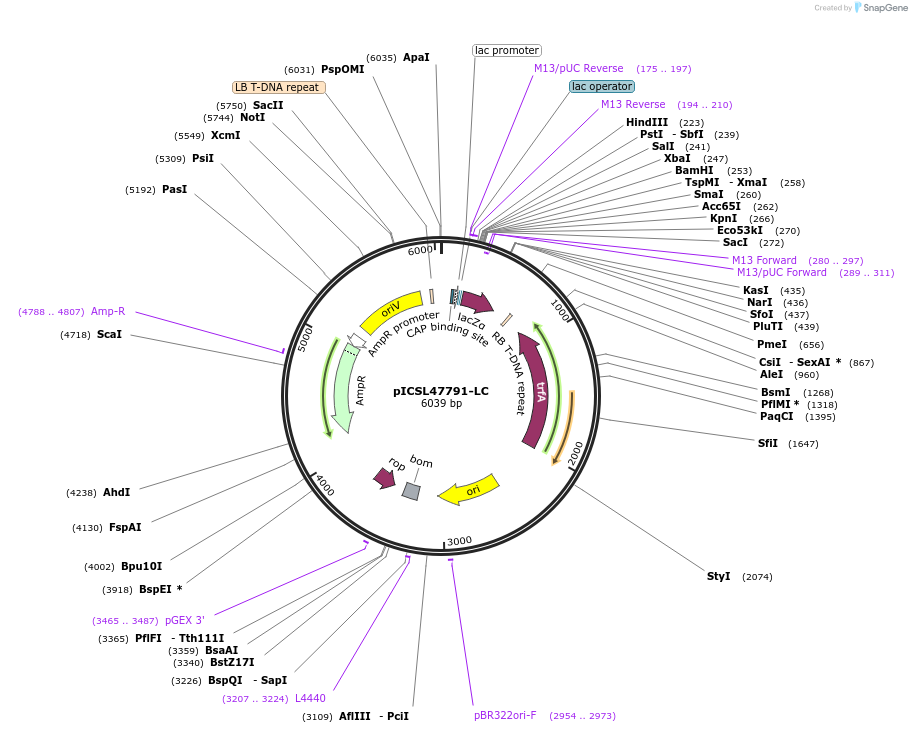
pICSL47791-LC
Plasmid#245718PurposeLevel 1 Position 7 FORWARD acceptor Low - copy (with pBR322 origin of replication) GGAG - GCTTDepositorTypeEmpty backboneUseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceJan. 2, 2026AvailabilityAcademic Institutions and Nonprofits only -
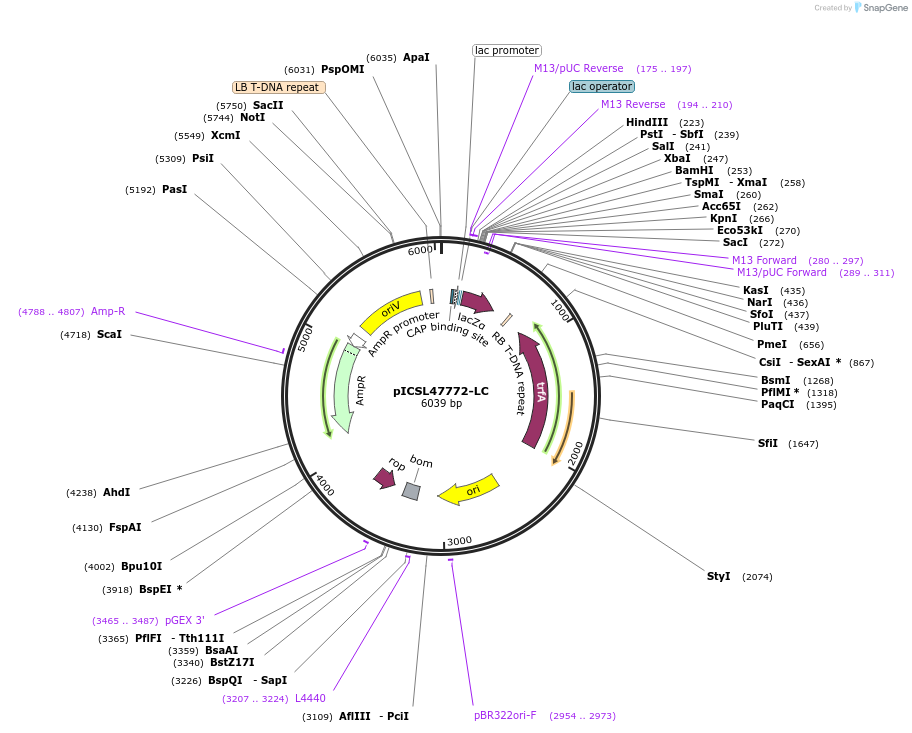
pICSL47772-LC
Plasmid#245716PurposeLevel 1 Position 5 FORWARD acceptor Low - copy (with pBR322 origin of replication) GGAG - GCTTDepositorTypeEmpty backboneUseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceJan. 