We narrowed to 40,628 results for: ANT
-
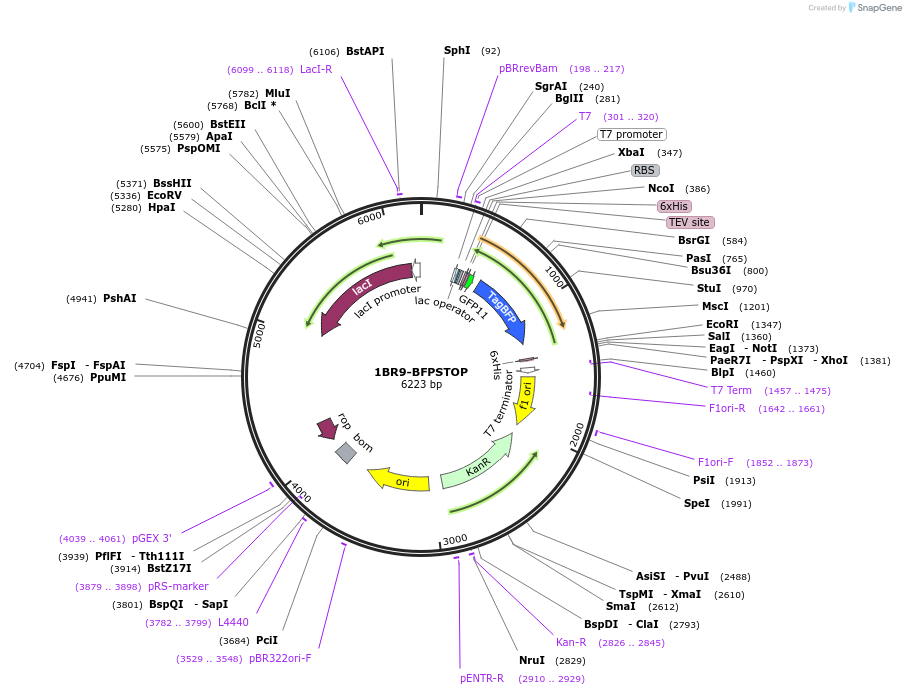
Plasmid#202055PurposeE. coli expression vector for N-ter GFP11 R9 tagged BFPDepositorInsertBFP
Tags6xHis and GFP11ExpressionBacterialPromoterT7 PromoterAvailable SinceAug. 29, 2023AvailabilityAcademic Institutions and Nonprofits only -
pDEST17-HO1(51-231)
Plasmid#203486PurposeFor recombinant protein production in Escherichia coli. His-tagged Arabidopsis thaliana HEME OXYGENASE 1 (HO1) without the plastid transit peptide under the control of the T7 promoter.DepositorInsertArabidopsis thaliana HEME OXYGENASE1 (51 aa – 231 aa)
ExpressionBacterial and PlantAvailable SinceAug. 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
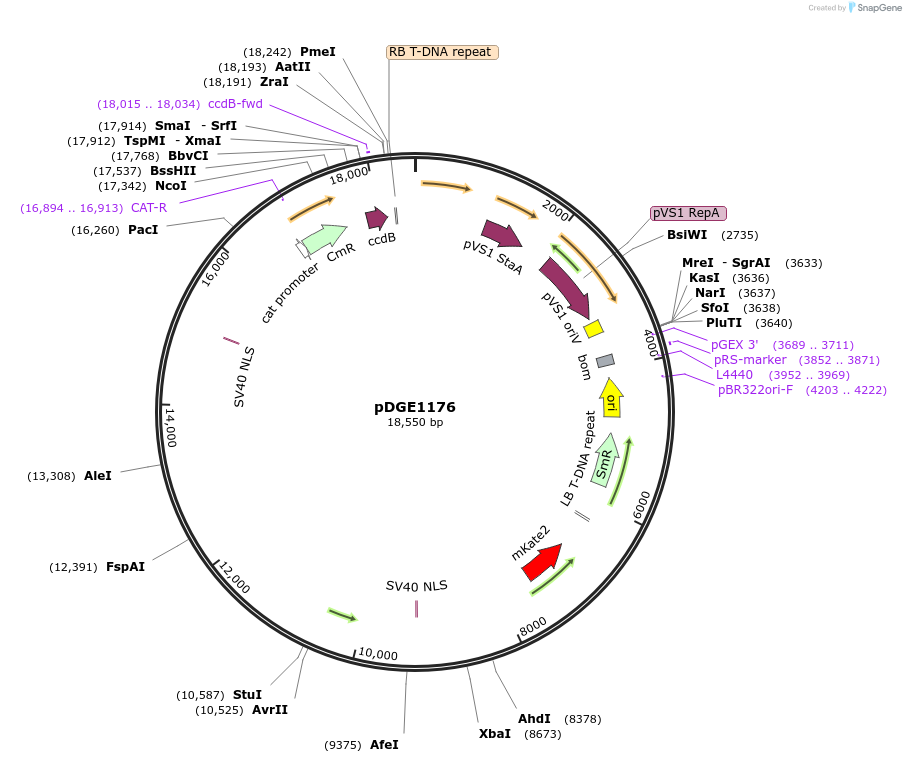
pDGE1176
Plasmid#202774PurposeCRISPR-KO in Arabidopsis thalianaDepositorInsertsFAST marker
pRPS5a:zCas9_tNbEU
ccdB cassette (BsaI-excisable)
UseCRISPRExpressionPlantAvailable SinceAug. 9, 2023AvailabilityAcademic Institutions and Nonprofits only -
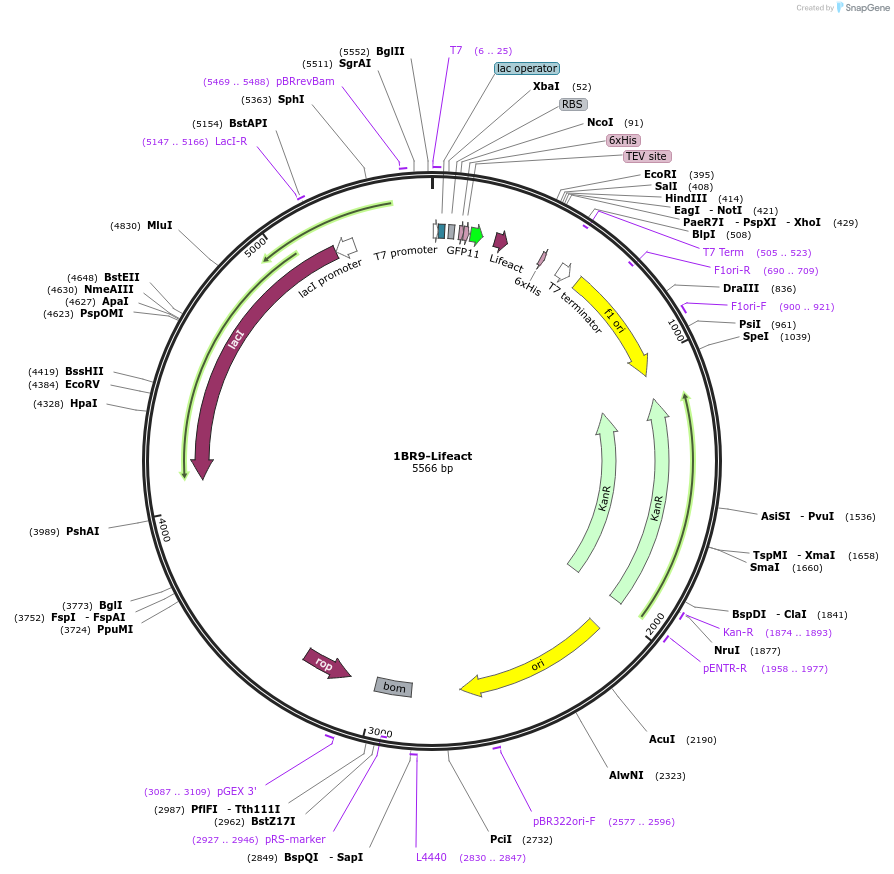
1BR9-Lifeact
Plasmid#193863PurposeE. coli expression vector for N-ter GFP11 and C-ter R9 tagged lifeact peptide.DepositorInsertLifeact
Tags6xHis, GFP11, and R9ExpressionBacterialPromoterT7 PromoterAvailable SinceAug. 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
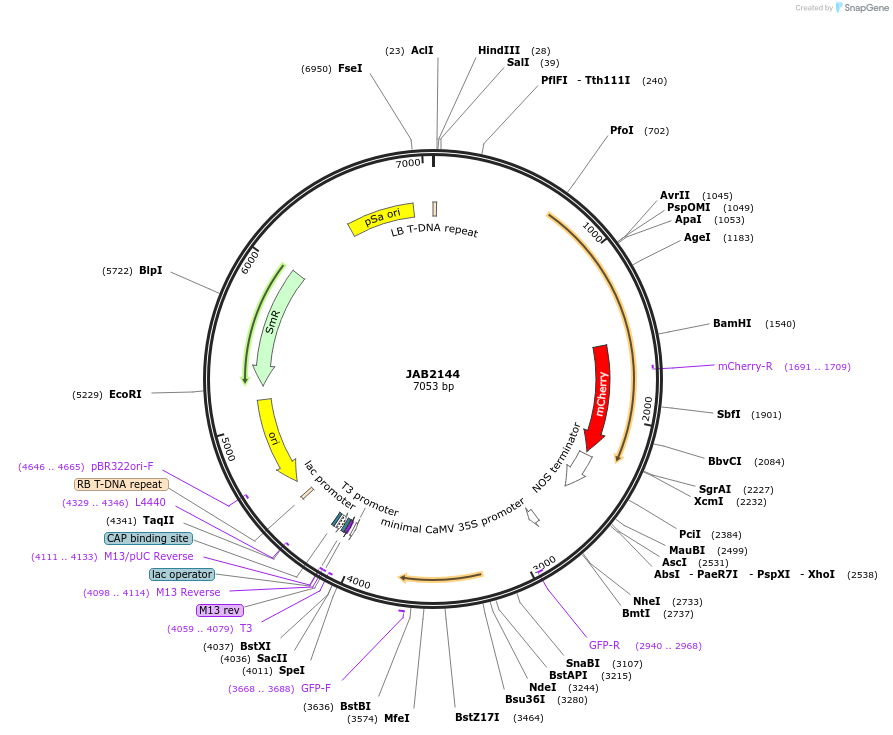
JAB2144
Plasmid#190344PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_6XAmtRO-2XPhlFO::GFPint-ADHt)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_6XAmtRO-2XPhlFO::GFPint-ADHt
