We narrowed to 13,384 results for: CAR
-
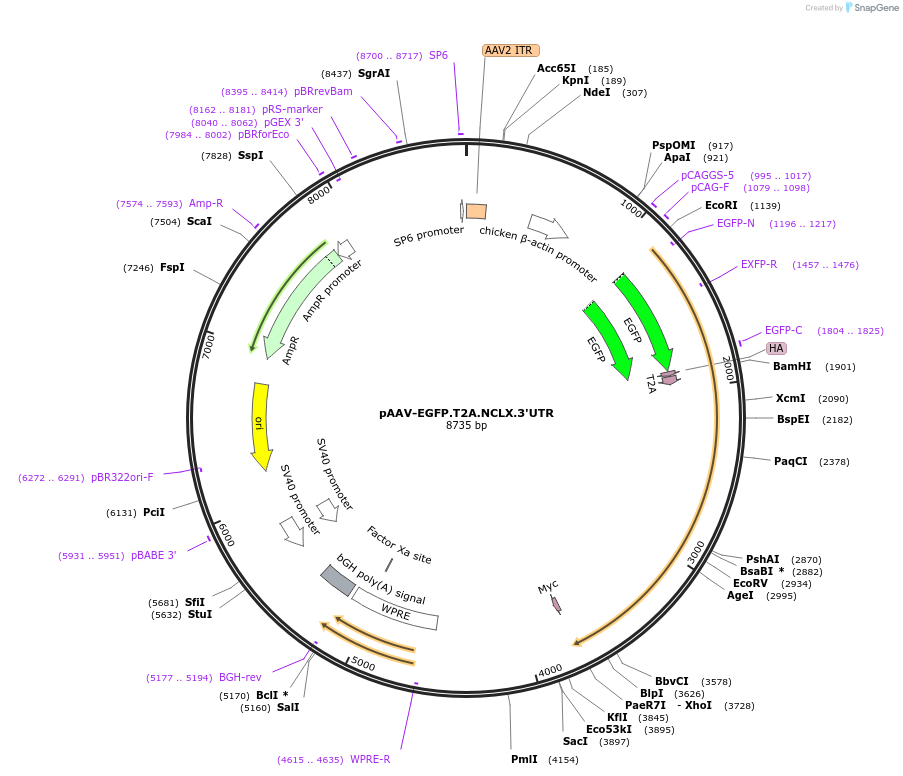
Plasmid#181872PurposeExpresses EGFP and murine NCLX (separated by a T2A cleavage site) under control of the synthetic CAG promoter (includes a large portion of the Slc8b1 3'UTR)DepositorInsertsUseAAV and AdenoviralTagsHA, T2A, and mycExpressionMammalianMutationincludes a large portion of the Slc8b1 3'UTR…Promotersynthetic hybrid CAG promoterAvailable SinceMarch 7, 2022AvailabilityAcademic Institutions and Nonprofits only
-
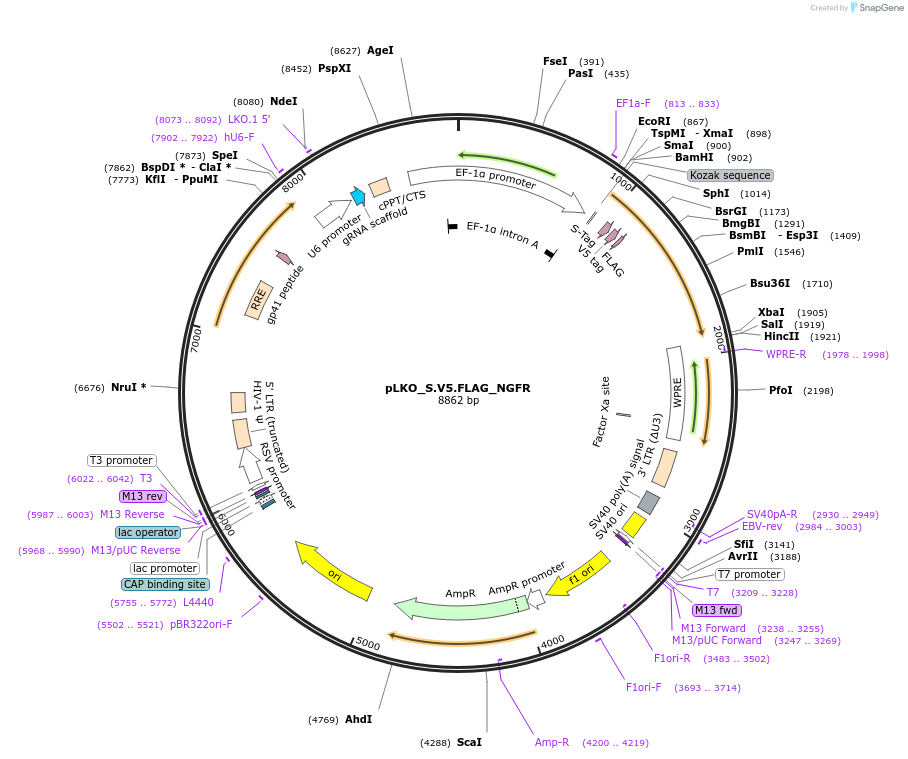
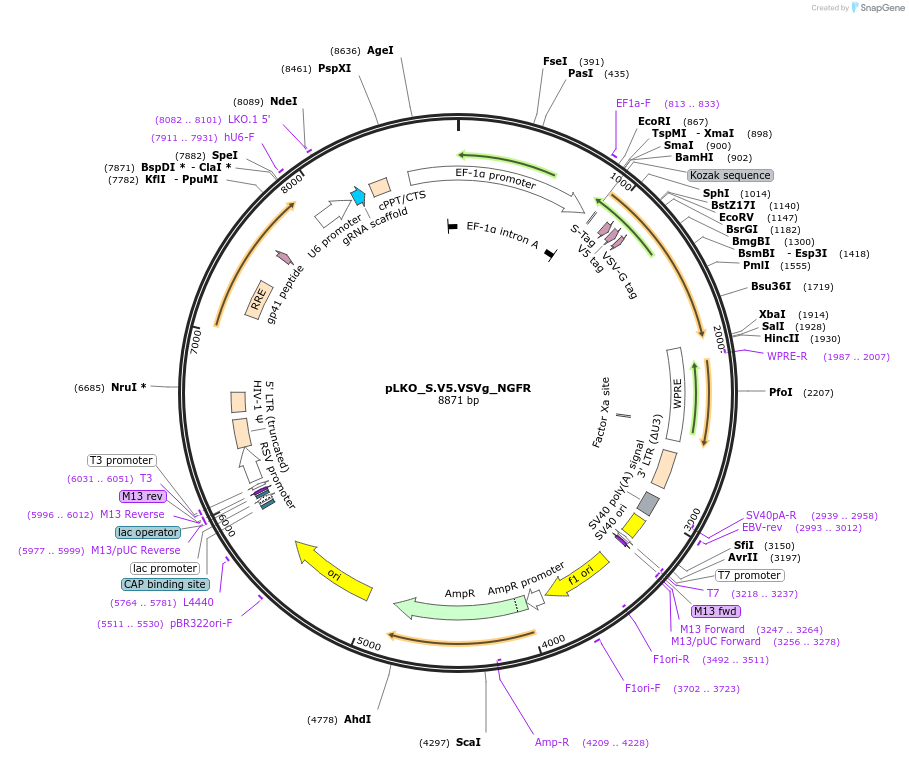
pLKO_S.V5.FLAG_NGFR
Plasmid#158337PurposeProtein Barcode (Pro-Code) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables phenotypic CRISPR screens at a single cell resolution (using cytometry).DepositorInsertPro-Code Tagged human dNGFR (NGFR Human)
UseCRISPR and LentiviralTagsS.V5.FLAGMutationTruncation of the signaling domainPromoterEF1aAvailable SinceSept. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
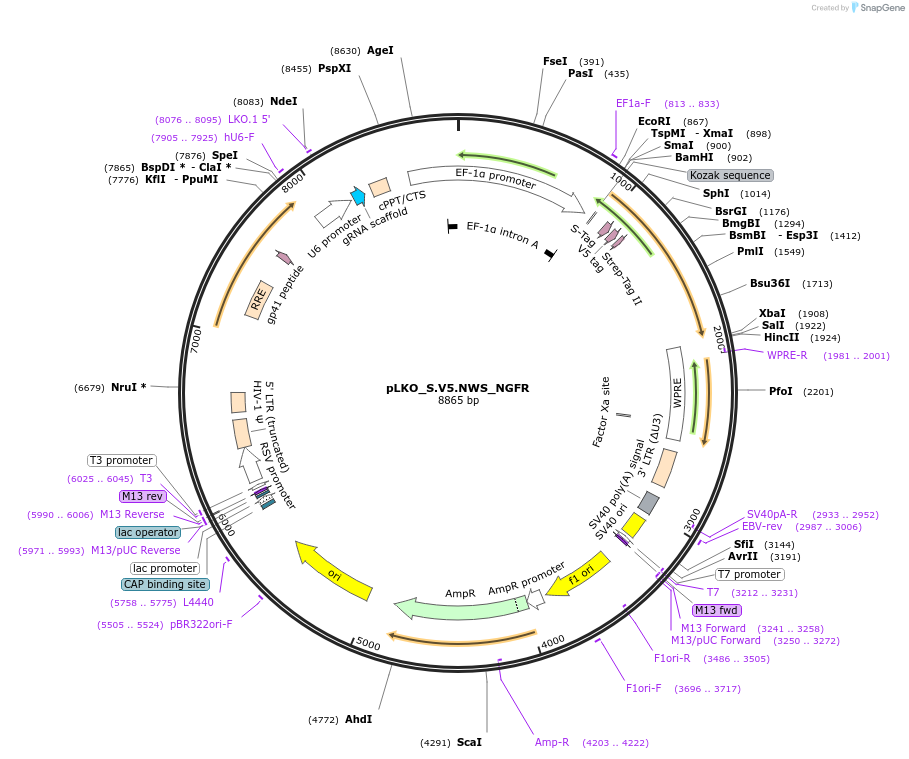
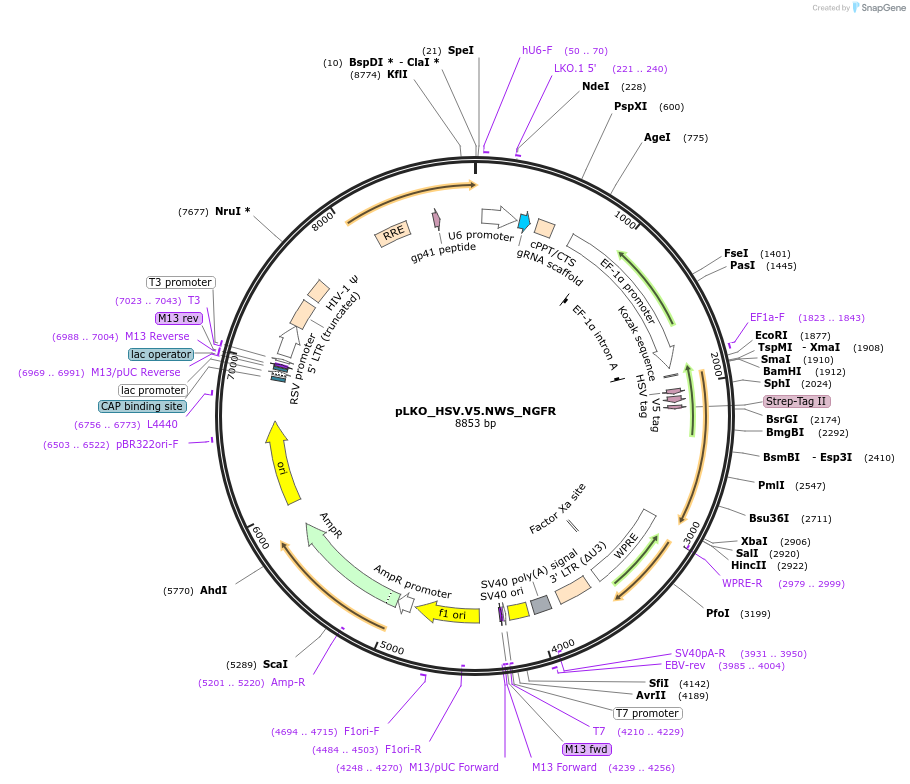
pLKO_S.V5.NWS_NGFR