2, 2026AvailabilityAcademic Institutions and Nonprofits only -
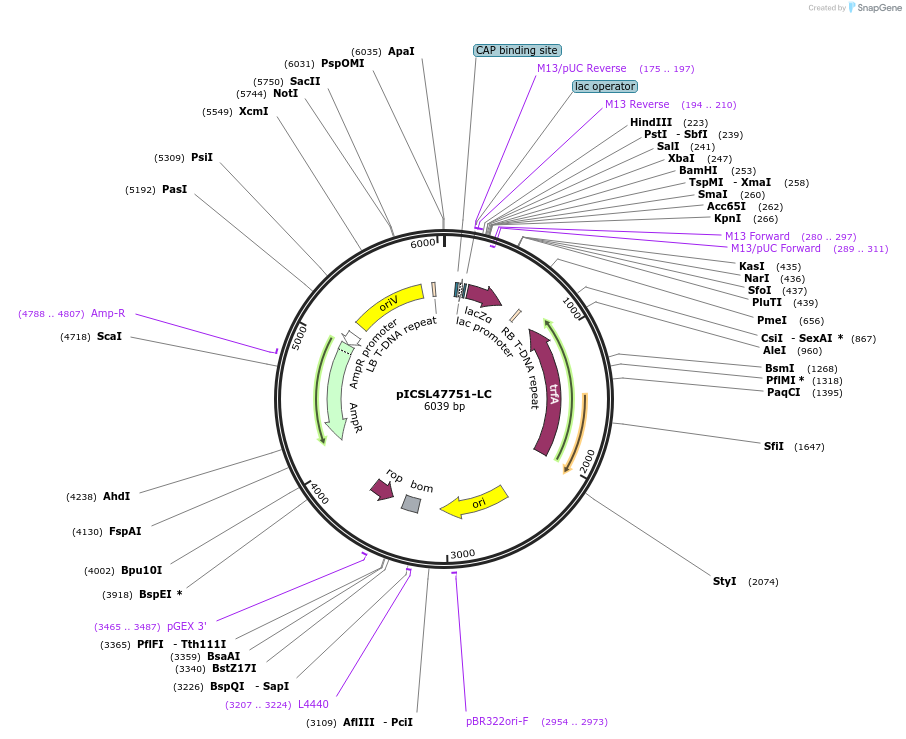
pICSL47751-LC
Plasmid#245714PurposeLevel 1 Position 3 FORWARD acceptor Low - copy (with pBR322 origin of replication) GGAG - GCTTDepositorTypeEmpty backboneUseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceJan. 2, 2026AvailabilityAcademic Institutions and Nonprofits only -
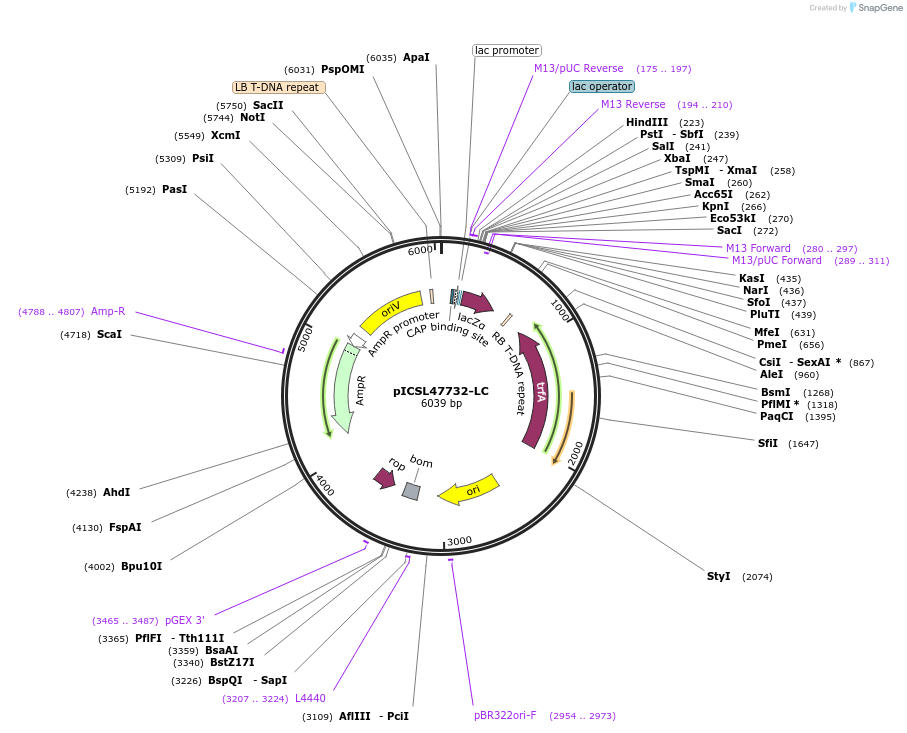
pICSL47732-LC