ExpressionPlantAvailable SinceJune 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
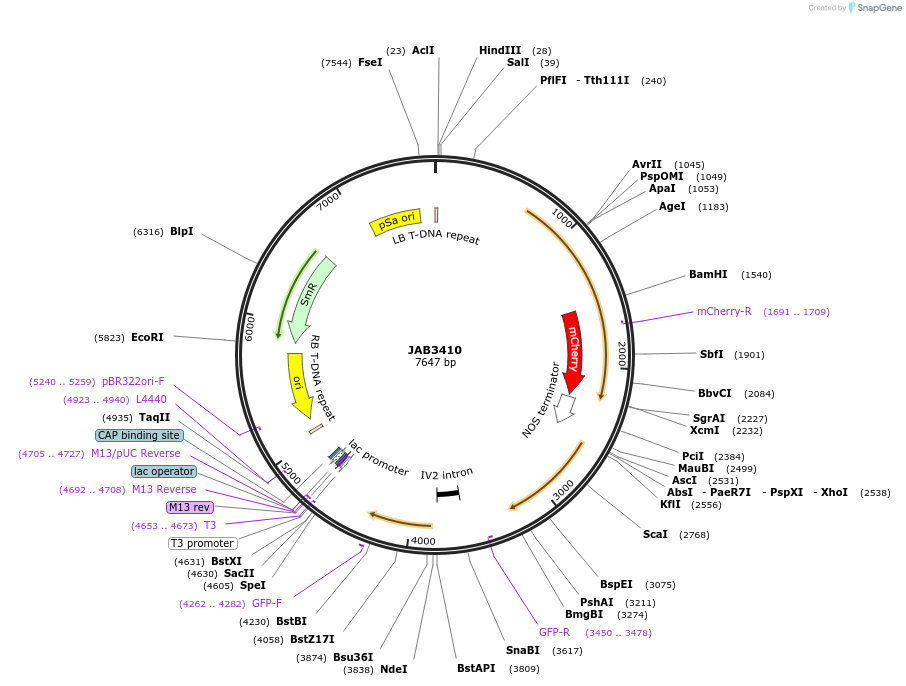
JAB3410
Plasmid#190410PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_proAA::GFPint-ADHter)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_proAA::GFPint-ADHter
ExpressionPlantAvailable SinceMay 23, 2023AvailabilityAcademic Institutions and Nonprofits only -
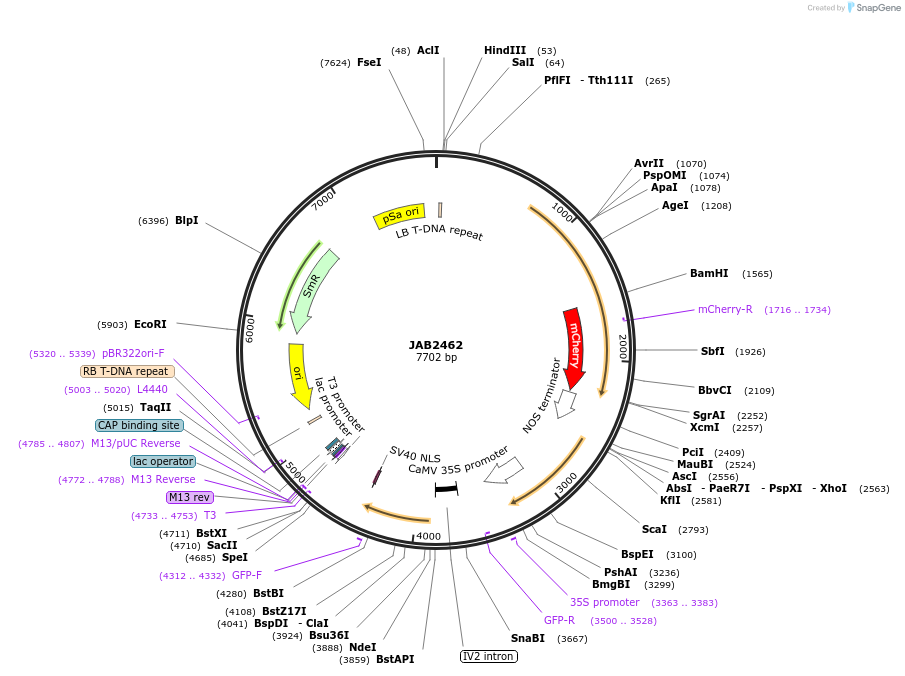
JAB2462
Plasmid#190369PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_pro35S-LmrAO::GFPint-NLS-ADHt)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_pro35S-LmrAO::GFPint-NLS-ADHt
ExpressionPlantAvailable SinceMay 23, 2023AvailabilityAcademic Institutions and Nonprofits only -
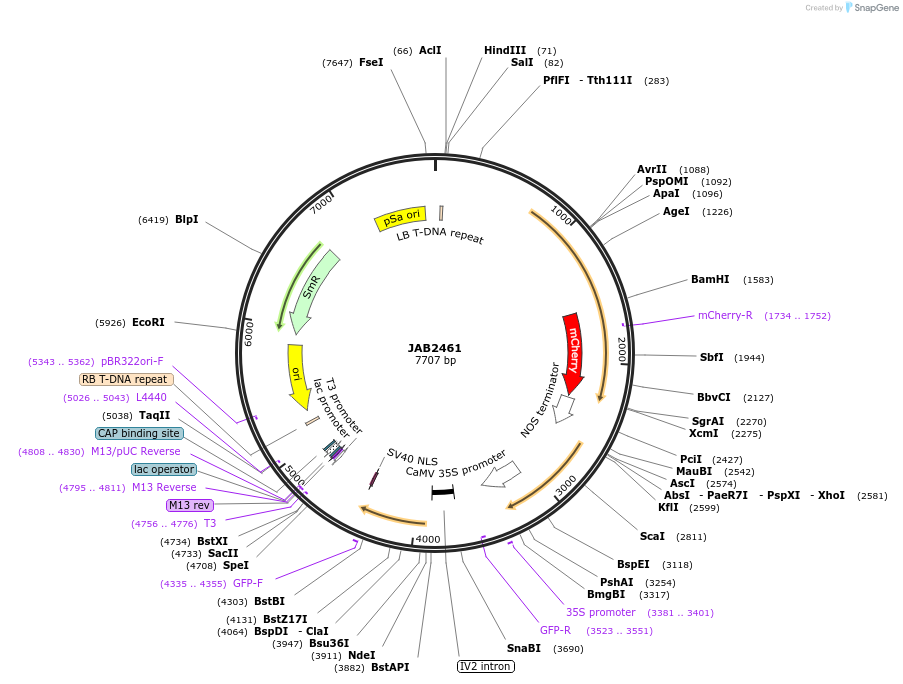
JAB2461
Plasmid#190368PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_pro35S-IcaRO::GFPint-NLS-ADHt)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_pro35S-IcaRO::GFPint-NLS-ADHt
ExpressionPlantAvailable SinceMay 23, 2023AvailabilityAcademic Institutions and Nonprofits only -
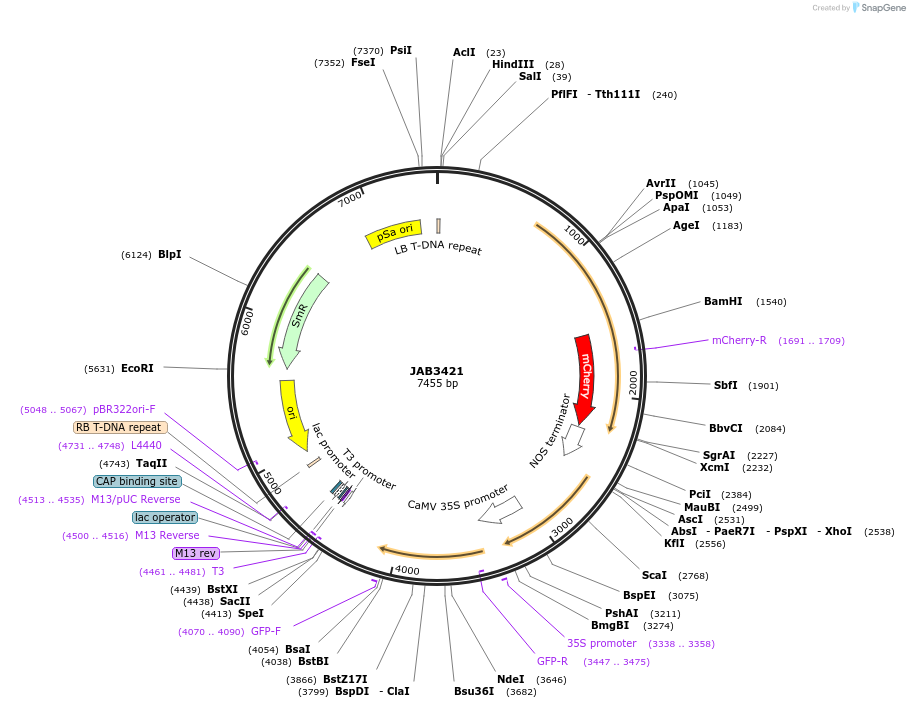
JAB3421
Plasmid#190411PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_pro35S::GFP-ADHt)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_pro35S::GFP-ADHt
ExpressionPlantAvailable SinceMay 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
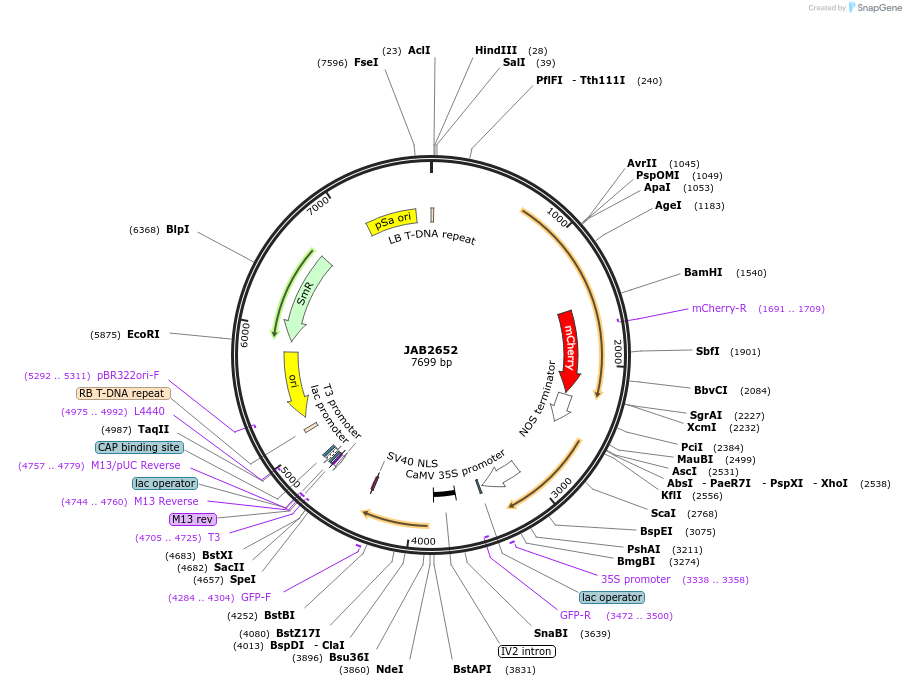
JAB2652
Plasmid#190392PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_pro35S-LacIO::GFPint-NLS-ADHt)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_pro35S-LacIO::GFPint-NLS-ADHt
