We narrowed to 691 results for: Bacillus
-
Plasmid#209980PurposeContains Level 0 Part: resistance cassette (cat) for the construction of Level 1 plasmidsDepositorInsertresistance cassette (cat)
ExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
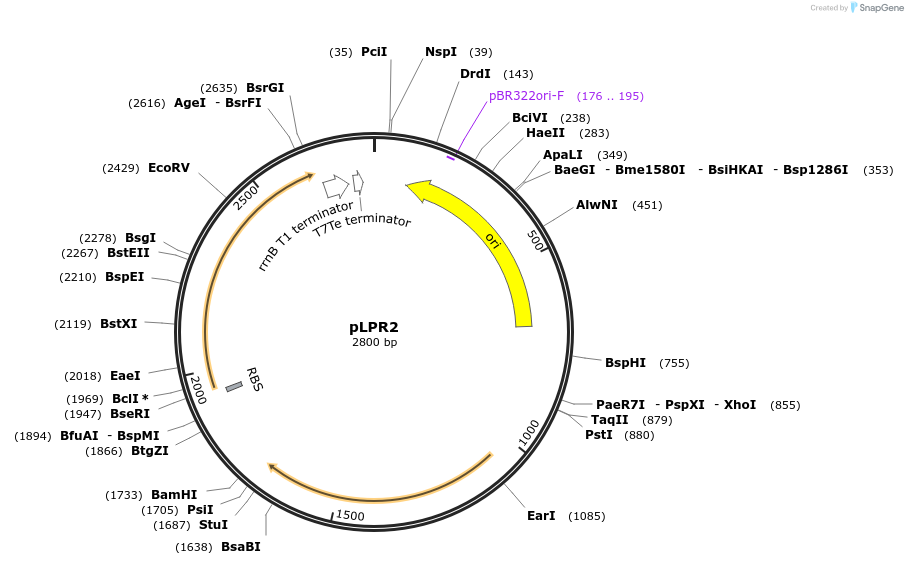
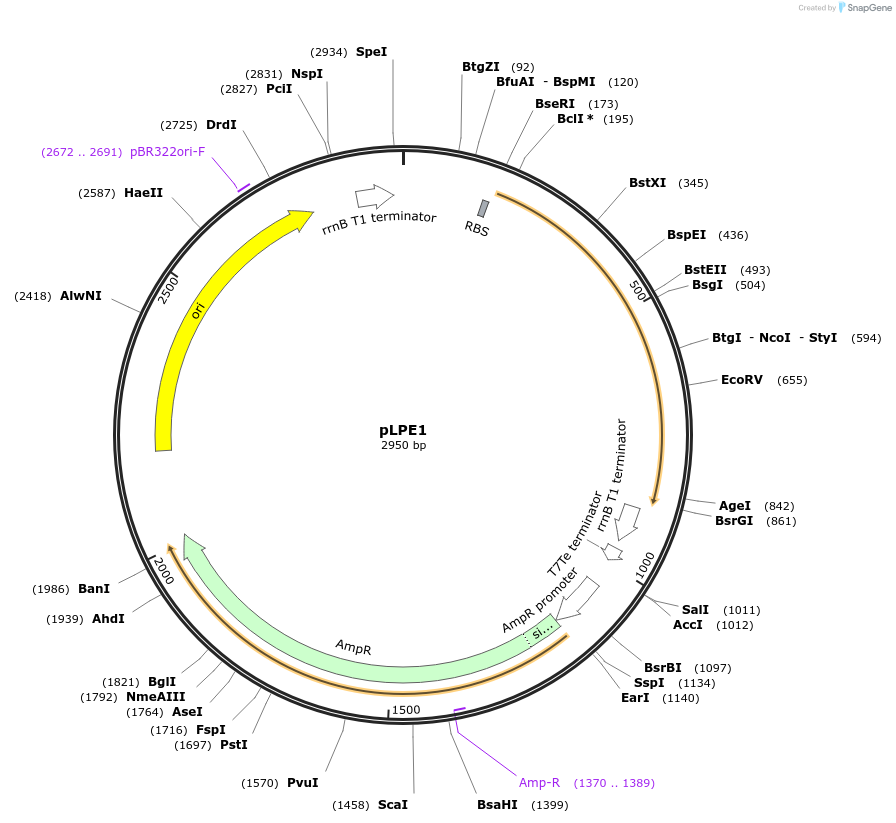
pLPE1
Plasmid#209981PurposeContains Level 0 Part: E. coli shuttle part (pMB1_ampR_B0017) for the construction of Level 1 plasmidsDepositorInsertE. coli shuttle part (pMB1_ampR_B0017)
ExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
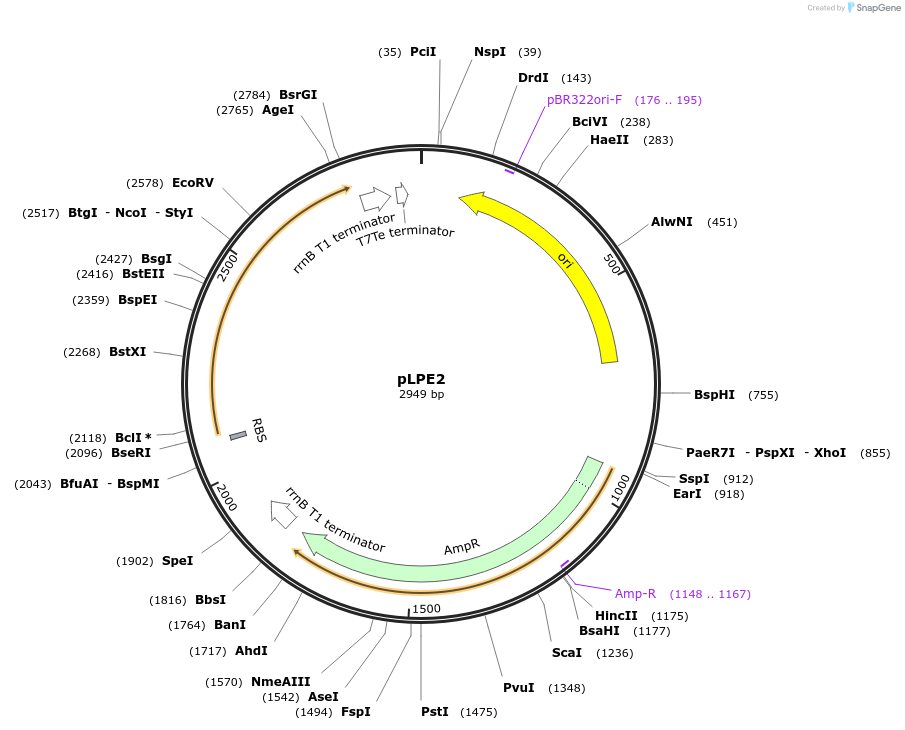
pLPE2
Plasmid#209982PurposeContains Level 0 Part: E. coli shuttle part (ampR_B0017) for the construction of Level 1 plasmidsDepositorInsertE. coli shuttle part (ampR_B0017)
ExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
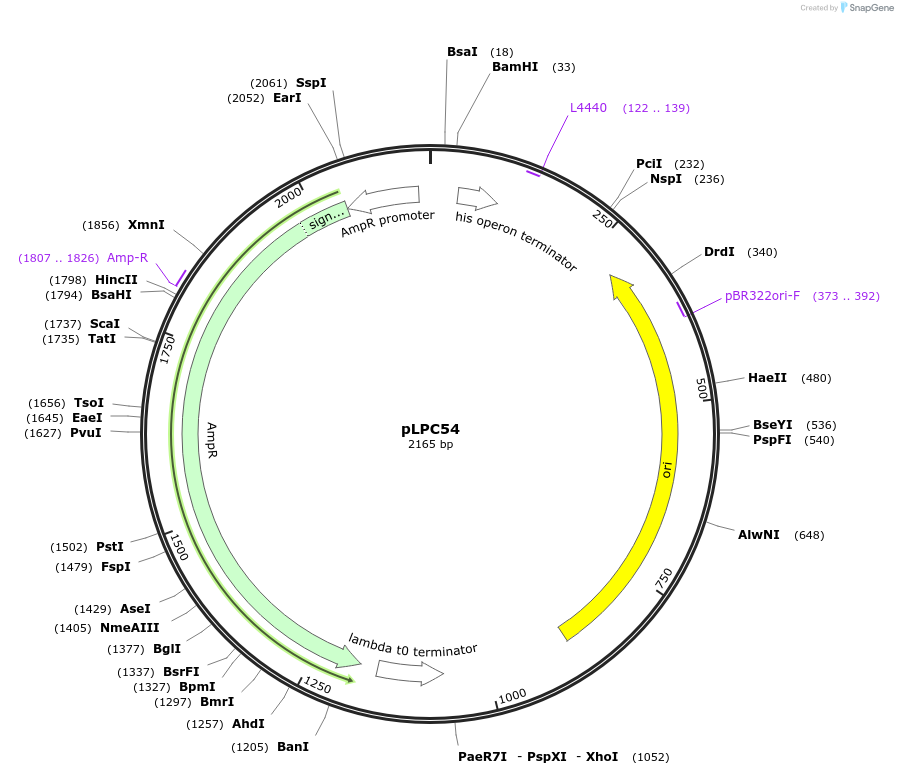
pLPC54
Plasmid#209983PurposeContains Level 0* Part: 5' Connector (5C_07_5C1C) for the construction of Level 2 plasmidsDepositorInsert5' Connector (5C_07_5C1C)
ExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
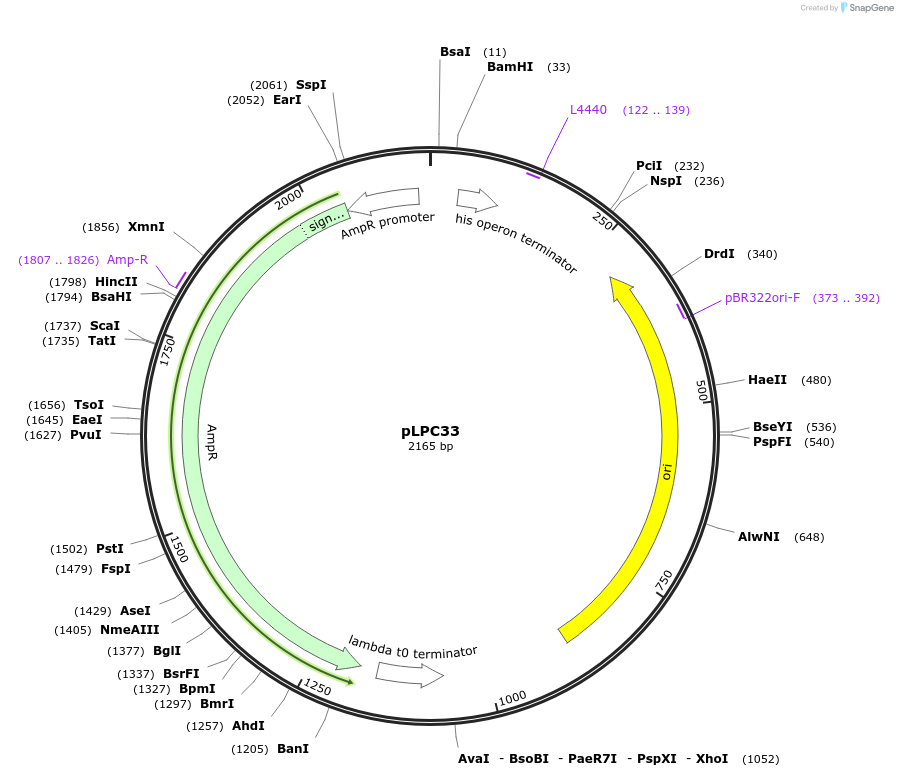
pLPC33
Plasmid#209984PurposeContains Level 0* Part: 3' Connector (3C_08_3C3) for the construction of Level 2 plasmidsDepositorInsert3' Connector (3C_08_3C3)
ExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
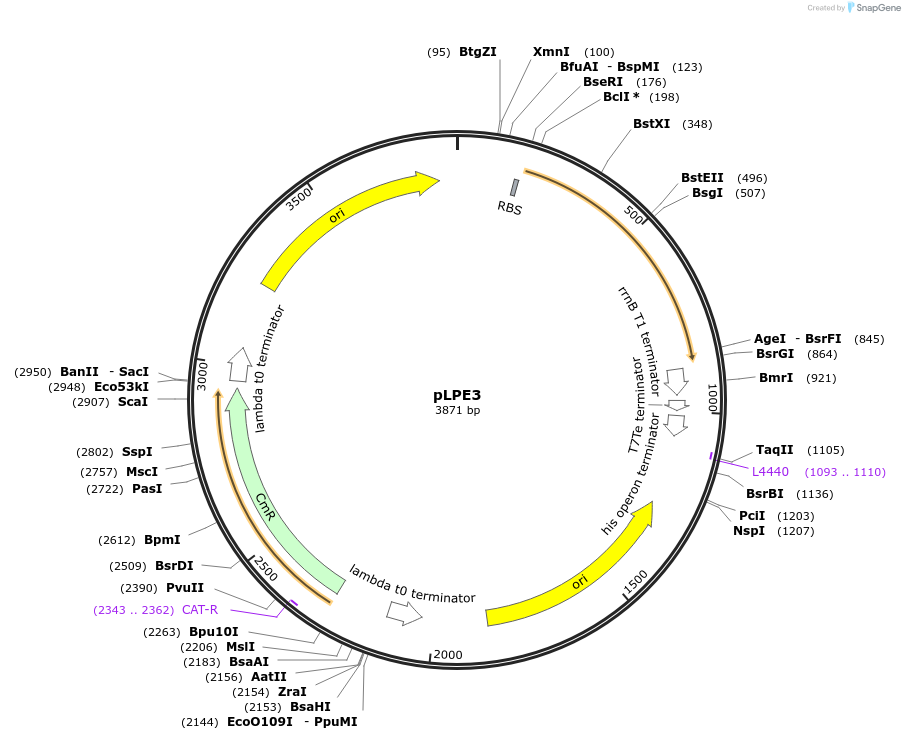
pLPE3