Plasmid#158338PurposeProtein Barcode (Pro-Code) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables phenotypic CRISPR screens at a single cell resolution (using cytometry).DepositorInsertPro-Code Tagged human dNGFR (NGFR Human)
UseCRISPR and LentiviralTagsS.V5.NWSMutationTruncation of the signaling domainPromoterEF1aAvailable SinceSept. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
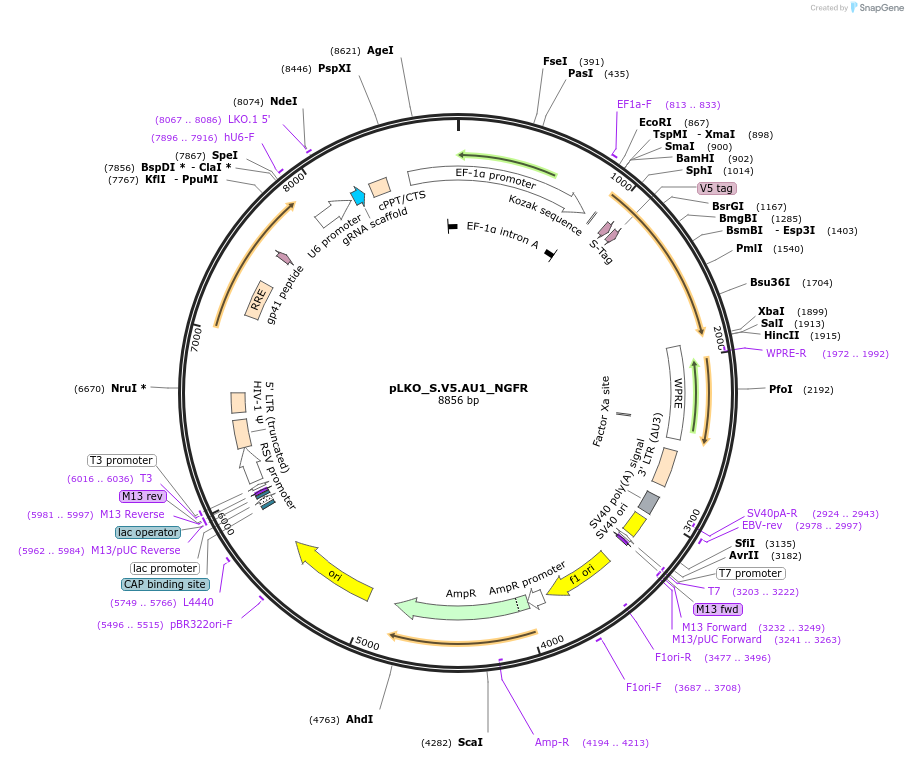
pLKO_S.V5.AU1_NGFR
Plasmid#158340PurposeProtein Barcode (Pro-Code) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables phenotypic CRISPR screens at a single cell resolution (using cytometry).DepositorInsertPro-Code Tagged human dNGFR (NGFR Human)
UseCRISPR and LentiviralTagsS.V5.AU1MutationTruncation of the signaling domainPromoterEF1aAvailable SinceSept. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
pLKO_S.V5.VSVg_NGFR
Plasmid#158341PurposeProtein Barcode (Pro-Code) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables phenotypic CRISPR screens at a single cell resolution (using cytometry).DepositorInsertPro-Code Tagged human dNGFR (NGFR Human)
UseCRISPR and LentiviralTagsS.V5.VSVgMutationTruncation of the signaling domainPromoterEF1aAvailable SinceSept. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