ExpressionPlantAvailable SinceMay 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
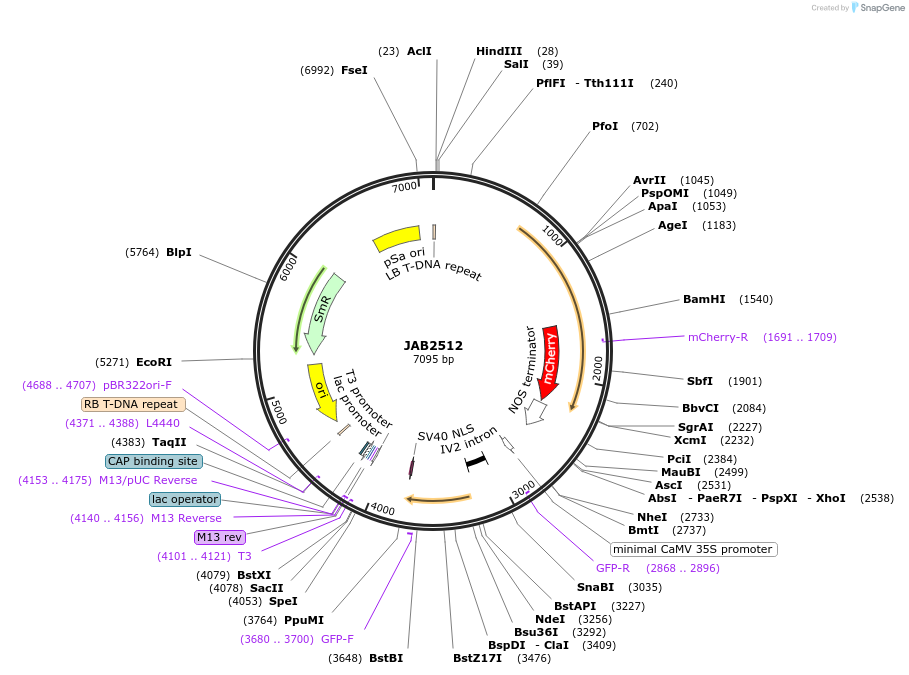
JAB2512
Plasmid#190381PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_6XBetIO-min35S::GFPint-ADHt)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_6XBetIO-min35S::GFPint-ADHt
ExpressionPlantAvailable SinceMay 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
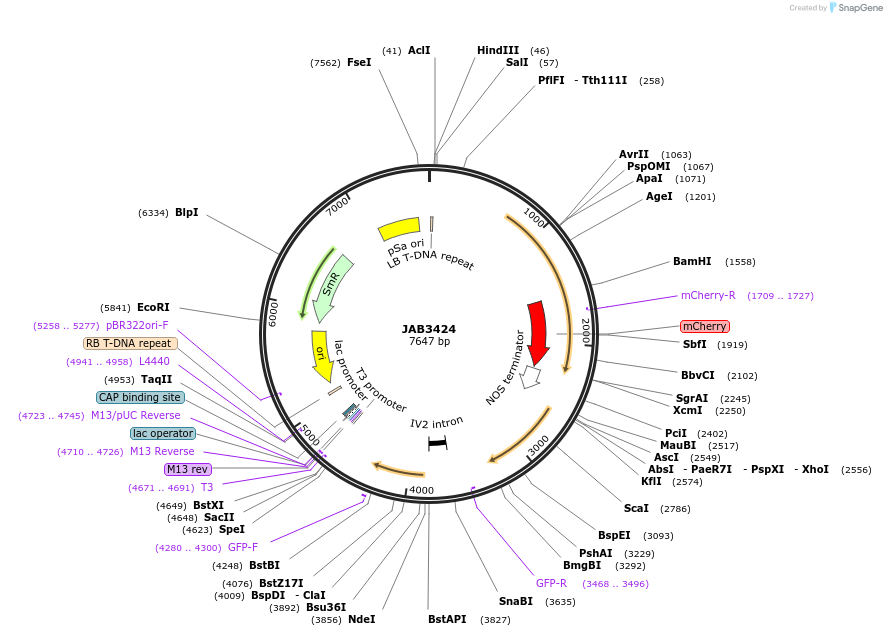
JAB3424
Plasmid#190414PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_proT::GFP-ADHt)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_proT::GFP-ADHt
ExpressionPlantAvailable SinceMay 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
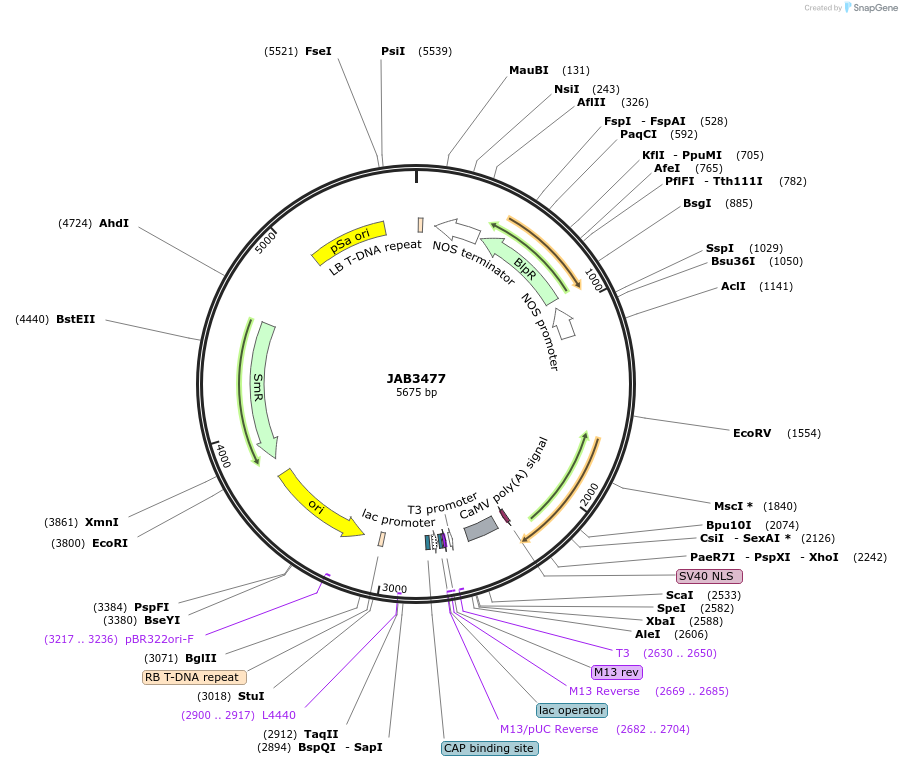
JAB3477
Plasmid#190416PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (6XPhlFO-3XAmtRO::TetR-NLS-19St)DepositorInsert6XPhlFO-3XAmtRO::TetR-NLS-19St
ExpressionPlantAvailable SinceMay 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
JAB3423
Plasmid#190413PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_proUBQ10::GFP-ADHt)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_proUBQ10::GFP-ADHt
ExpressionPlantAvailable SinceMay 19, 2023AvailabilityAcademic Institutions and Nonprofits only -
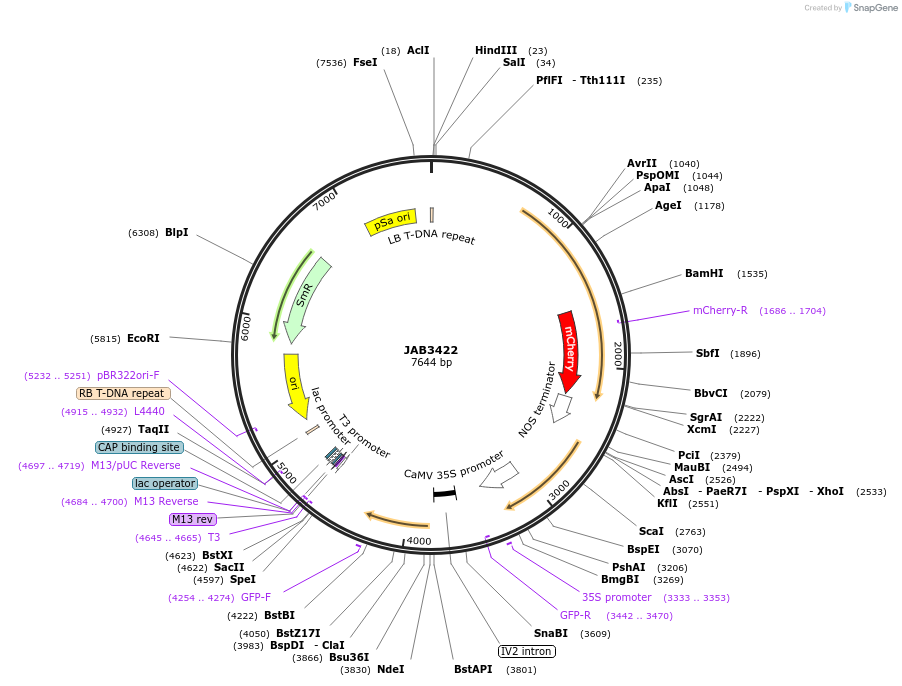
JAB3422
Plasmid#190412PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_pro35S::GFPint-ADHt)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_pro35S::GFPint-ADHt
ExpressionPlantAvailable SinceMay 19, 2023AvailabilityAcademic Institutions and Nonprofits only -
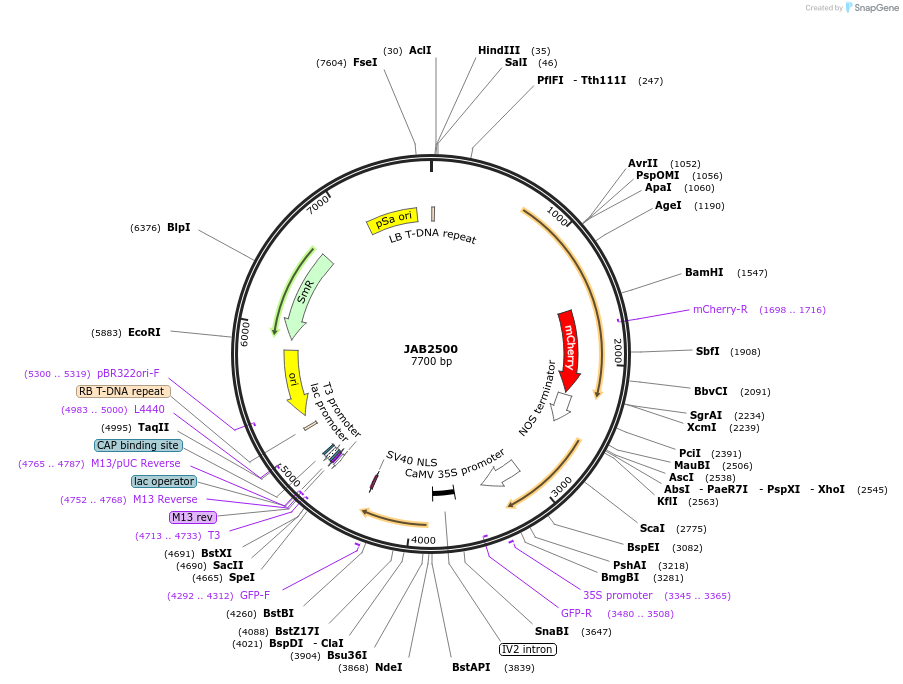
JAB2500
Plasmid#190379PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_pro35S-BetIO::GFPint-NLS_ADHt)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_pro35S-BetIO::GFPint-NLS_ADHt
ExpressionPlantAvailable SinceMay 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
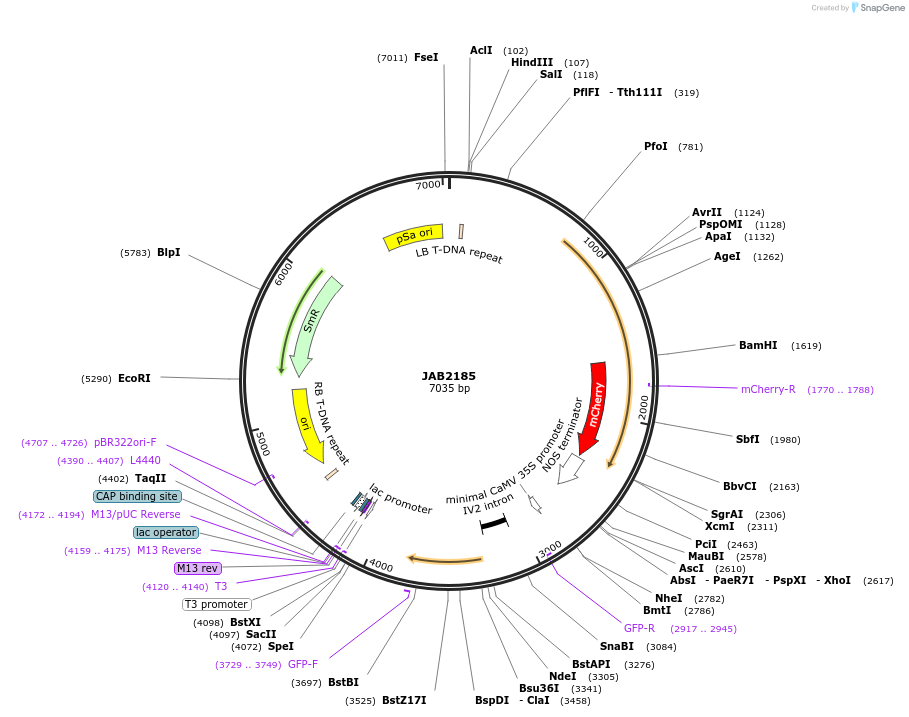
JAB2185
Plasmid#190354PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_6XMcbRO-min35S::GFPint-ADHt)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_6XMcbRO-min35S::GFPint-ADHt
ExpressionPlantAvailable SinceMay 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
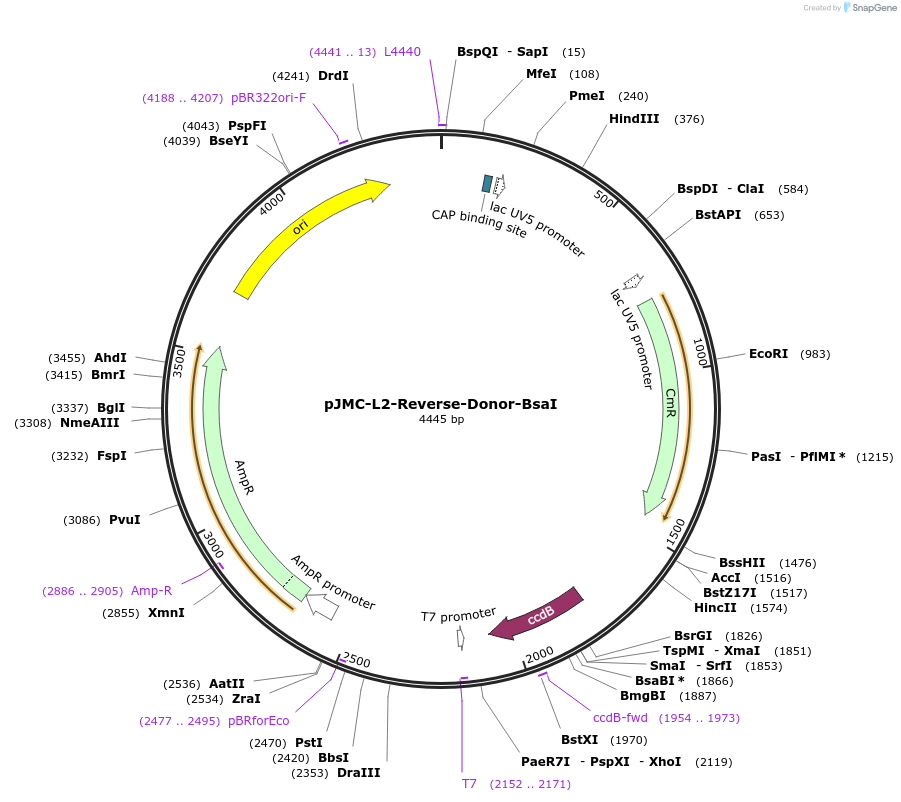
pJMC-L2-Reverse-Donor-BsaI
Plasmid#197843PurposeDonor vector to restore L2 destination via BsaI (reverse orientation)DepositorInsertNone
UseUnspecifiedAvailable SinceMay 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
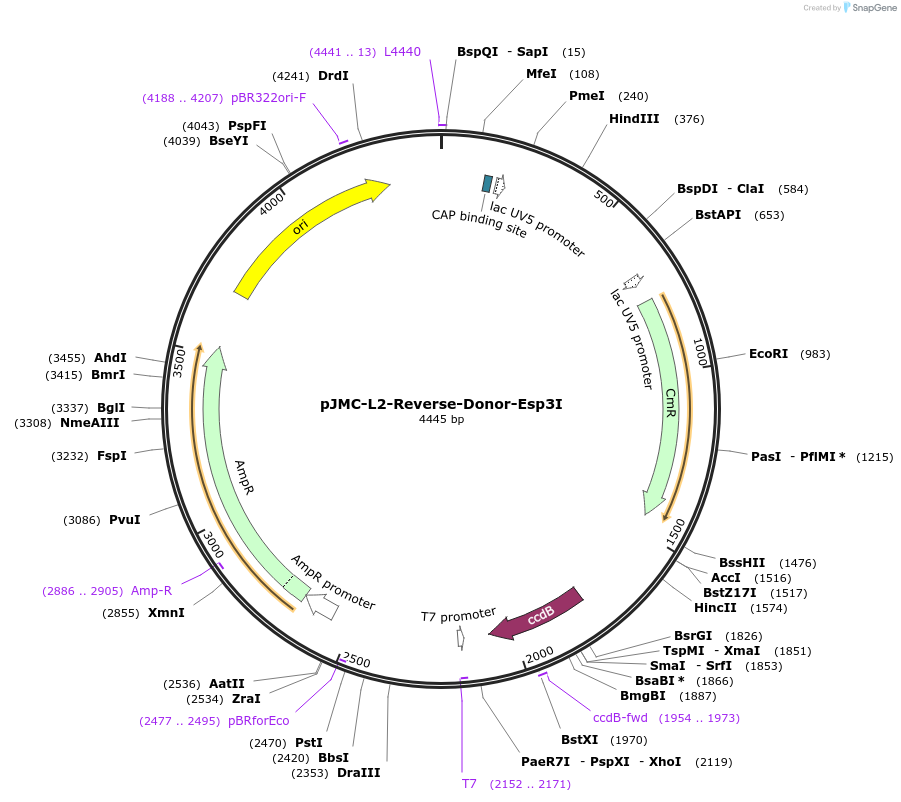
pJMC-L2-Reverse-Donor-Esp3I
Plasmid#197844PurposeDonor vector to restore L2 destination via Esp3I (reverse orientation)DepositorInsertNone
UseUnspecifiedAvailable SinceMay 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
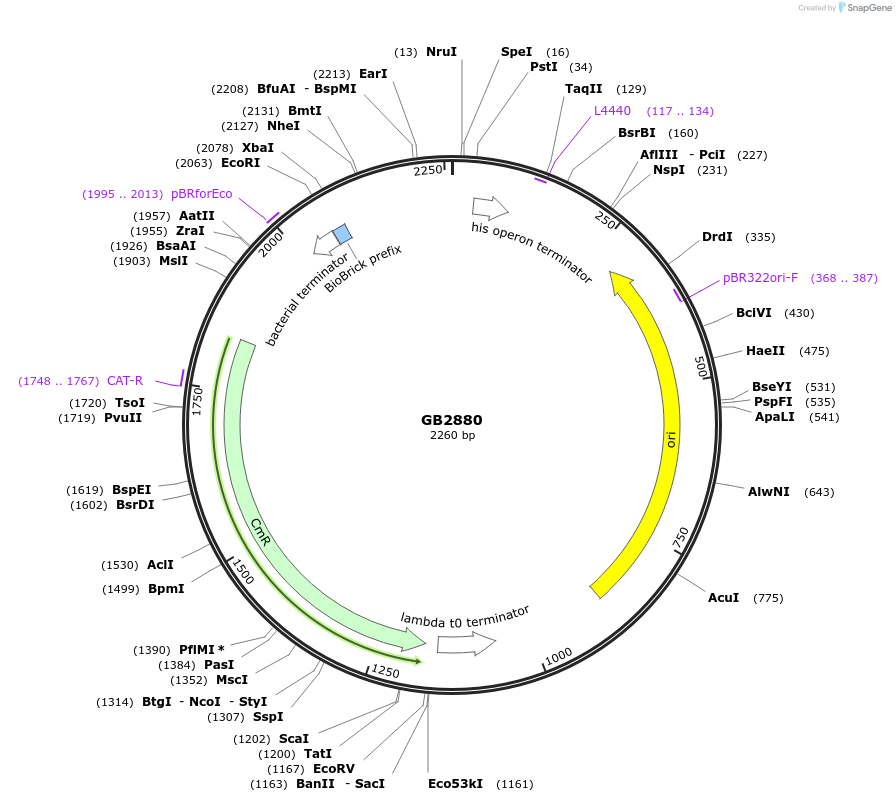
GB2880
Plasmid#193117PurposeA2 Proximal promoter sequence consisting of the target sequences for the gRNA1 (GB1838) and gRNA2 (GB1839) flanked by random sequences.DepositorInsertGB_SynP (A2) G1aG2b.6
UseSynthetic BiologyAvailable SinceMay 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
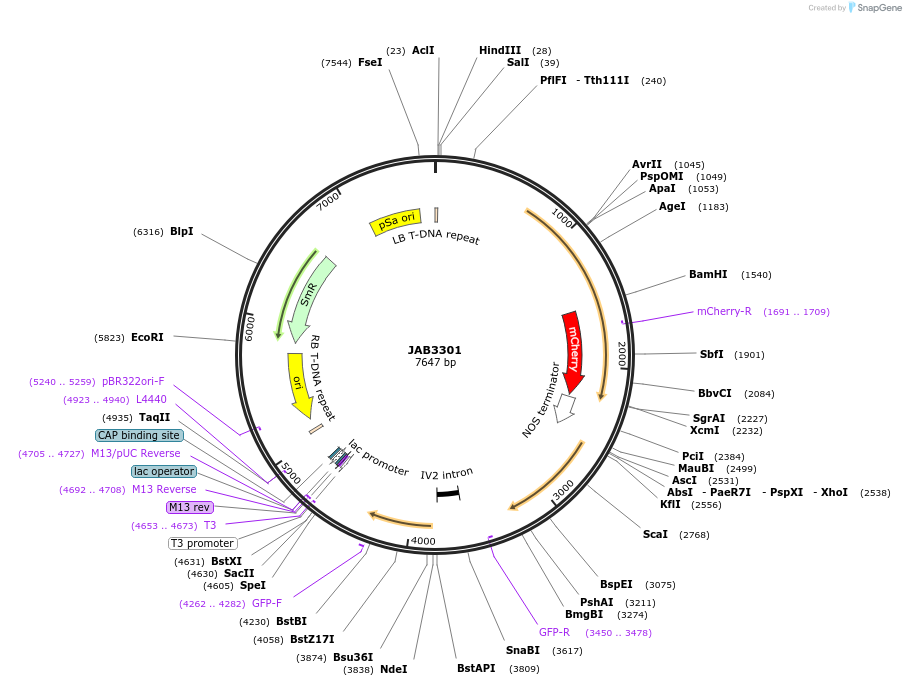
JAB3301
Plasmid#190405PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_pro35S-AmtRO(internal)::GFPint-ADHter)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_pro35S-AmtRO(internal)::GFPint-ADHter