Plasmid#209985PurposeContains Level 0* Part: E. coli shuttle part (pMB1_cat) for the construction of Level 2 plasmidsDepositorInsertE. coli shuttle part (pMB1_cat)
ExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
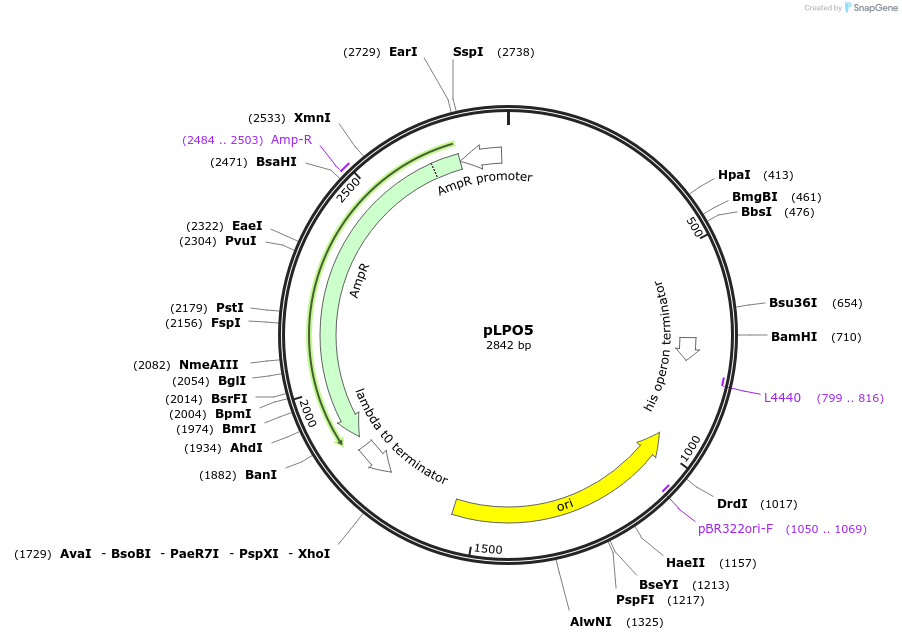
pLPO5
Plasmid#209987PurposeContains Level 0* Part: replication origin (256rep) for the construction of Level 2 plasmidsDepositorInsertreplication origin (256_rep)
ExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
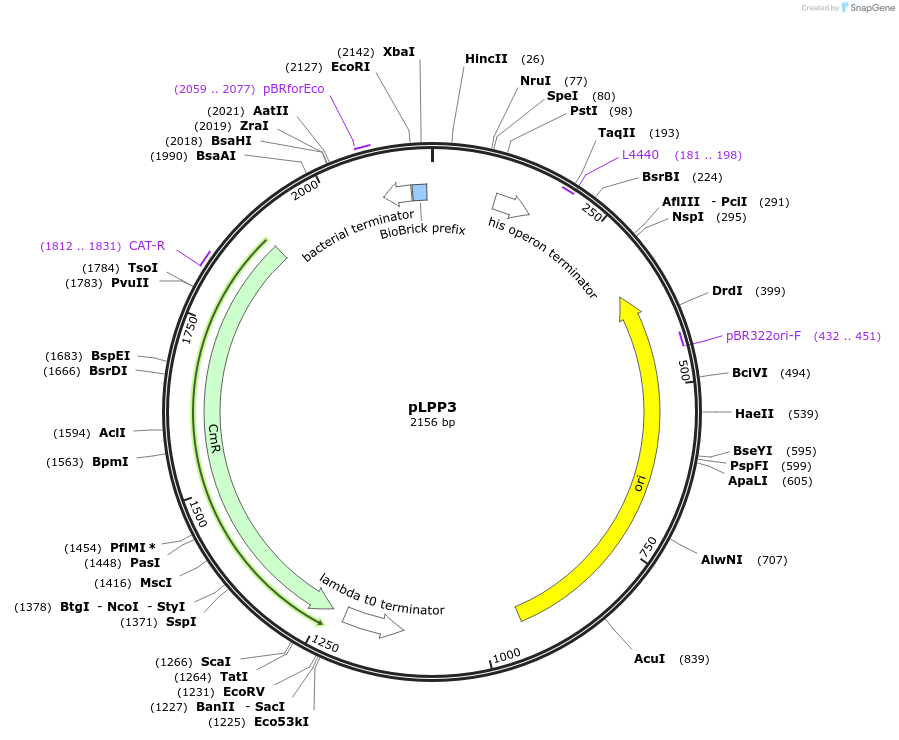
pLPP3
Plasmid#209965PurposeContains Level 0 Part: constitutive Promoter (P25) for the construction of Level 1 plasmidsDepositorInsertPromoter (P25)
ExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