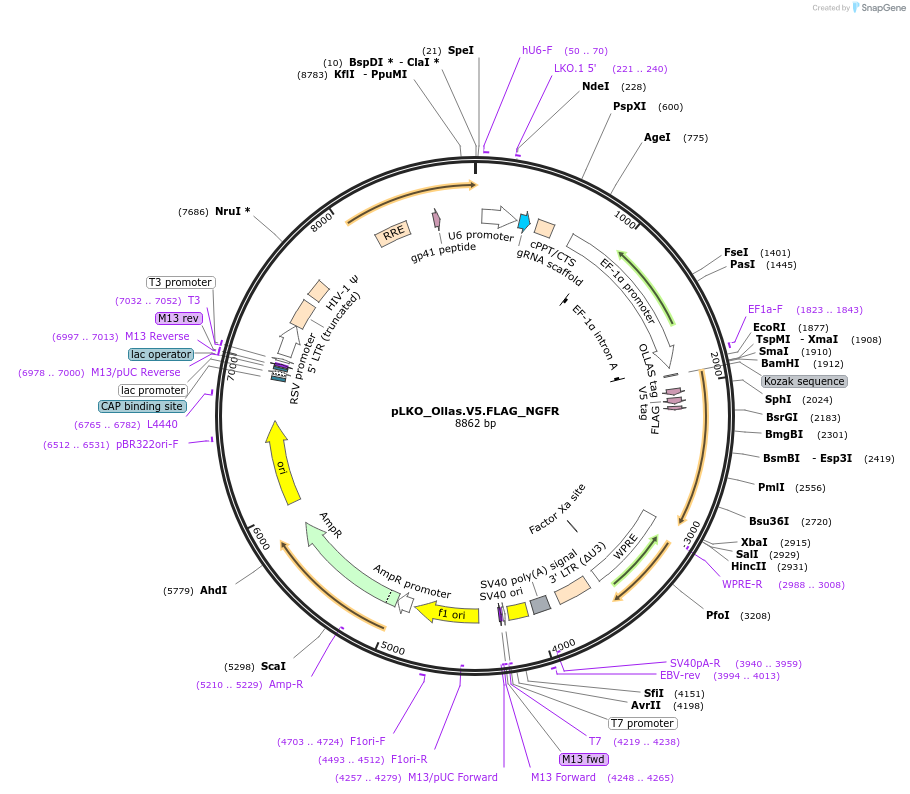
pLKO_Ollas.V5.FLAG_NGFR
Plasmid#158316PurposeProtein Barcode (Pro-Code) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables phenotypic CRISPR screens at a single cell resolution (using cytometry).DepositorInsertPro-Code Tagged human dNGFR (NGFR Human)
UseCRISPR and LentiviralTagsOllas.V5.FLAGMutationTruncation of the signaling domainPromoterEF1aAvailable SinceSept. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
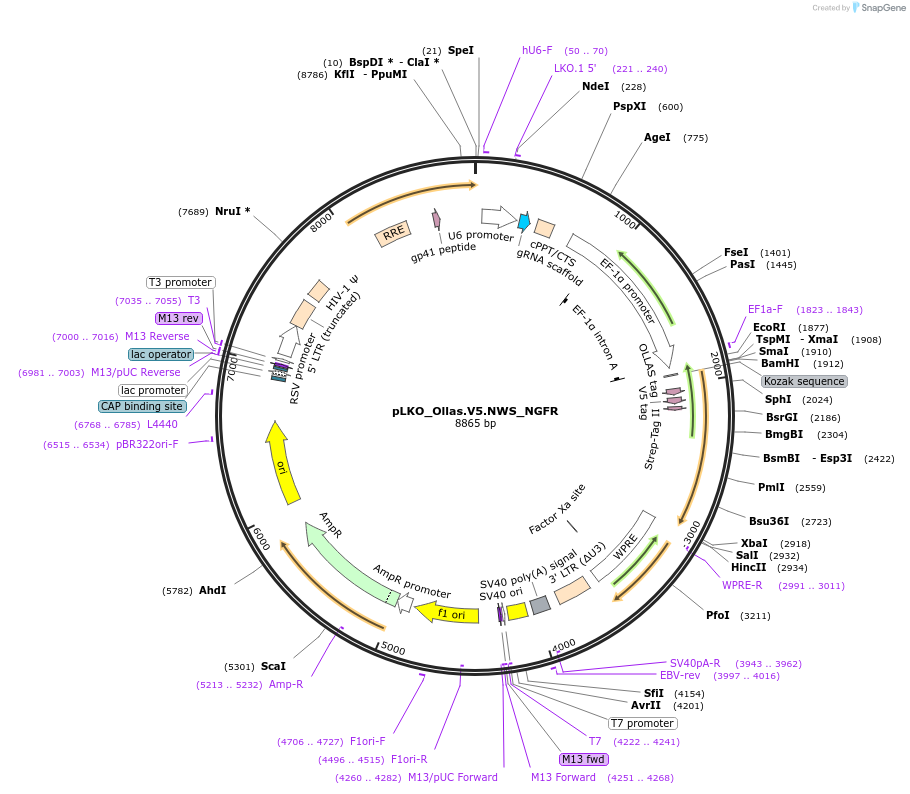
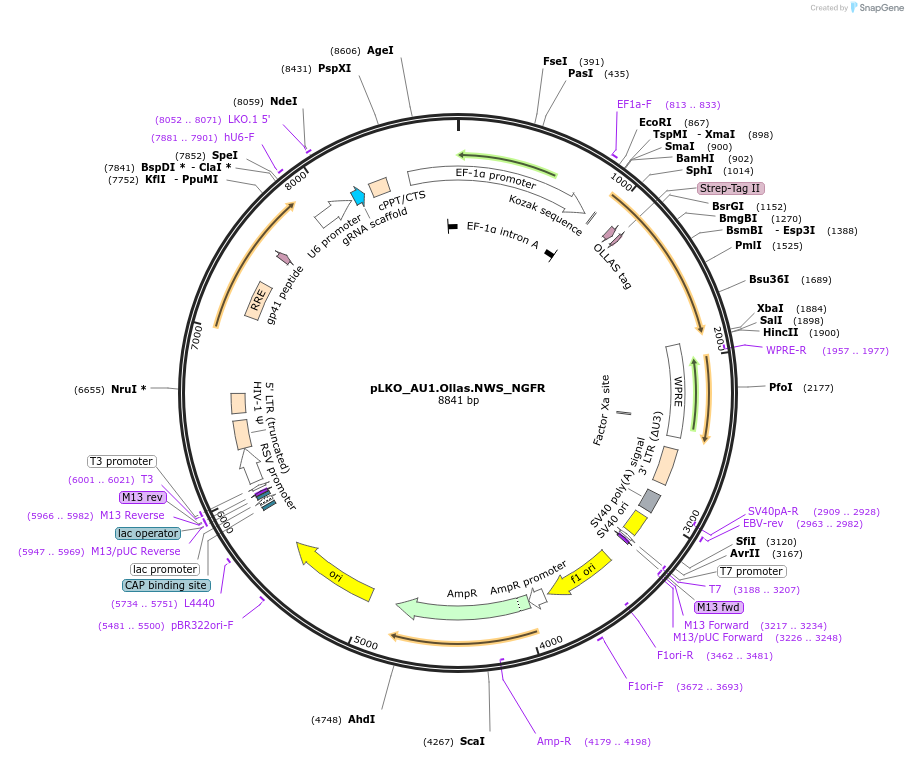
pLKO_Ollas.V5.NWS_NGFR
Plasmid#158317PurposeProtein Barcode (Pro-Code) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables phenotypic CRISPR screens at a single cell resolution (using cytometry).DepositorInsertPro-Code Tagged human dNGFR (NGFR Human)
UseCRISPR and LentiviralTagsOllas.V5.NWSMutationTruncation of the signaling domainPromoterEF1aAvailable SinceSept. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
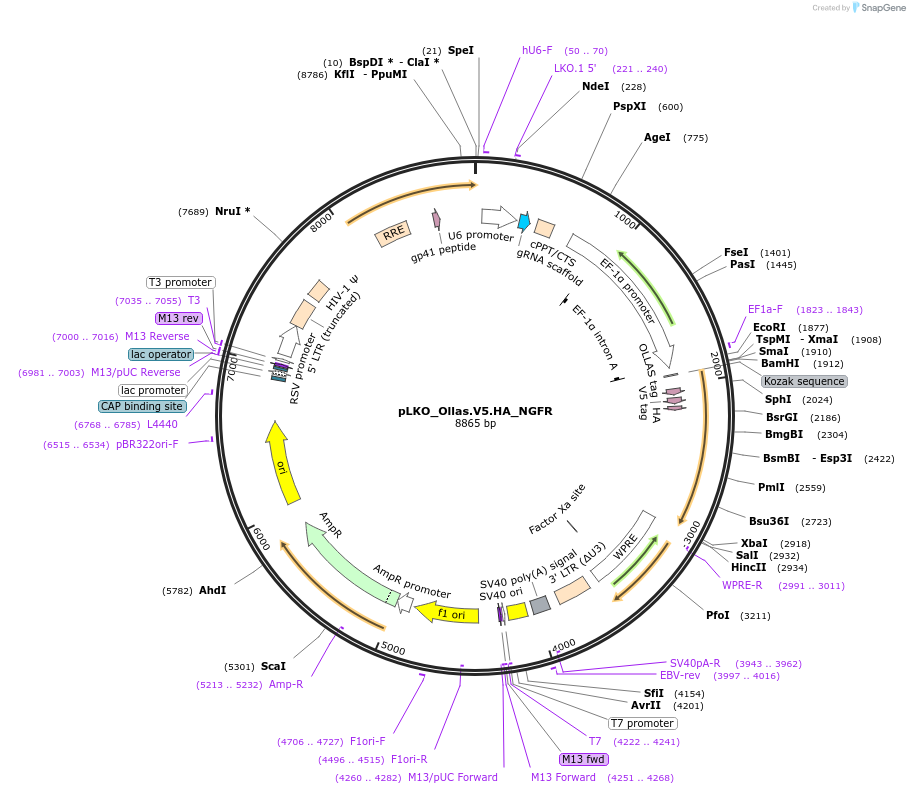
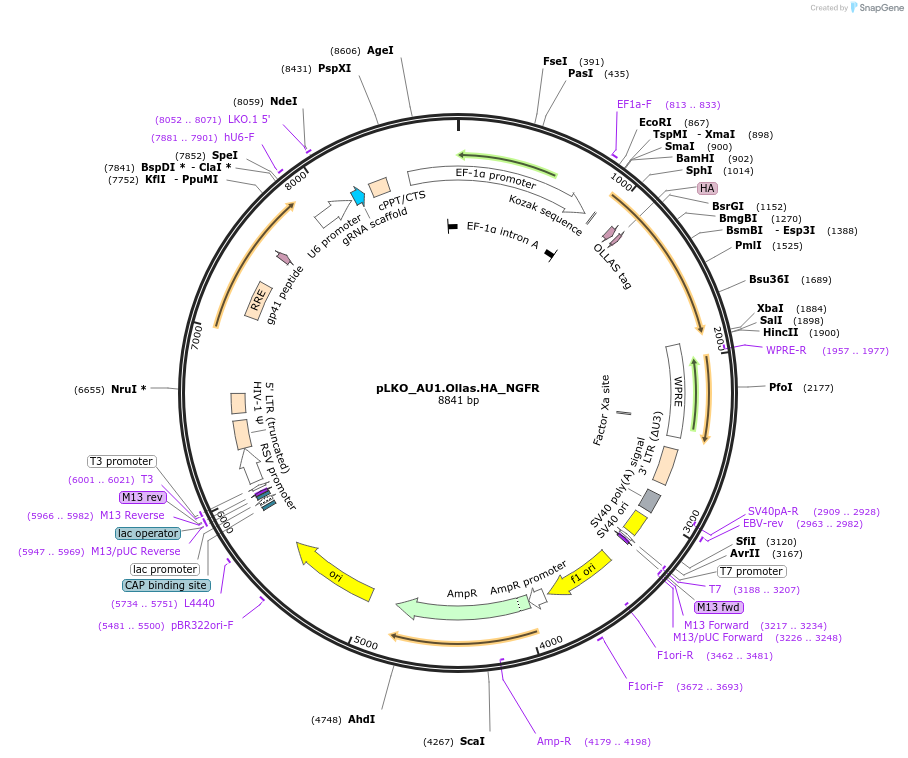
pLKO_Ollas.V5.HA_NGFR