ExpressionPlantAvailable SinceApril 26, 2023AvailabilityAcademic Institutions and Nonprofits only -
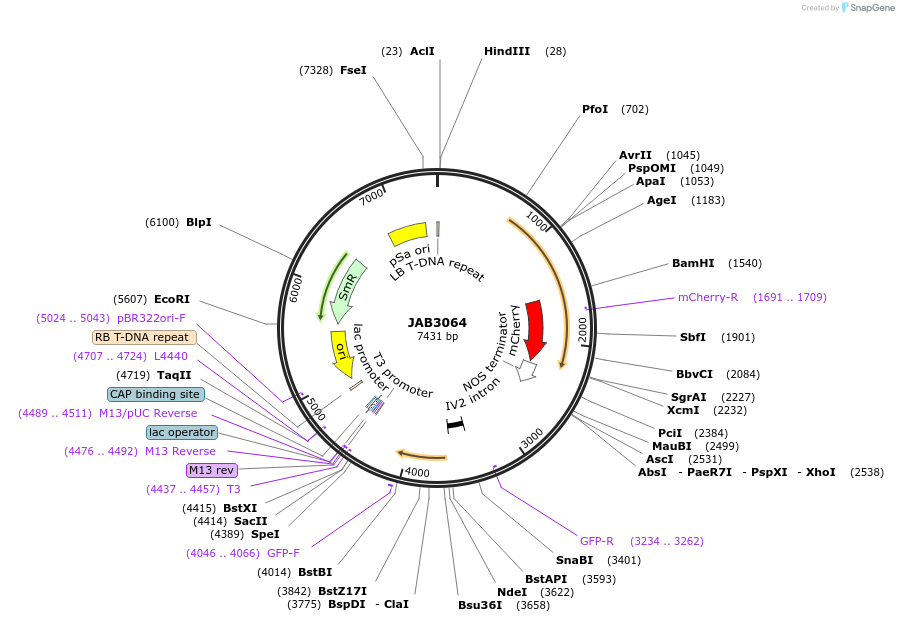
JAB3064
Plasmid#190400PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_proUBQ10s::GFPint-ADHt)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_proUBQ10s::GFPint-ADHt
ExpressionPlantAvailable SinceApril 26, 2023AvailabilityAcademic Institutions and Nonprofits only -
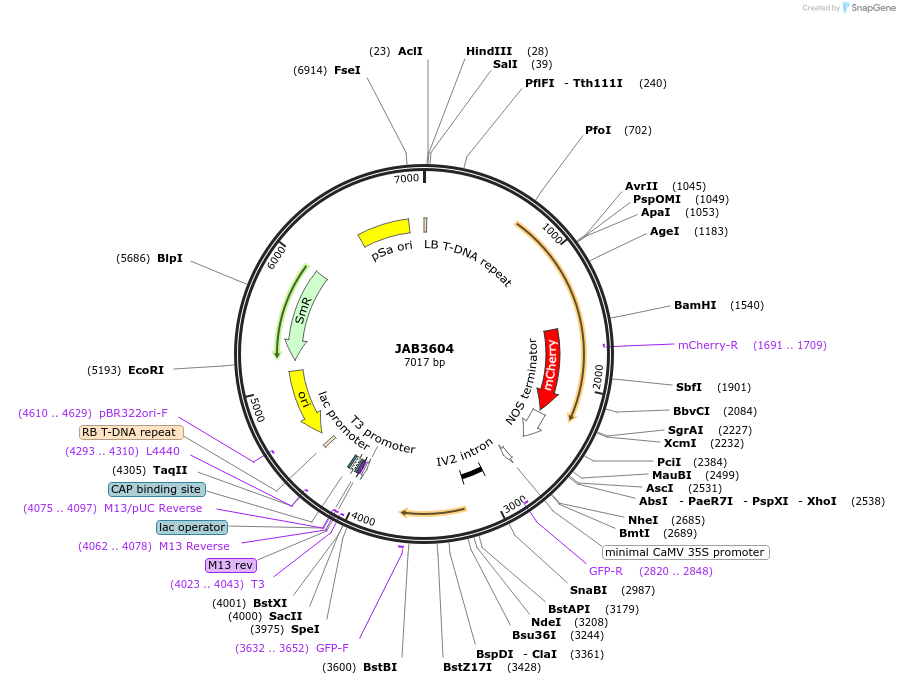
JAB3604
Plasmid#190423PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_3XPhlFO::GFPint-ADHt)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_3XPhlFO::GFPint-ADHt
ExpressionPlantAvailable SinceApril 25, 2023AvailabilityAcademic Institutions and Nonprofits only -
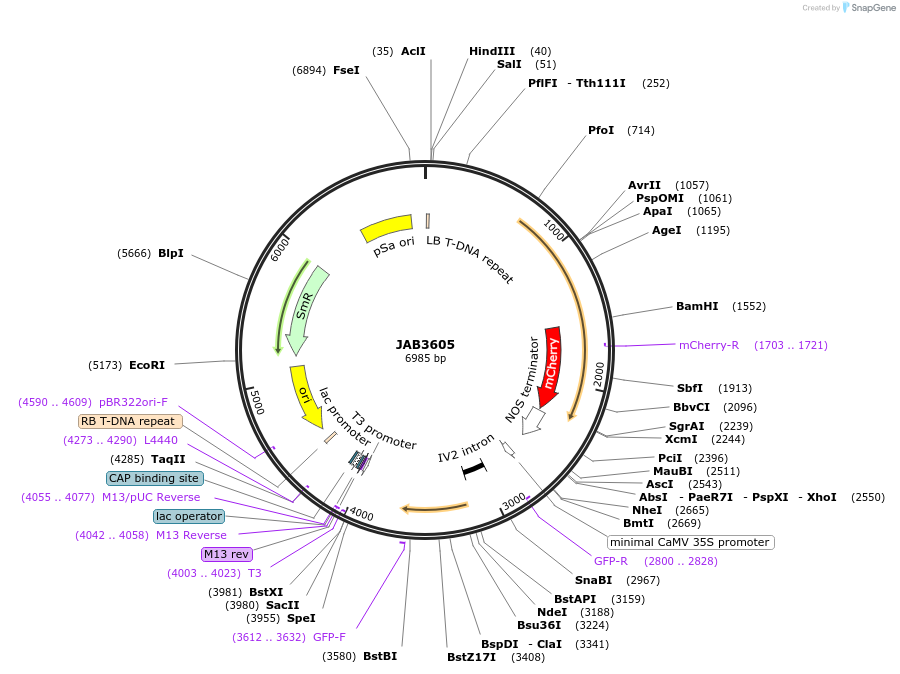
JAB3605
Plasmid#190424PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_2XPhlFO::GFPint-ADHt)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_2XPhlFO::GFPint-ADHt
ExpressionPlantAvailable SinceApril 25, 2023AvailabilityAcademic Institutions and Nonprofits only -
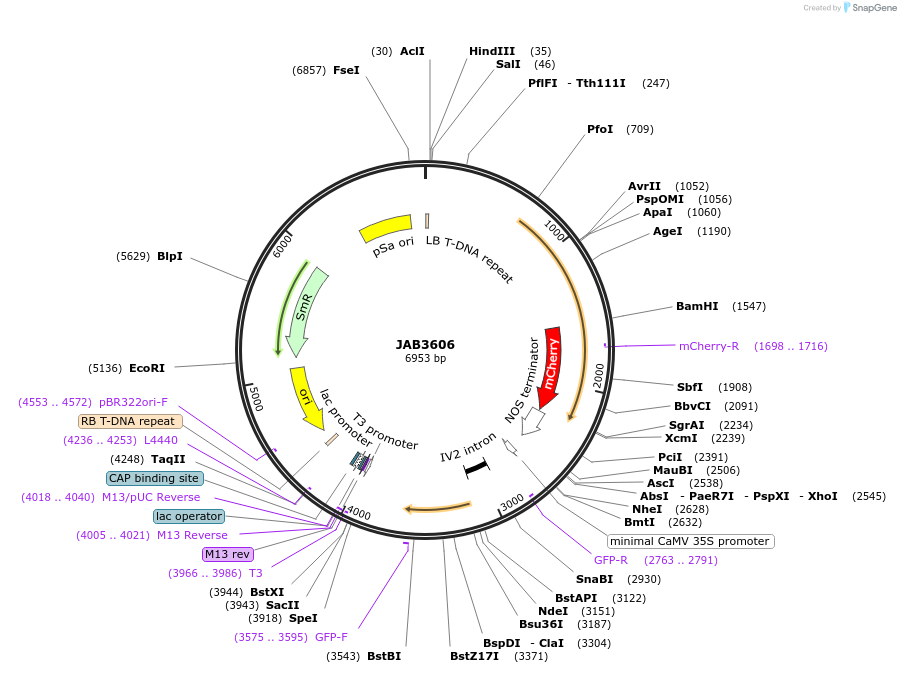
JAB3606
Plasmid#190425PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_1XPhlFO::GFPint-ADHt)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_1XPhlFO::GFPint-ADHt
ExpressionPlantAvailable SinceApril 25, 2023AvailabilityAcademic Institutions and Nonprofits only -
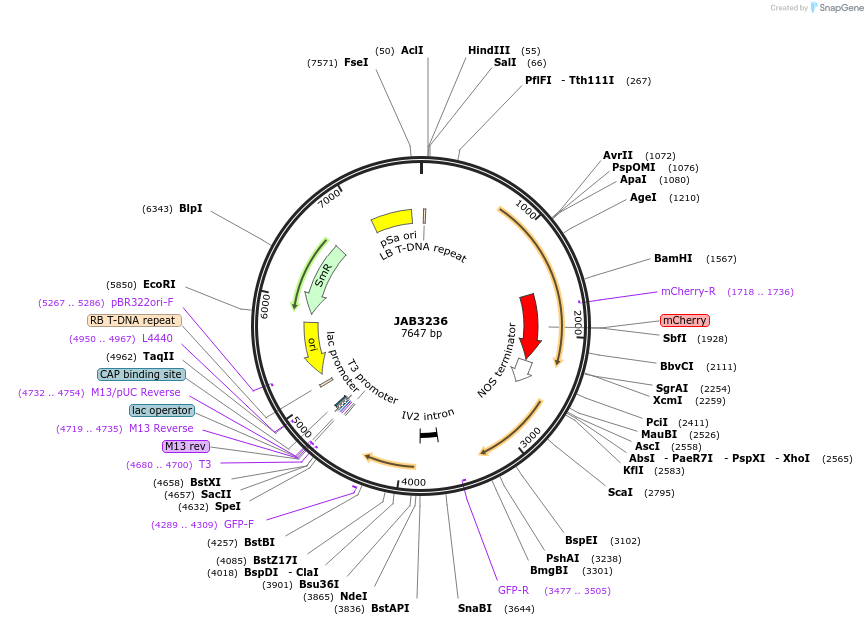
JAB3236
