We narrowed to 81,061 results for: myc
-
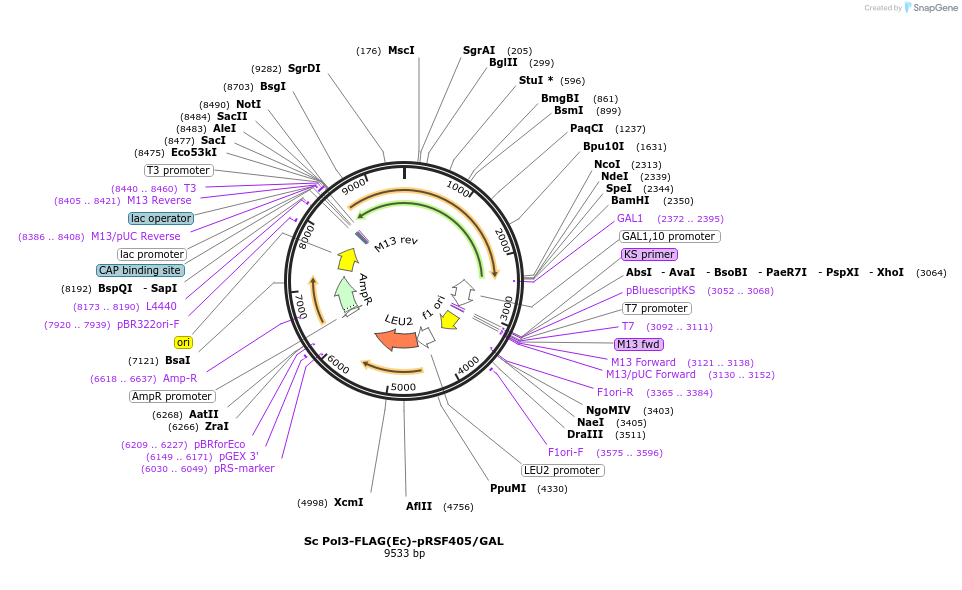
Plasmid#239198PurposeOverexpress Sc Pol3(C-terminal 3X FLAG tag) when integrated into yeast (S. cer)DepositorInsertPol3 (POL3 Budding Yeast)
Tags3X FLAGExpressionYeastMutationE. coli codon bias (synthetic gene), but same ami…Available SinceJune 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
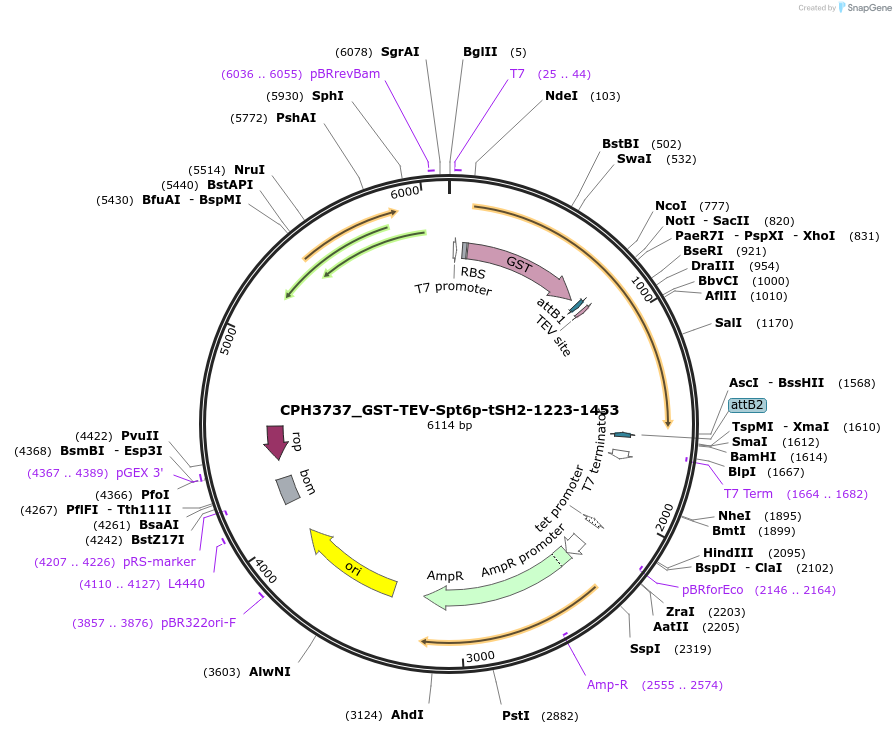
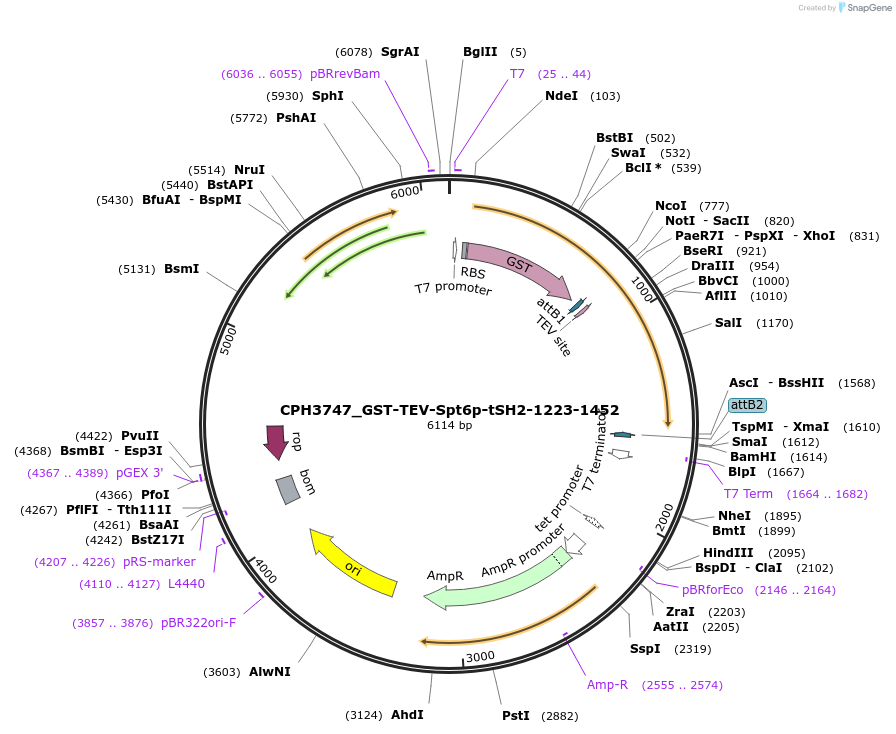
CPH3737_GST-TEV-Spt6p-tSH2-1223-1453