Plasmid#158318PurposeProtein Barcode (Pro-Code) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables phenotypic CRISPR screens at a single cell resolution (using cytometry).DepositorInsertPro-Code Tagged human dNGFR (NGFR Human)
UseCRISPR and LentiviralTagsOllas.V5.HAMutationTruncation of the signaling domainPromoterEF1aAvailable SinceSept. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
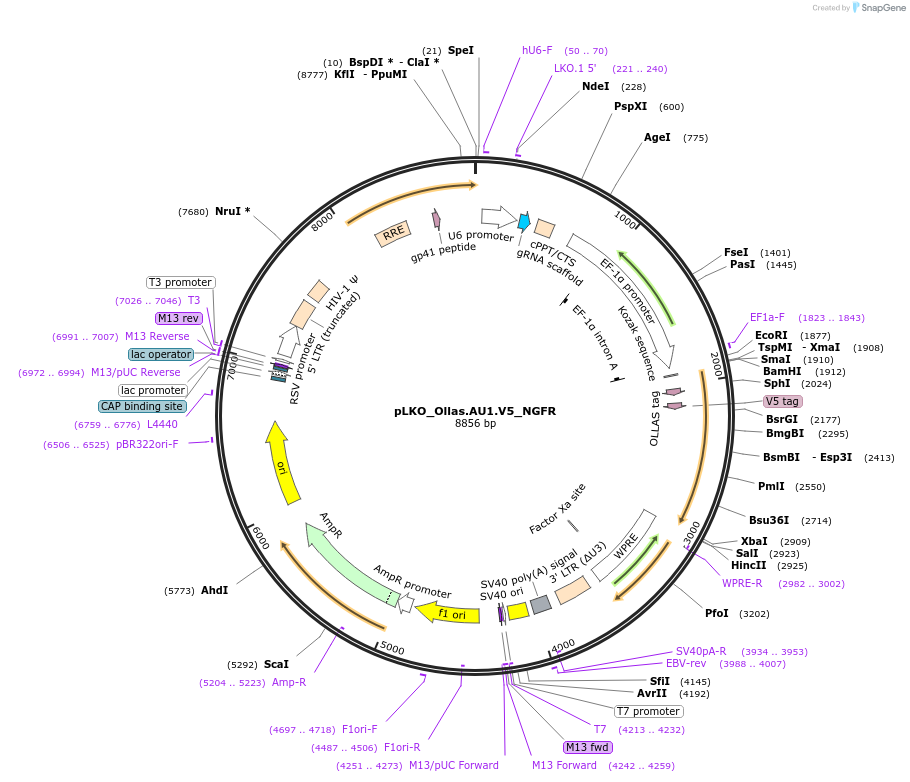
pLKO_Ollas.AU1.V5_NGFR
Plasmid#158319PurposeProtein Barcode (Pro-Code) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables phenotypic CRISPR screens at a single cell resolution (using cytometry).DepositorInsertPro-Code Tagged human dNGFR (NGFR Human)
UseCRISPR and LentiviralTagsOllas.AU1.V5MutationTruncation of the signaling domainPromoterEF1aAvailable SinceSept. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
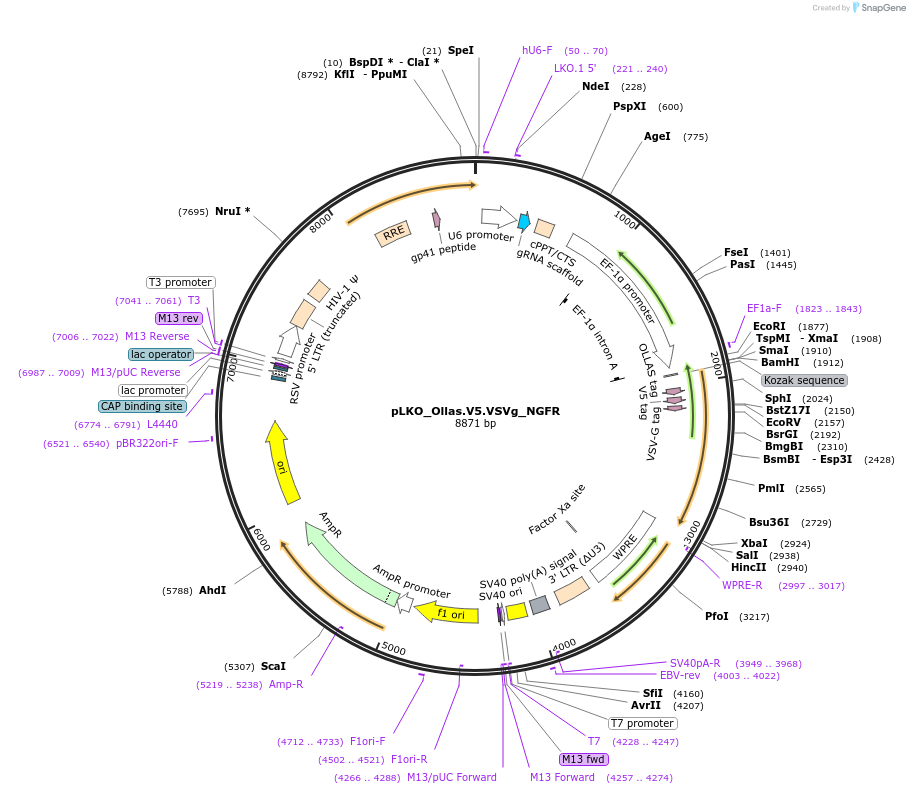
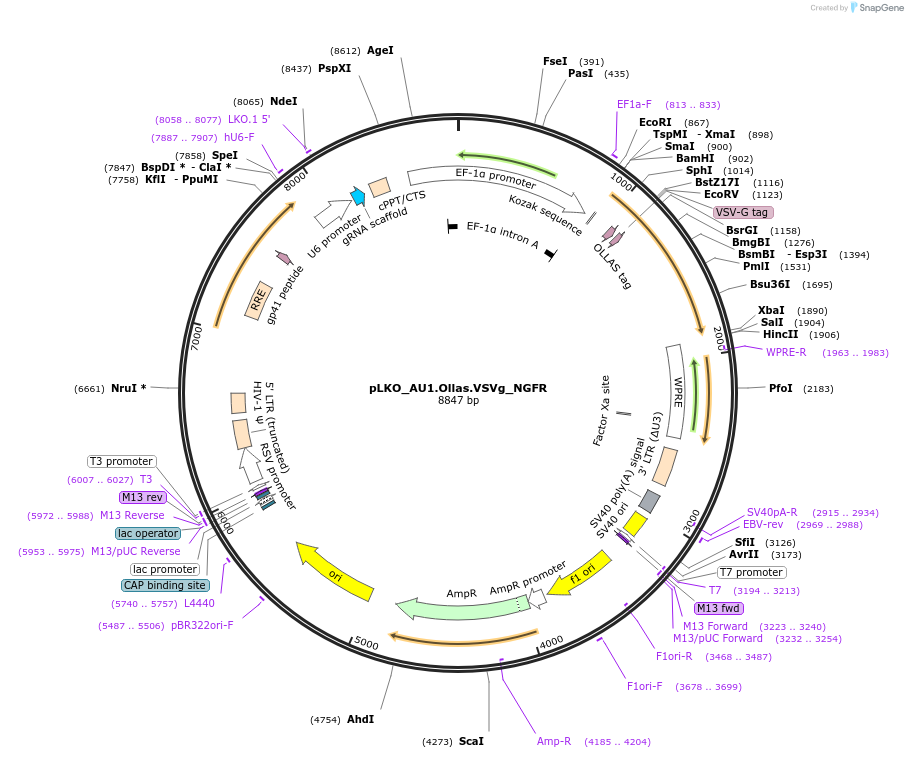
pLKO_Ollas.V5.VSVg_NGFR
Plasmid#158320PurposeProtein Barcode (Pro-Code) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables phenotypic CRISPR screens at a single cell resolution (using cytometry).DepositorInsertPro-Code Tagged human dNGFR (NGFR Human)