Plasmid#190401PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_proTT*::GFPint_ADHt)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_proTT*::GFPint_ADHt
ExpressionPlantAvailable SinceApril 25, 2023AvailabilityAcademic Institutions and Nonprofits only -
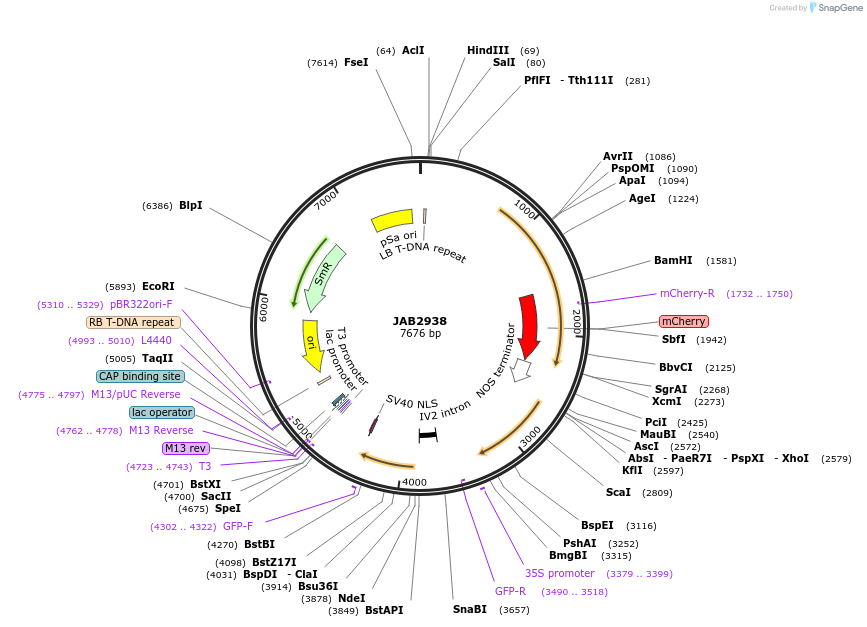
JAB2938
Plasmid#190395PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_proTT::GFPint-NLS_ADHt)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_proTT::GFPint-NLS_ADHt
ExpressionPlantAvailable SinceApril 7, 2023AvailabilityAcademic Institutions and Nonprofits only -
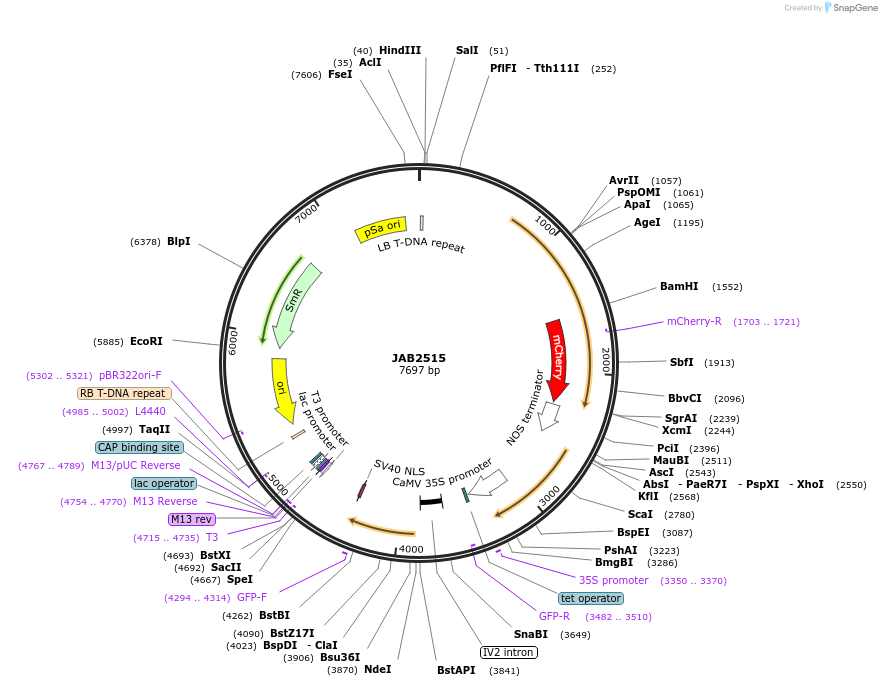
JAB2515
Plasmid#190383PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_pro35S-TetRO::GFPint-NLS-ADHt)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_pro35S-TetRO::GFPint-NLS-ADHt
ExpressionPlantAvailable SinceApril 4, 2023AvailabilityAcademic Institutions and Nonprofits only -
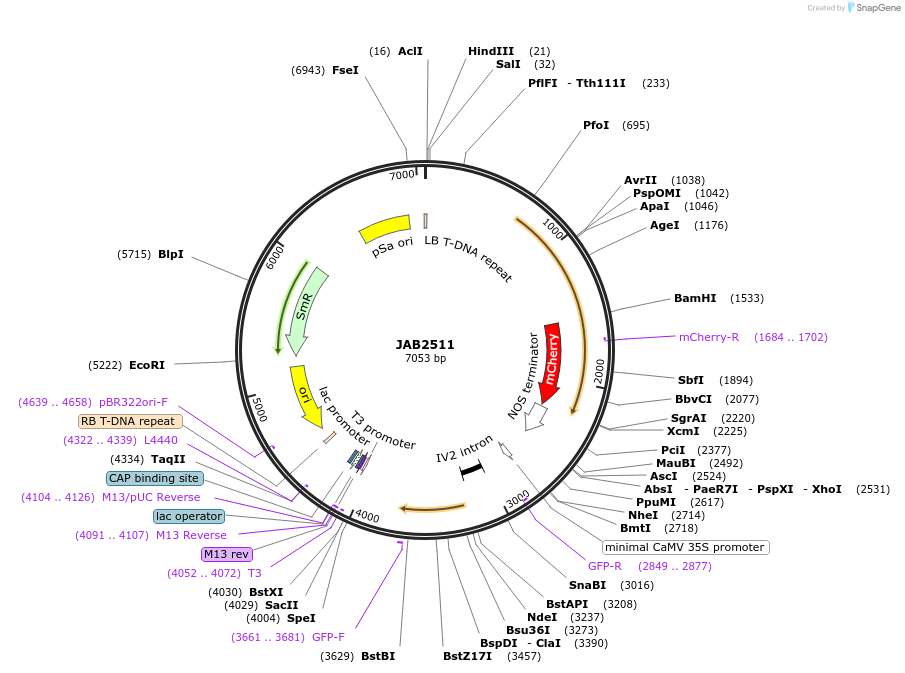
JAB2511
Plasmid#190380PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_6XRpaRO-min35S::GFPint-ADHt)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_6XRpaRO-min35S::GFPint-ADHt
ExpressionPlantAvailable SinceApril 4, 2023AvailabilityAcademic Institutions and Nonprofits only -
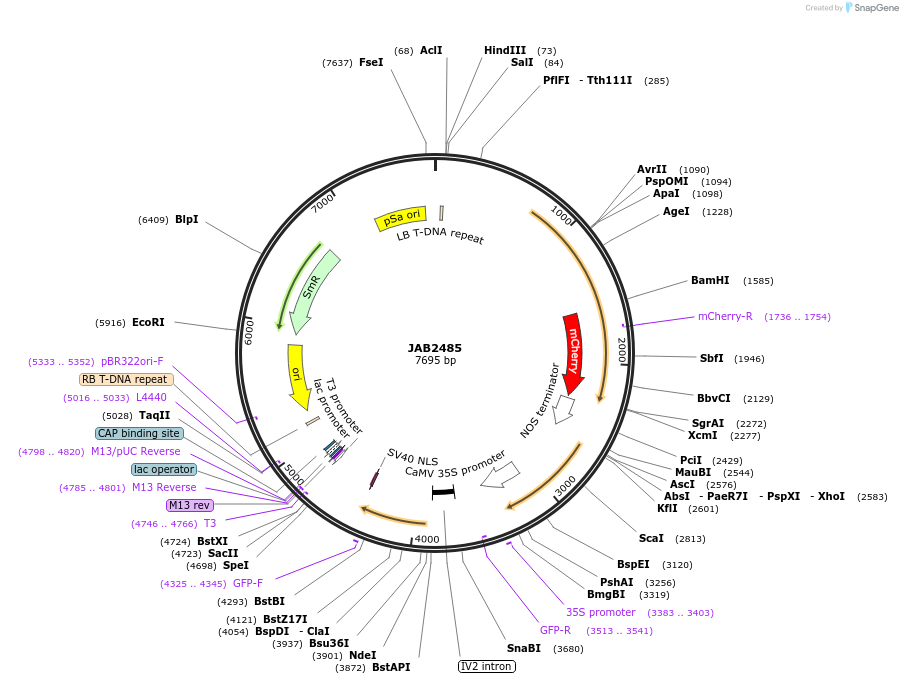
JAB2485
Plasmid#190371PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_pro35S-McbRO::GFPint-NLS-ADHt)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_pro35S-McbRO::GFPint-NLS-ADHt
ExpressionPlantAvailable SinceApril 4, 2023AvailabilityAcademic Institutions and Nonprofits only -
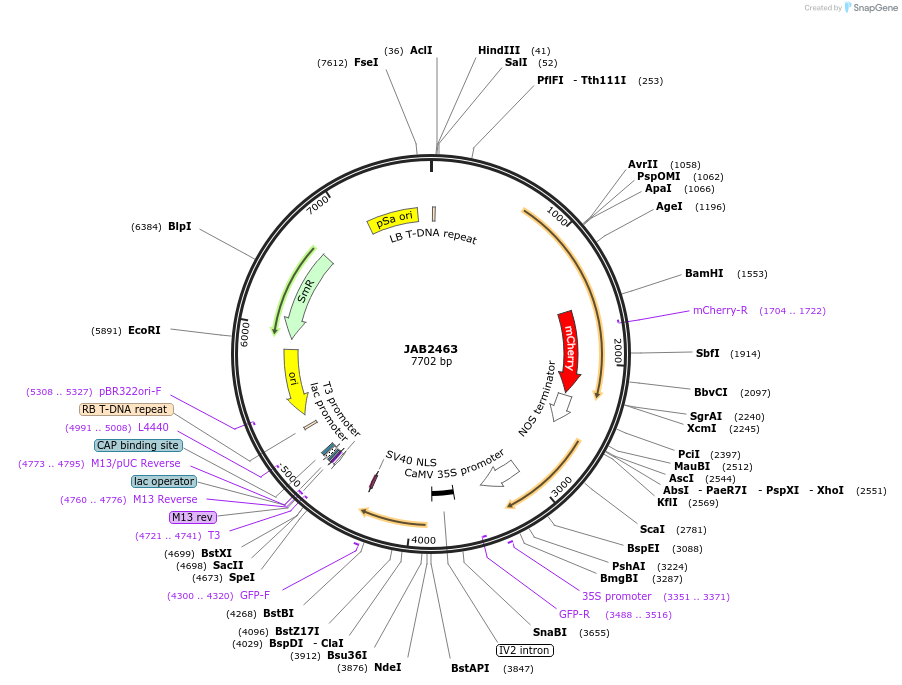
JAB2463