Plasmid#245712PurposeLevel 1 Position 1 FORWARD acceptor Low - copy (with pBR322 origin of replication) GGAG - GCTTDepositorTypeEmpty backboneUseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceJan. 2, 2026AvailabilityAcademic Institutions and Nonprofits only -
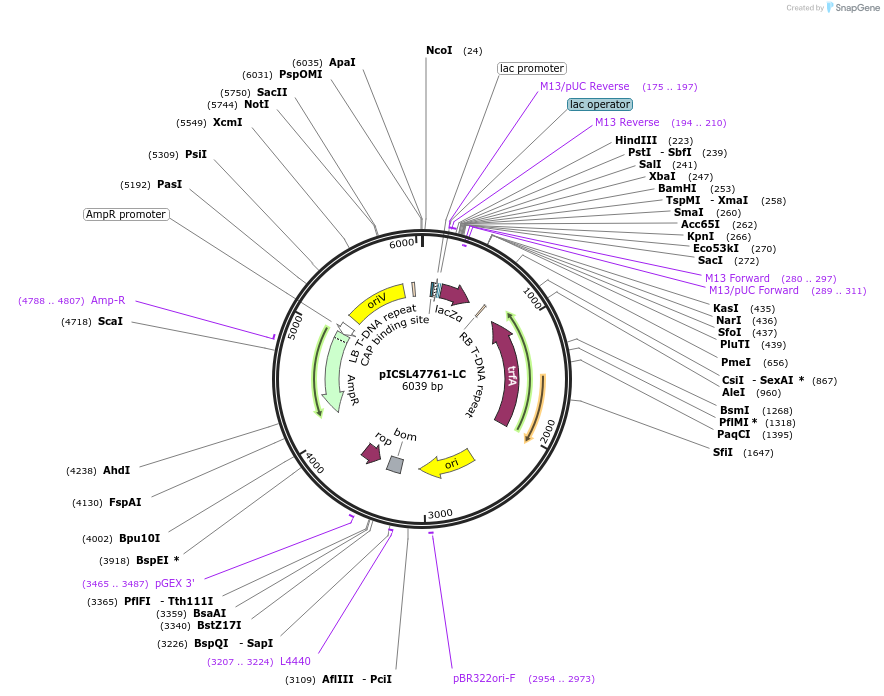
pICSL47761-LC
Plasmid#245715PurposeLevel 1 Position 4 FORWARD acceptor Low - copy (with pBR322 origin of replication) GGAG - GCTTDepositorTypeEmpty backboneUseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceJan. 2, 2026AvailabilityAcademic Institutions and Nonprofits only