UseCRISPR and LentiviralTagsOllas.V5.VSVgMutationTruncation of the signaling domainPromoterEF1aAvailable SinceSept. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
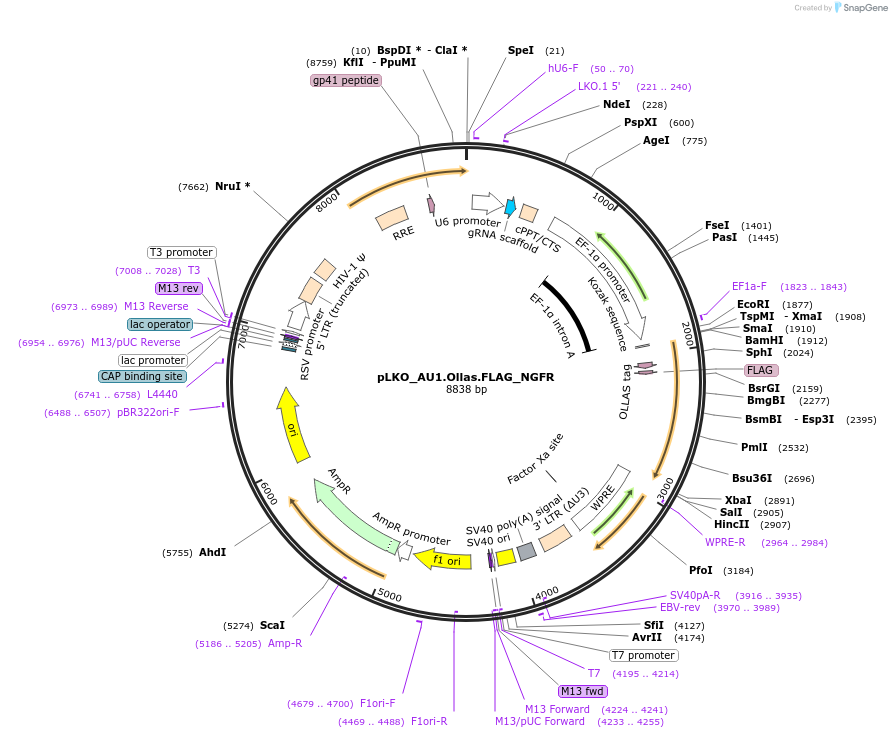
pLKO_AU1.Ollas.FLAG_NGFR
Plasmid#158323PurposeProtein Barcode (Pro-Code) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables phenotypic CRISPR screens at a single cell resolution (using cytometry).DepositorInsertPro-Code Tagged human dNGFR (NGFR Human)
UseCRISPR and LentiviralTagsAU1.Ollas.FLAGMutationTruncation of the signaling domainPromoterEF1aAvailable SinceSept. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
pLKO_AU1.Ollas.NWS_NGFR
Plasmid#158326PurposeProtein Barcode (Pro-Code) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables phenotypic CRISPR screens at a single cell resolution (using cytometry).DepositorInsertPro-Code Tagged human dNGFR (NGFR Human)
UseCRISPR and LentiviralTagsAU1.Ollas.NWSMutationTruncation of the signaling domainPromoterEF1aAvailable SinceSept. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
pLKO_AU1.Ollas.HA_NGFR