Plasmid#190370PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_pro35S-QacRO::GFPint-NLS-ADHt)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_pro35S-QacRO::GFPint-NLS-ADHt
ExpressionPlantAvailable SinceApril 4, 2023AvailabilityAcademic Institutions and Nonprofits only -
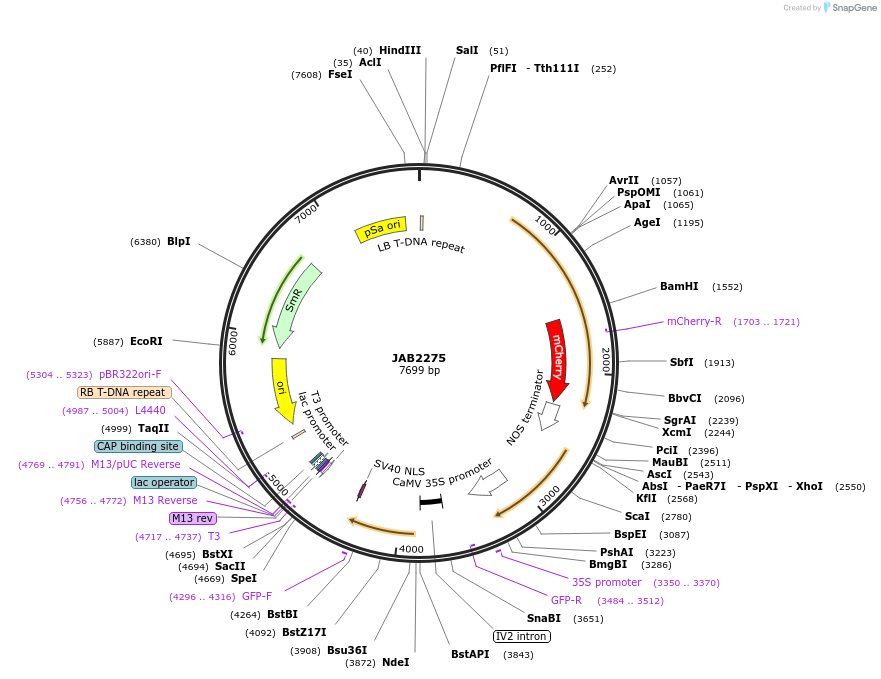
JAB2275
Plasmid#190361PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_pro35S-AmtRO::GFPint-NLS-ADHt)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_pro35S-AmtRO::GFPint-NLS-ADHt
ExpressionPlantAvailable SinceApril 4, 2023AvailabilityAcademic Institutions and Nonprofits only -
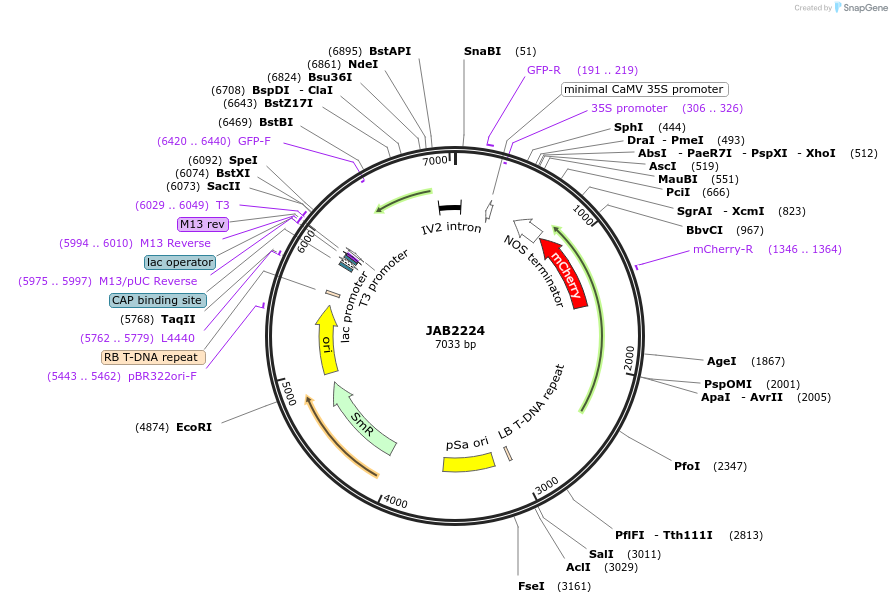
JAB2224
Plasmid#190358PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_proG1090::GFPint-ADHterm)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_proG1090::GFPint-ADHterm
ExpressionPlantAvailable SinceApril 4, 2023AvailabilityAcademic Institutions and Nonprofits only -
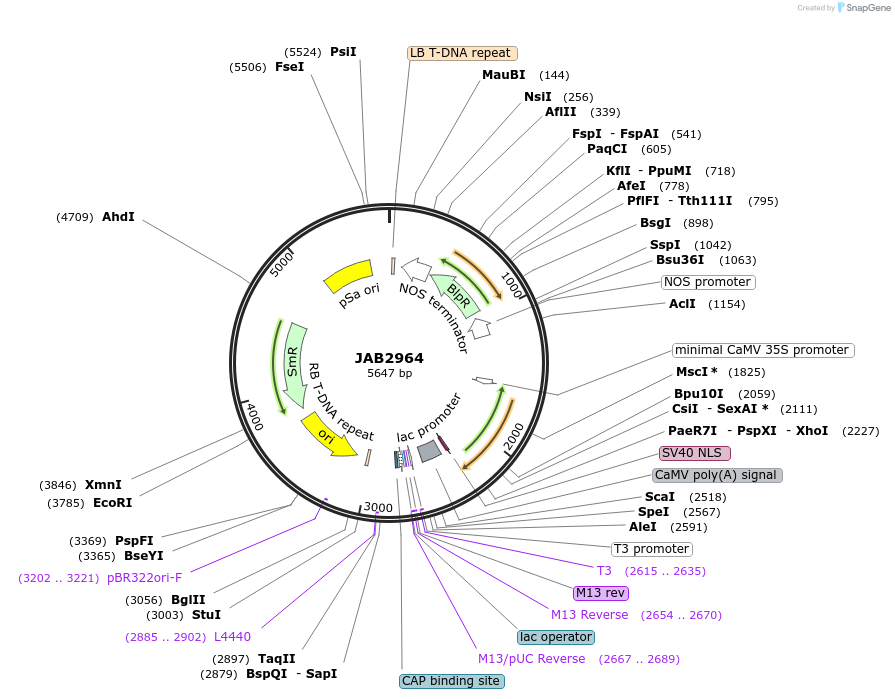
JAB2964
Plasmid#190398PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (6XAmtRO-2XPhlFO::TetR-NLS-19St)DepositorInsert6XAmtRO-2XPhlFO::TetR-NLS-19St
ExpressionPlantAvailable SinceMarch 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
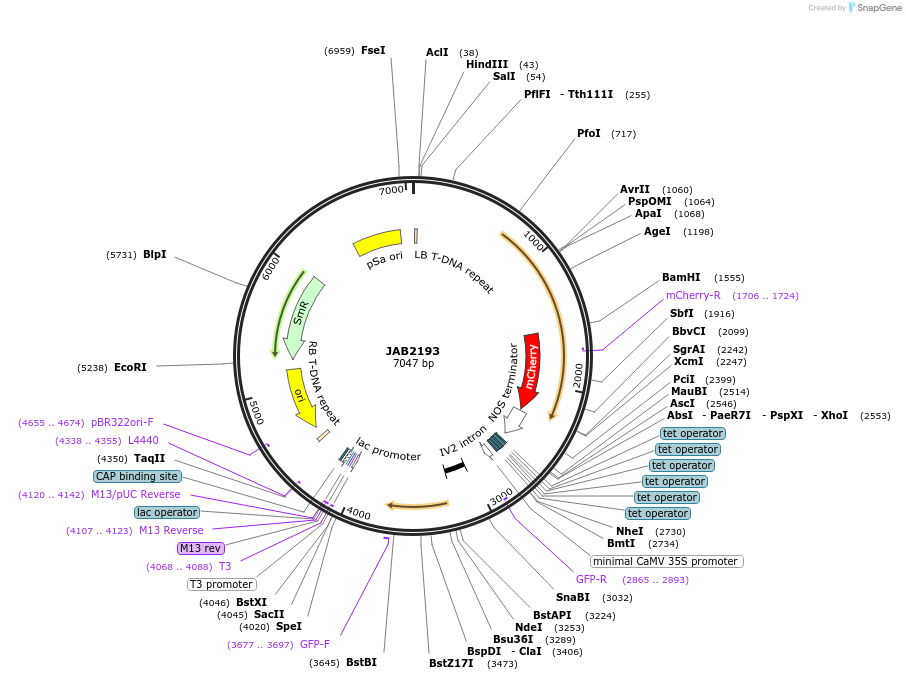
JAB2193
Plasmid#190357PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_6XTetRO-min35S::GFPint-ADHt)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_6XTetRO-min35S::GFPint-ADHt
ExpressionPlantAvailable SinceMarch 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
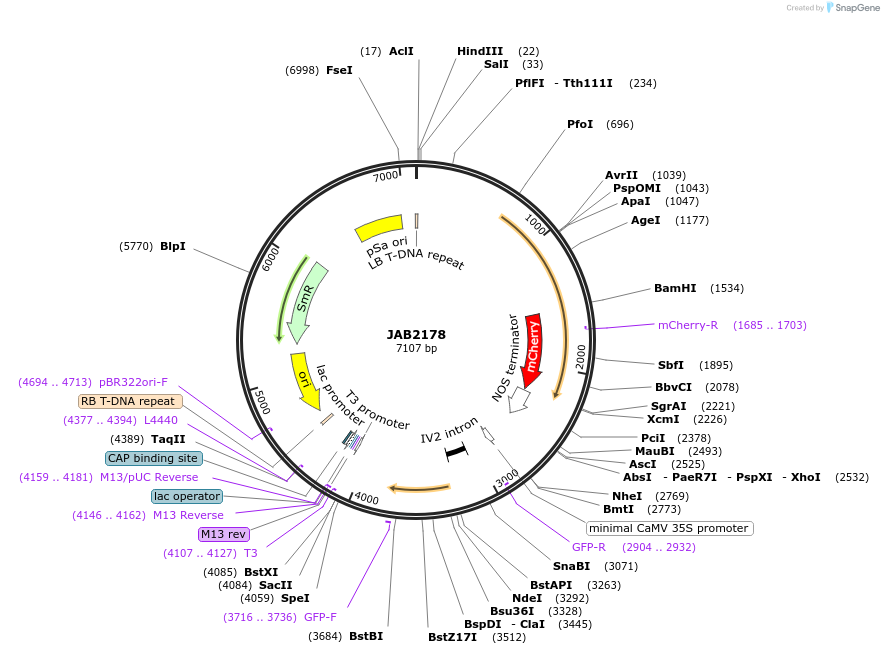
JAB2178