pLPP7
Plasmid#209969PurposeContains Level 0 Part: autoinducible Promoter (PfabZ) for the construction of Level 1 plasmidsDepositorInsertPromoter (P_fabZ)
ExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
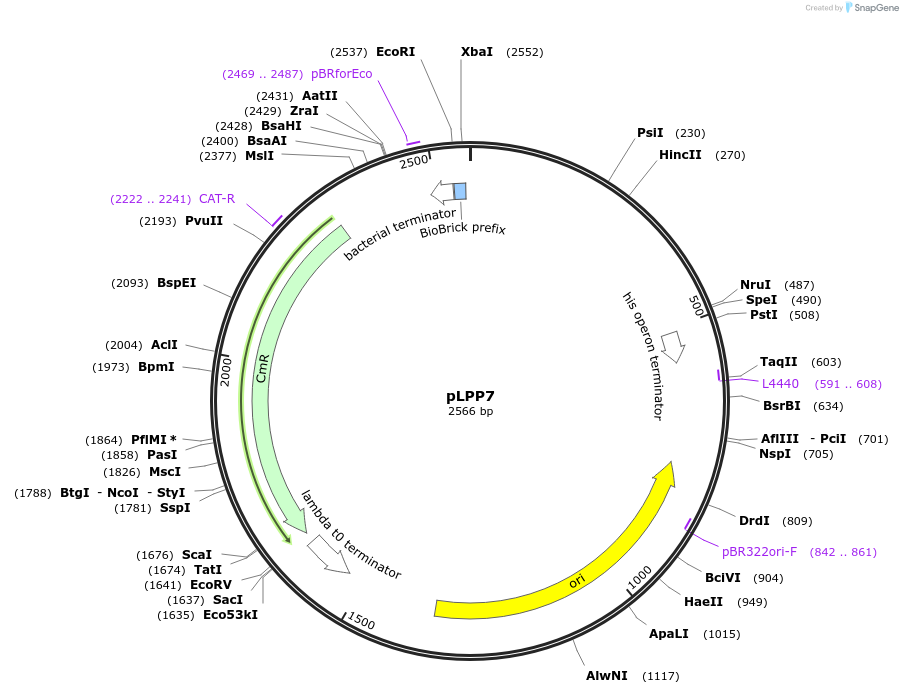
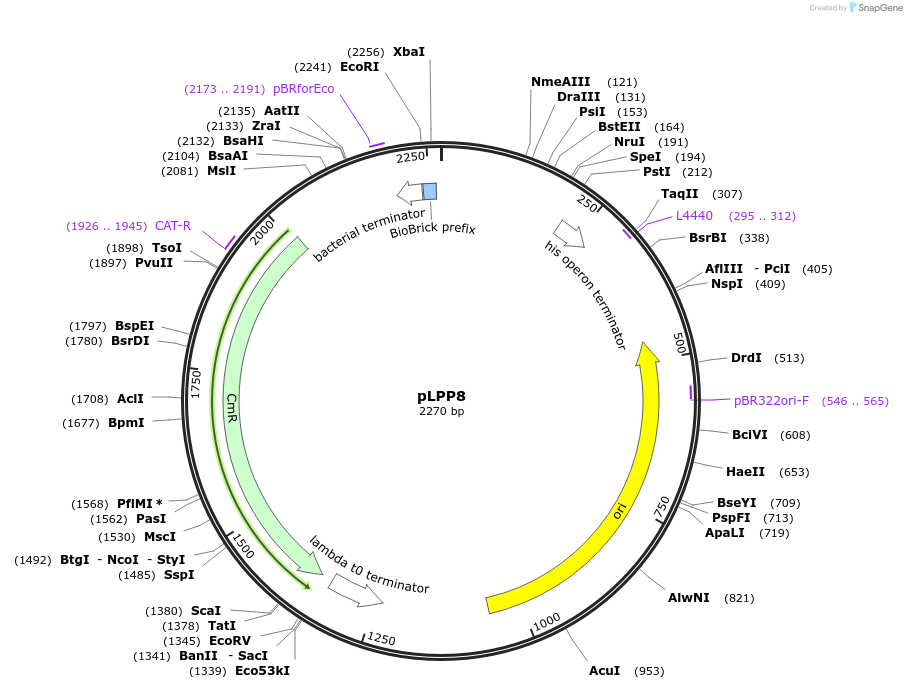
pLPP8
Plasmid#209970PurposeContains Level 0 Part: autoinducible Promoter (PgadB) for the construction of Level 1 plasmidsDepositorInsertPromoter (P_gadB)
ExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
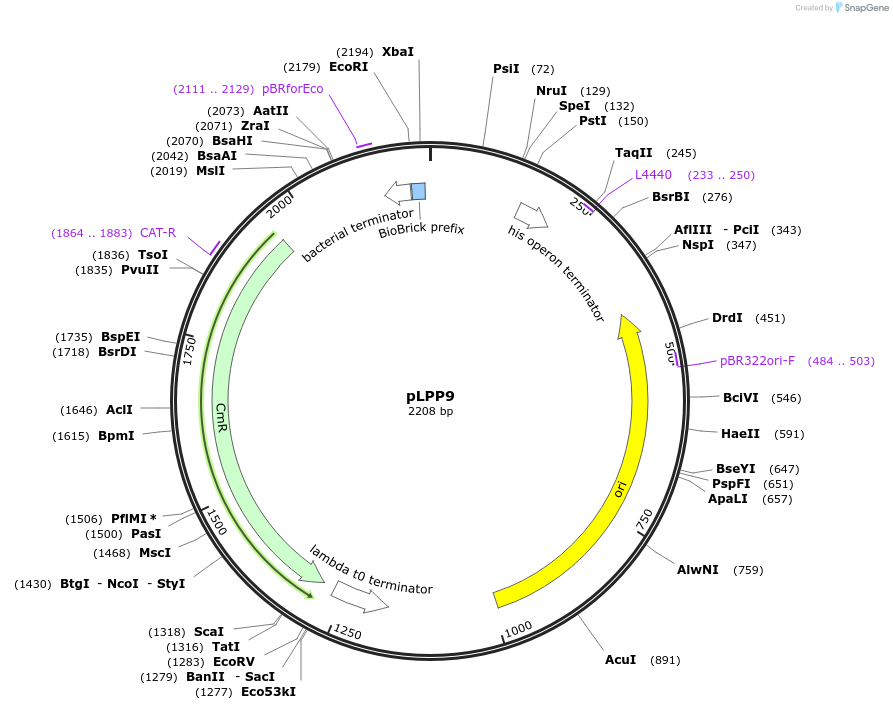
pLPP9
Plasmid#209971PurposeContains Level 0 Part: constitutive Promoter (PldhD) for the construction of Level 1 plasmidsDepositorInsertPromoter (P_ldhD)
ExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
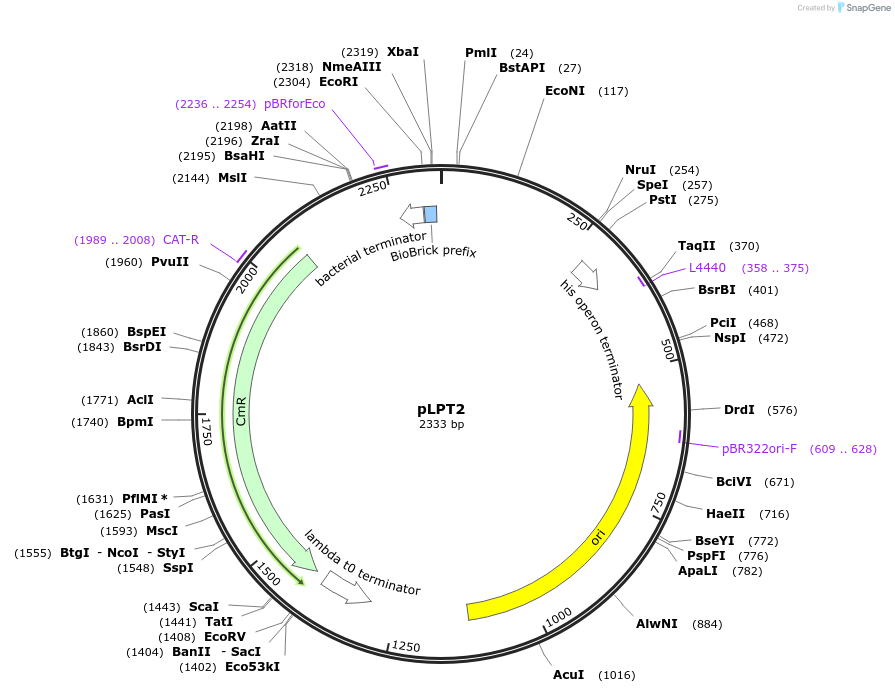
pLPT2
Plasmid#209974PurposeContains Level 0 Part: Terminator (TpepN) for the construction of Level 1 plasmidsDepositorInsertTerminator (T_pepN)
ExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
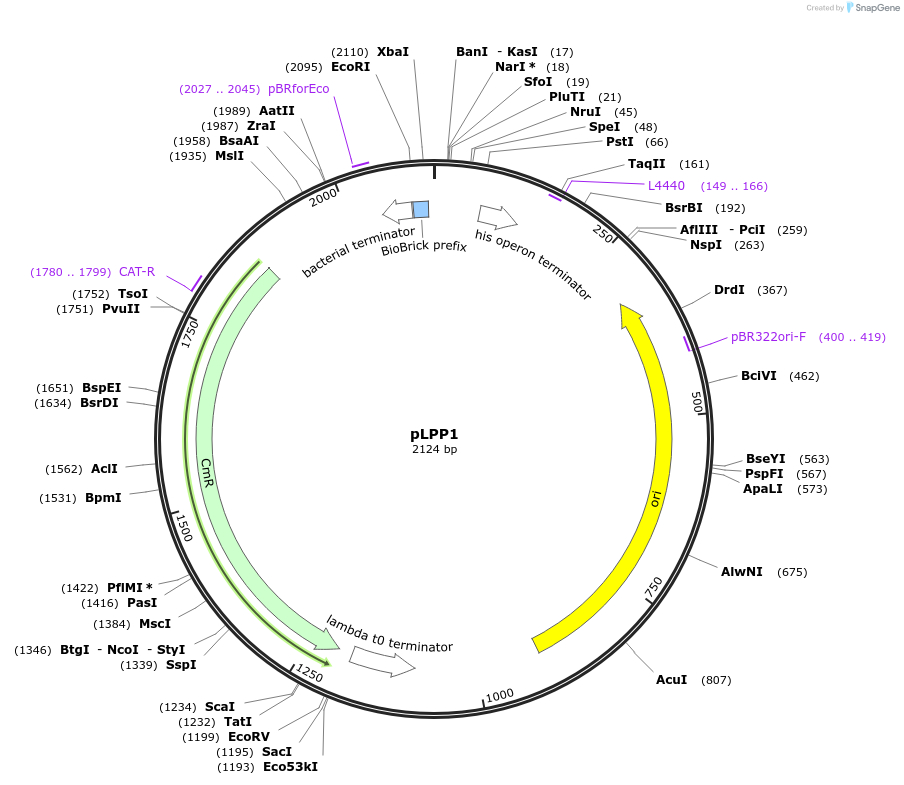
pLPP1
Plasmid#209963PurposeContains Level 0 Part: mock / negative control Promoter (P-nc) for the construction of Level 1 plasmidsDepositorInsertPromoter (mock)
ExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
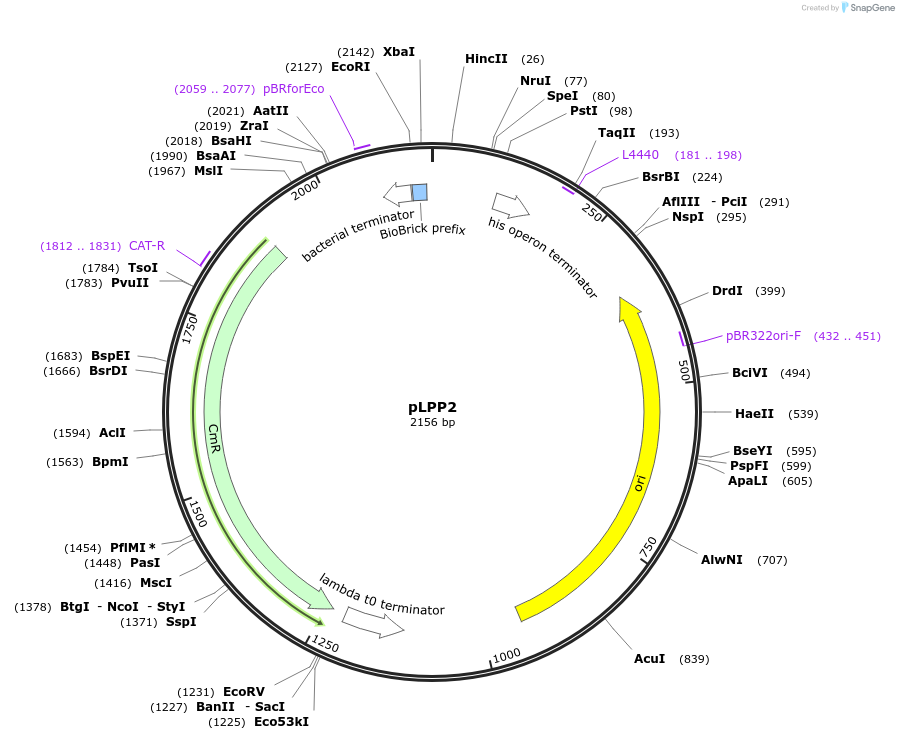
pLPP2
Plasmid#209964PurposeContains Level 0 Part: constitutive Promoter (P48) for the construction of Level 1 plasmidsDepositorInsertPromoter (P48)
ExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
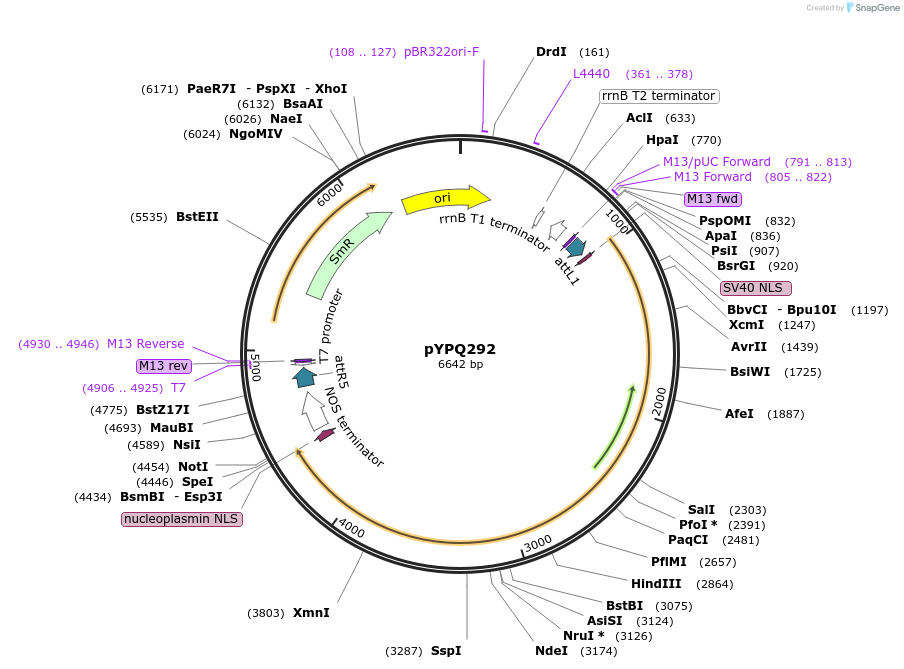
pYPQ292
Plasmid#129672PurposeGateway entry clone for CRISPR-Cas12b systemsDepositorInsertAaCas12b
UseCRISPR; Gateway compatible aacas12b entry cloneExpressionPlantMutationAaCas12b is rice codon optimizedAvailable SinceMarch 18, 2020AvailabilityAcademic Institutions and Nonprofits only -
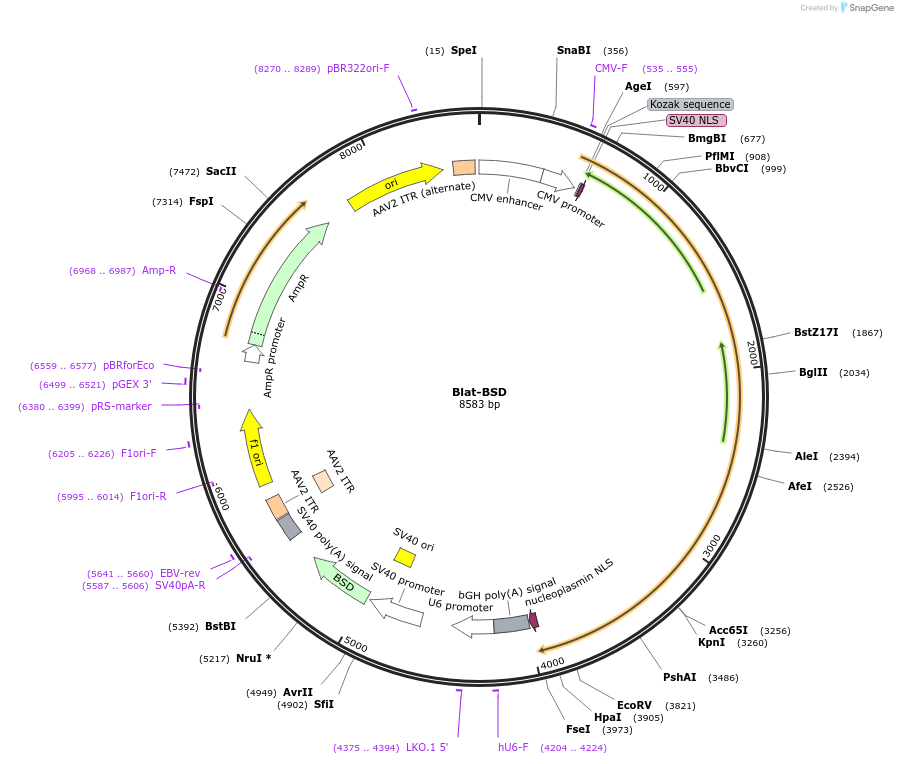
Blat-BSD
Plasmid#163791PurposeThe plasmid expresses human codon-optimized BlatCas9, gRNA expression elements and blasticidin resistence gene.DepositorTypeEmpty backboneUseAAV and CRISPRExpressionMammalianAvailable SinceMarch 18, 2021AvailabilityAcademic Institutions and Nonprofits only -