Plasmid#158328PurposeProtein Barcode (Pro-Code) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables phenotypic CRISPR screens at a single cell resolution (using cytometry).DepositorInsertPro-Code Tagged human dNGFR (NGFR Human)
UseCRISPR and LentiviralTagsAU1.Ollas.HAMutationTruncation of the signaling domainPromoterEF1aAvailable SinceSept. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
pLKO_AU1.Ollas.VSVg_NGFR
Plasmid#158330PurposeProtein Barcode (Pro-Code) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables phenotypic CRISPR screens at a single cell resolution (using cytometry).DepositorInsertPro-Code Tagged human dNGFR (NGFR Human)
UseCRISPR and LentiviralTagsAU1.Ollas.VSVgMutationTruncation of the signaling domainPromoterEF1aAvailable SinceSept. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
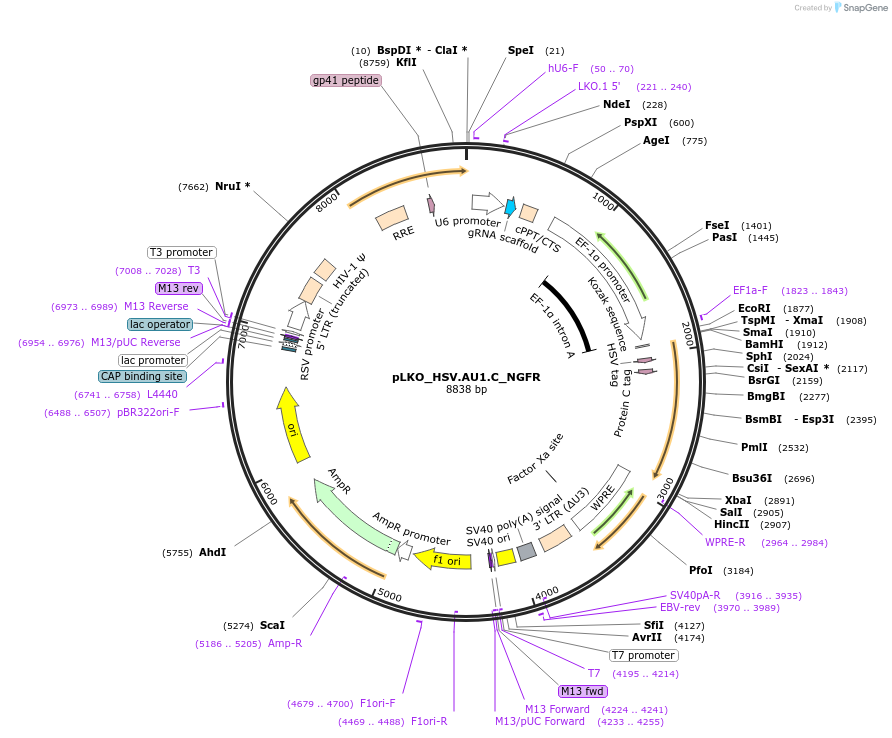
pLKO_HSV.AU1.C_NGFR
Plasmid#158286PurposeProtein Barcode (Pro-Code) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables phenotypic CRISPR screens at a single cell resolution (using cytometry).DepositorInsertPro-Code Tagged human dNGFR (NGFR Human)
UseCRISPR and LentiviralTagsHSV.AU1.CMutationTruncation of the signaling domainPromoterEF1aAvailable SinceSept. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