Plasmid#190351PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_6XIcaRO-min35S::GFPint-ADHt)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_6XIcaRO-min35S::GFPint-ADHt
ExpressionPlantAvailable SinceMarch 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
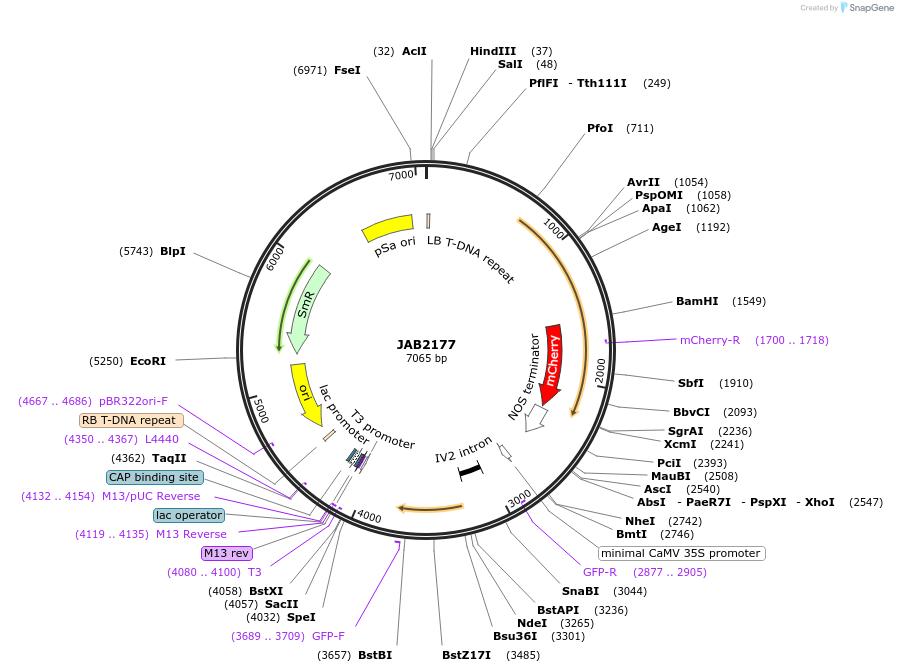
JAB2177
Plasmid#190350PurposePlasmids encode parts for syntetic genetic circuit construction in eudicot plants (proUBQ10::PIP2A-mCherry-NOSt_6XAmtRO-min35S::GFPint-ADHt)DepositorInsertproUBQ10::PIP2A-mCherry-NOSt_6XAmtRO-min35S::GFPint-ADHt
ExpressionPlantAvailable SinceMarch 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
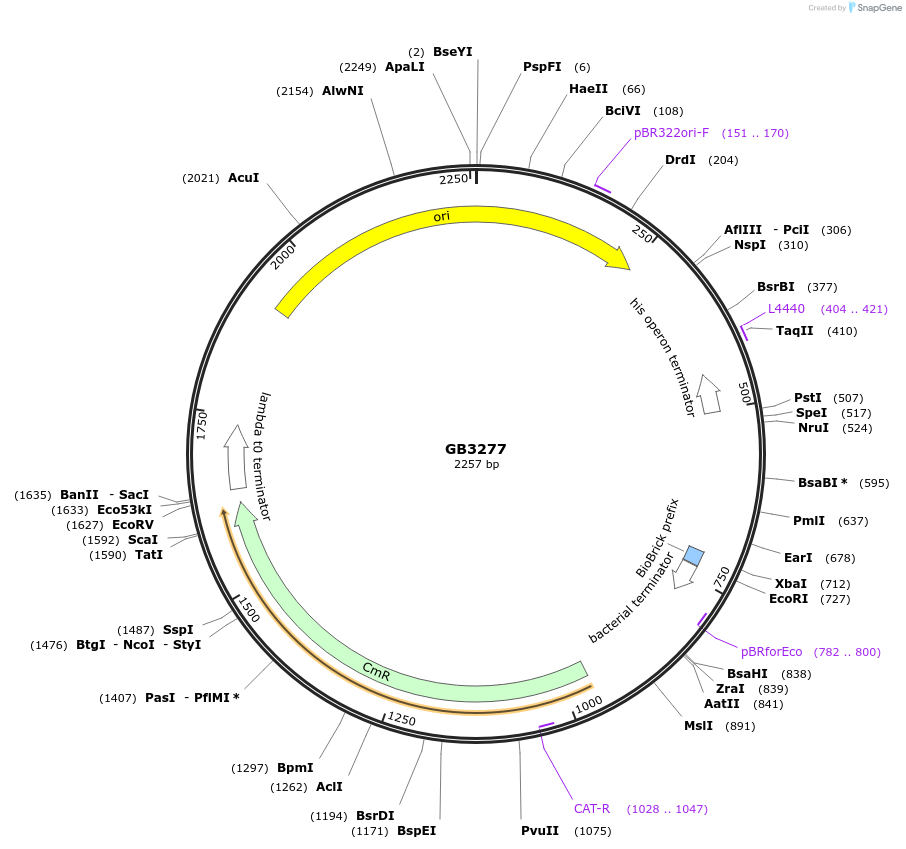
GB3277
Plasmid#193131PurposeA2 Proximal promoter sequence consisting of three times the target sequence for the gRNA1 (GB1838) flanked by random sequences.DepositorInsertGB_SynP (A2) G1abc.4
UseSynthetic BiologyAvailable SinceMarch 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
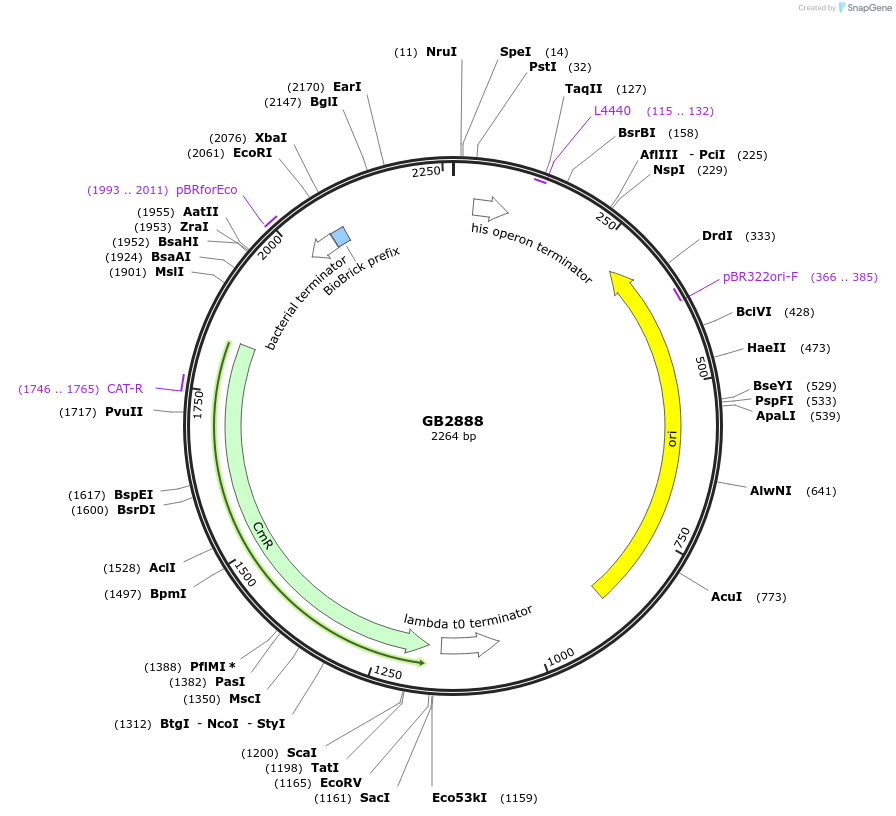
GB2888
Plasmid#193126PurposeA2 Proximal promoter sequence consisting of two times the target sequence for the gRNA1 (GB1838) flanked by random sequences.DepositorInsertGB_SynP (A2) G1ab.4
UseSynthetic BiologyAvailable SinceJan. 20, 2023AvailabilityAcademic Institutions and Nonprofits only -
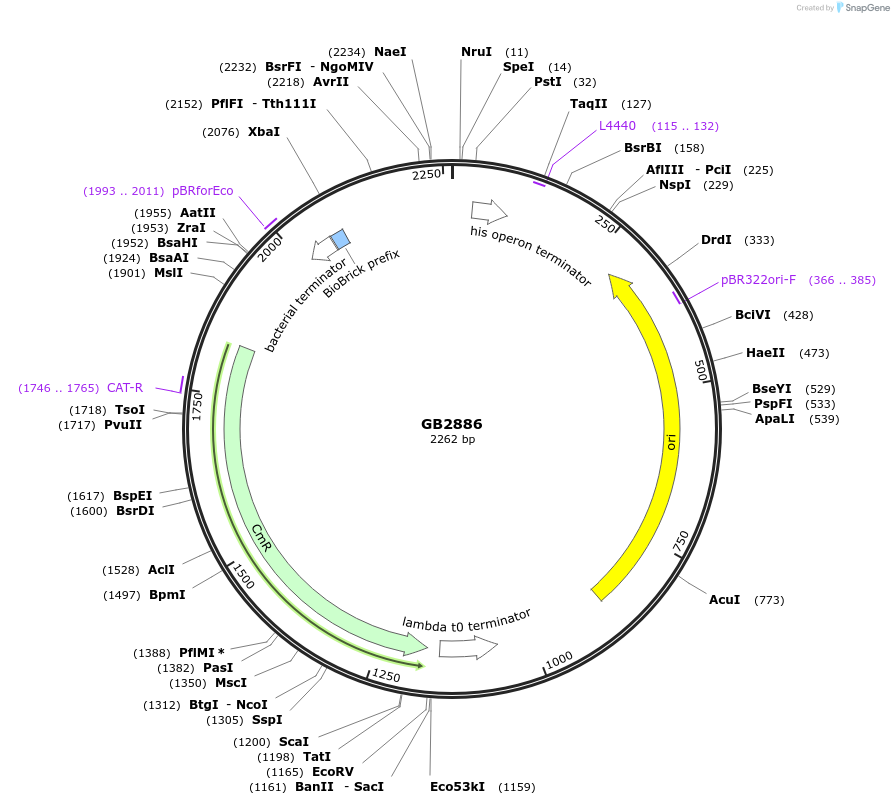
GB2886
Plasmid#193124PurposeA2 Proximal promoter sequence consisting of two times the target sequence for the gRNA1 (GB1838) flanked by random sequences.DepositorInsertGB_SynP (A2) G1ab.3
UseSynthetic BiologyAvailable SinceJan. 18, 2023AvailabilityAcademic Institutions and Nonprofits only