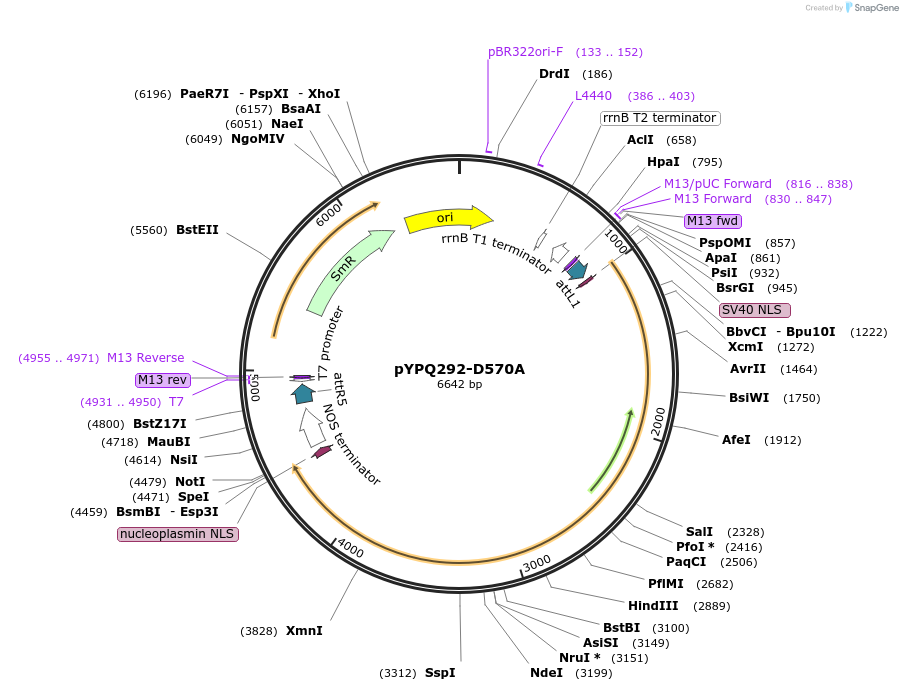
pYPQ292-D570A
Plasmid#129679PurposeGateway entry clone for CRISPR-Cas12b systemsDepositorInsertAaCas12b
UseCRISPR; Gateway compatible aacas12b entry cloneExpressionPlantMutationD570A; AaCas12b is rice codon optimizedAvailable SinceMarch 18, 2020AvailabilityAcademic Institutions and Nonprofits only -
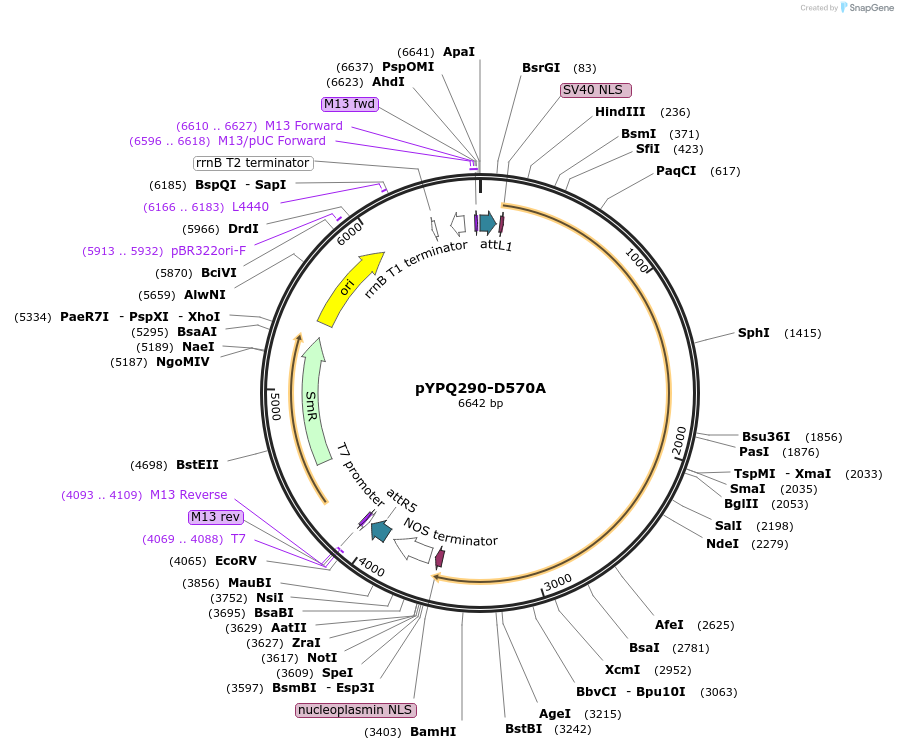
pYPQ290-D570A
Plasmid#129673PurposeGateway entry clone for CRISPR-Cas12b systemsDepositorInsertAacCas12b
UseCRISPR; Gateway compatible aaccas12b-d570a entry …ExpressionPlantMutationD570A; AacCas12b is rice codon optimizedAvailable SinceMarch 18, 2020AvailabilityAcademic Institutions and Nonprofits only -