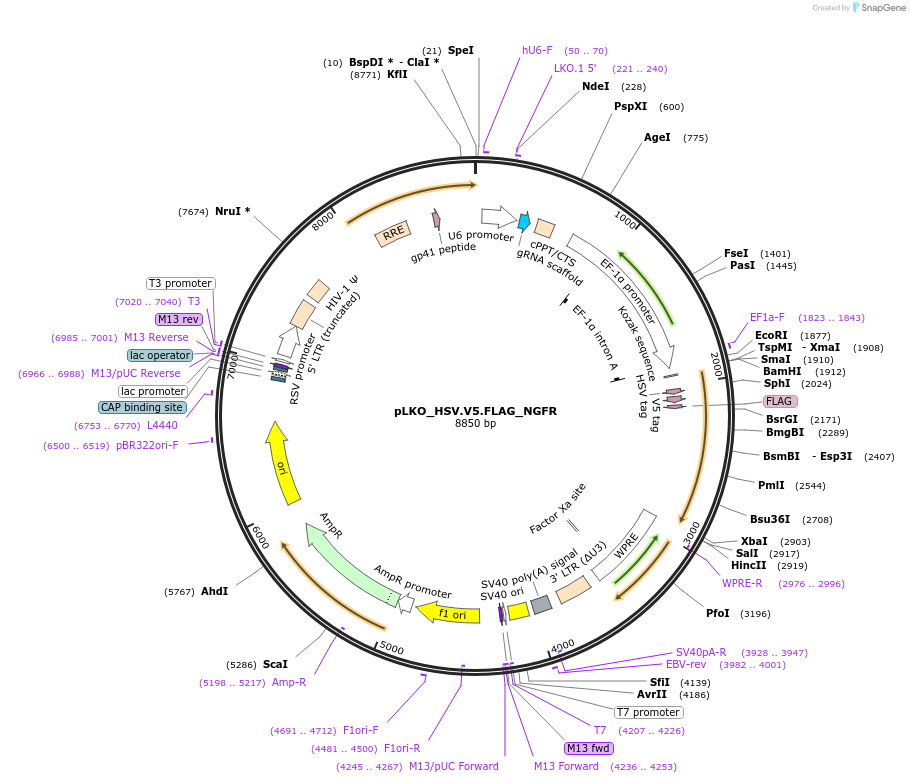
pLKO_HSV.V5.FLAG_NGFR
Plasmid#158288PurposeProtein Barcode (Pro-Code) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables phenotypic CRISPR screens at a single cell resolution (using cytometry).DepositorInsertPro-Code Tagged human dNGFR (NGFR Human)
UseCRISPR and LentiviralTagsHSV.V5.FLAGMutationTruncation of the signaling domainPromoterEF1aAvailable SinceSept. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
pLKO_HSV.V5.NWS_NGFR
Plasmid#158289PurposeProtein Barcode (Pro-Code) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables phenotypic CRISPR screens at a single cell resolution (using cytometry).DepositorInsertPro-Code Tagged human dNGFR (NGFR Human)
UseCRISPR and LentiviralTagsHSV.V5.NWSMutationTruncation of the signaling domainPromoterEF1aAvailable SinceSept. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
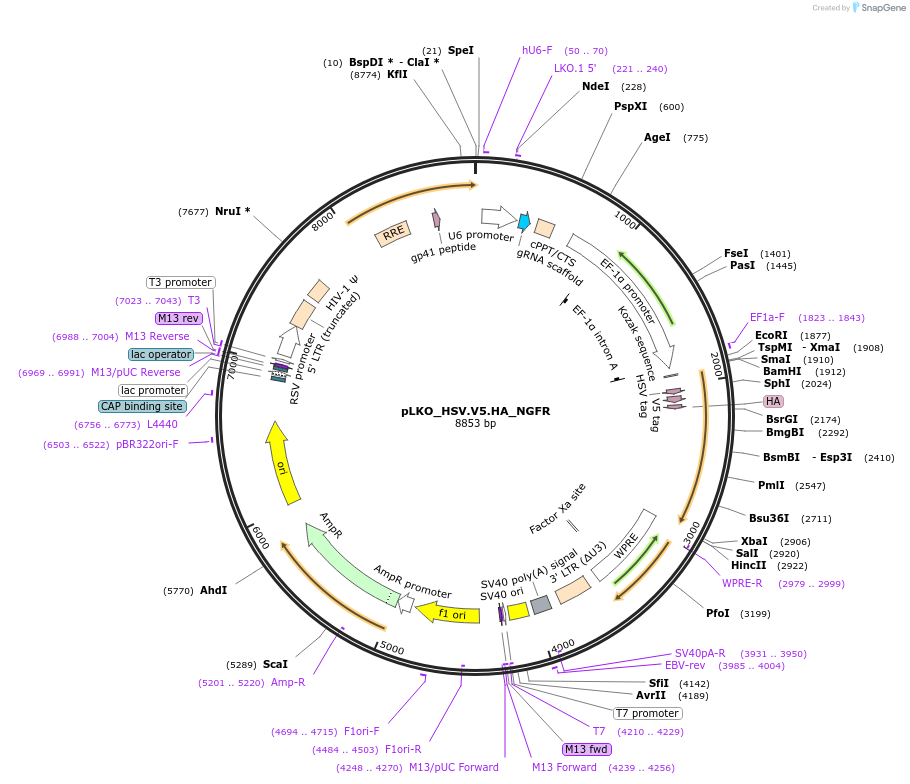
pLKO_HSV.V5.HA_NGFR
Plasmid#158290PurposeProtein Barcode (Pro-Code) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables phenotypic CRISPR screens at a single cell resolution (using cytometry).DepositorInsertPro-Code Tagged human dNGFR (NGFR Human)
UseCRISPR and LentiviralTagsHSV.V5.HAMutationTruncation of the signaling domainPromoterEF1aAvailable SinceSept. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
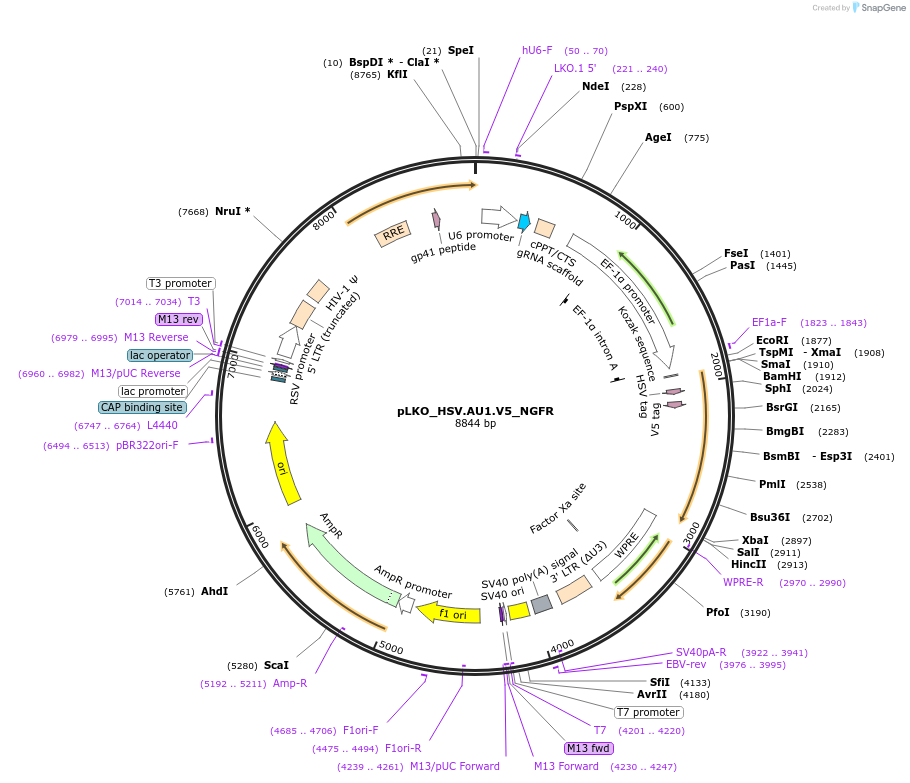
pLKO_HSV.AU1.V5_NGFR
Plasmid#158291PurposeProtein Barcode (Pro-Code) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables phenotypic CRISPR screens at a single cell resolution (using cytometry).DepositorInsertPro-Code Tagged human dNGFR (NGFR Human)
UseCRISPR and LentiviralTagsHSV.AU1.V5MutationTruncation of the signaling domainPromoterEF1aAvailable SinceSept. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
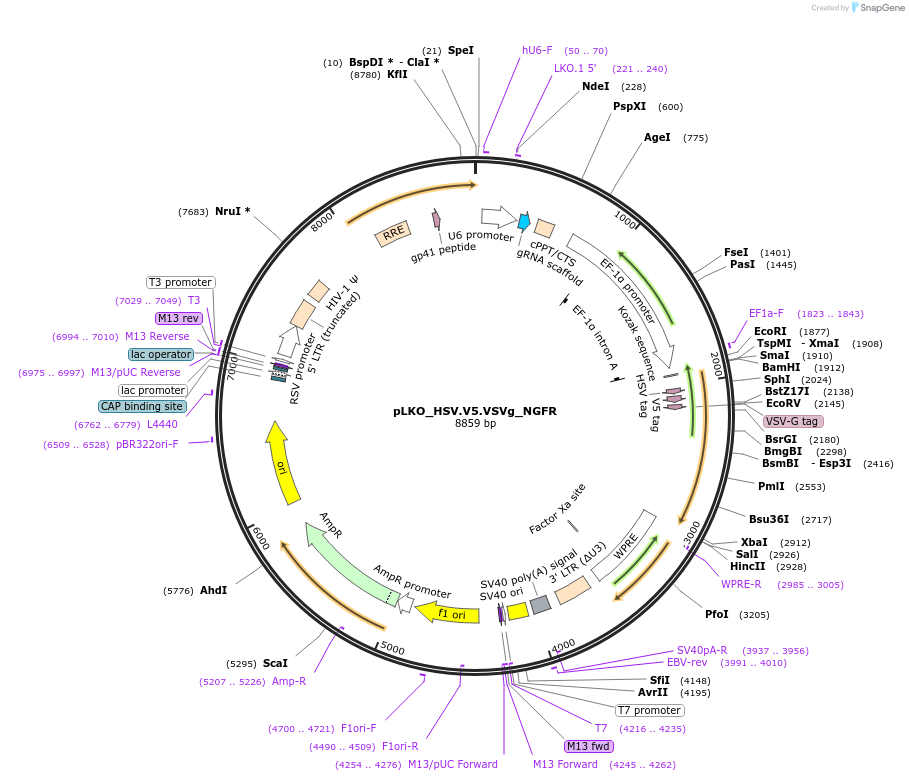
pLKO_HSV.V5.VSVg_NGFR
Plasmid#158292PurposeProtein Barcode (Pro-Code) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables phenotypic CRISPR screens at a single cell resolution (using cytometry).DepositorInsertPro-Code Tagged human dNGFR (NGFR Human)
UseCRISPR and LentiviralTagsHSV.V5.VSVgMutationTruncation of the signaling domainPromoterEF1aAvailable SinceSept. 3, 2021AvailabilityAcademic Institutions and Nonprofits only