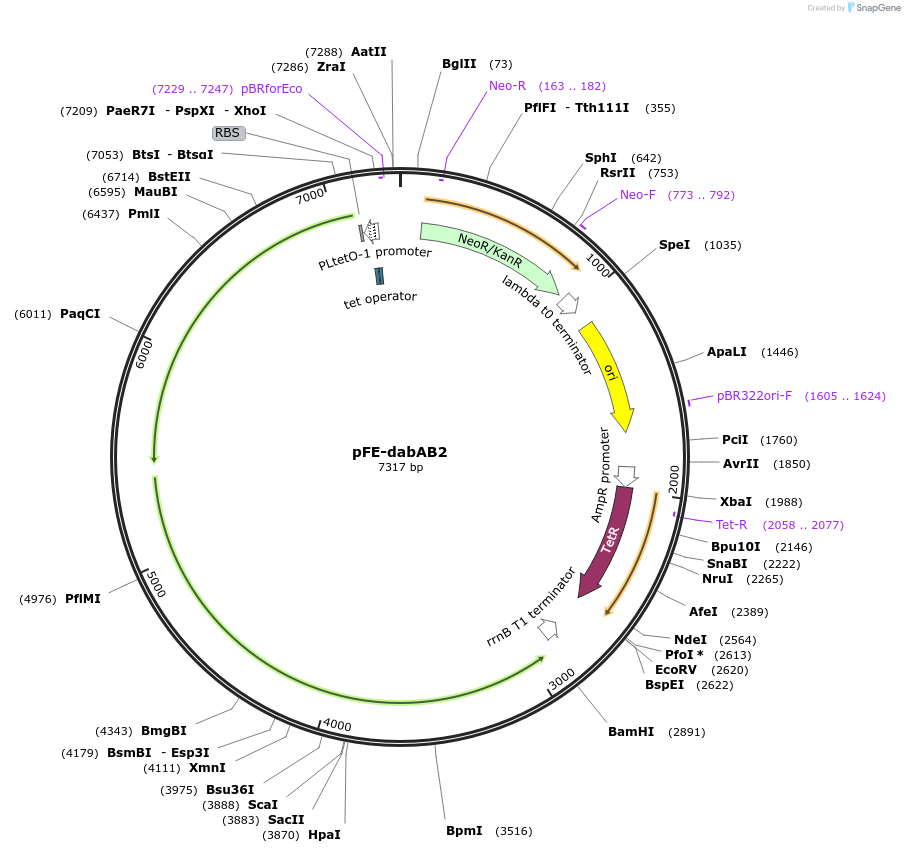
pFE-dabAB2
Plasmid#133002PurposepFE carrying both the dabA2 (Uniprot: D0KWS7) and dabB2 genes (Uniprot: D0KWS8)DepositorInsertsdabB2
dabA2
ExpressionBacterialPromotertetRAvailable SinceNov. 11, 2019AvailabilityAcademic Institutions and Nonprofits only -
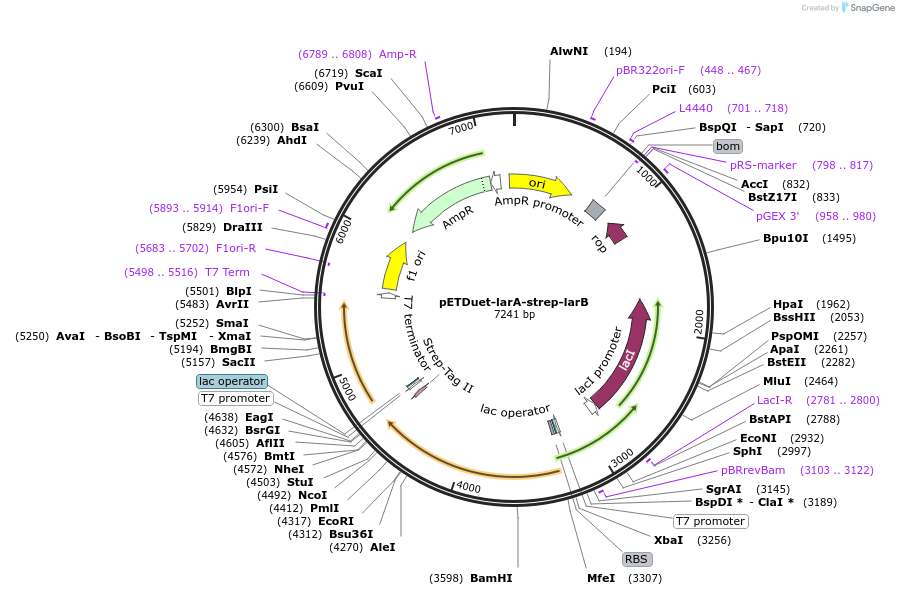
pETDuet-larA-strep-larB
Plasmid#232792PurposeExpression of L. plantarum larA-strep and larB genes in E. coliDepositorInsertsLactate racemase
Pyridinium-3,5-biscarboxylic acid mononucleotide synthase
TagsStrep-tag IIExpressionBacterialAvailable SinceSept. 11, 2025AvailabilityAcademic Institutions and Nonprofits only