Plasmid#227087PurposeBacterial expression of K1355A/K1435A double-mutant GST-tagged Spt6 tSH2 domain (1223-1453)DepositorInsertspt6-tsh2 (SPT6 Budding Yeast)
TagsGST-TEV_cleavage_siteExpressionBacterialMutationK1355A, K1435APromoterT7Available SinceApril 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
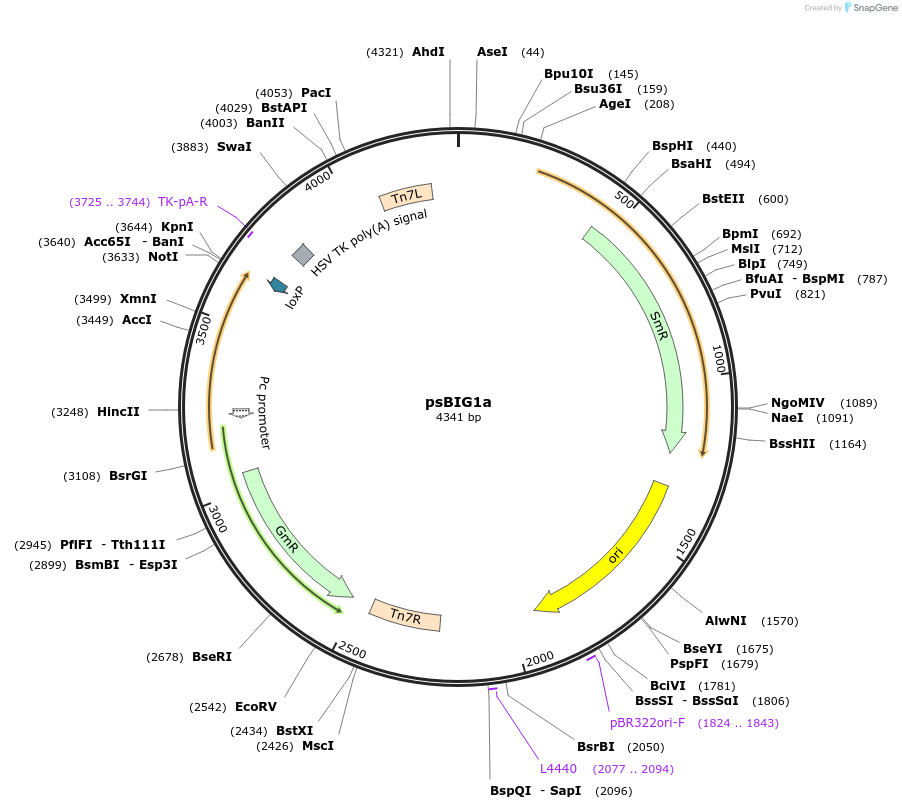
psBIG1a
Plasmid#229957PurposebigMamAct is evolution of biGBac for mammalian cells. psBIG plasmids are recipient of multi-assembly step by Gibson cloningDepositorTypeEmpty backboneUseCRISPRExpressionMammalianAvailable SinceFeb. 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
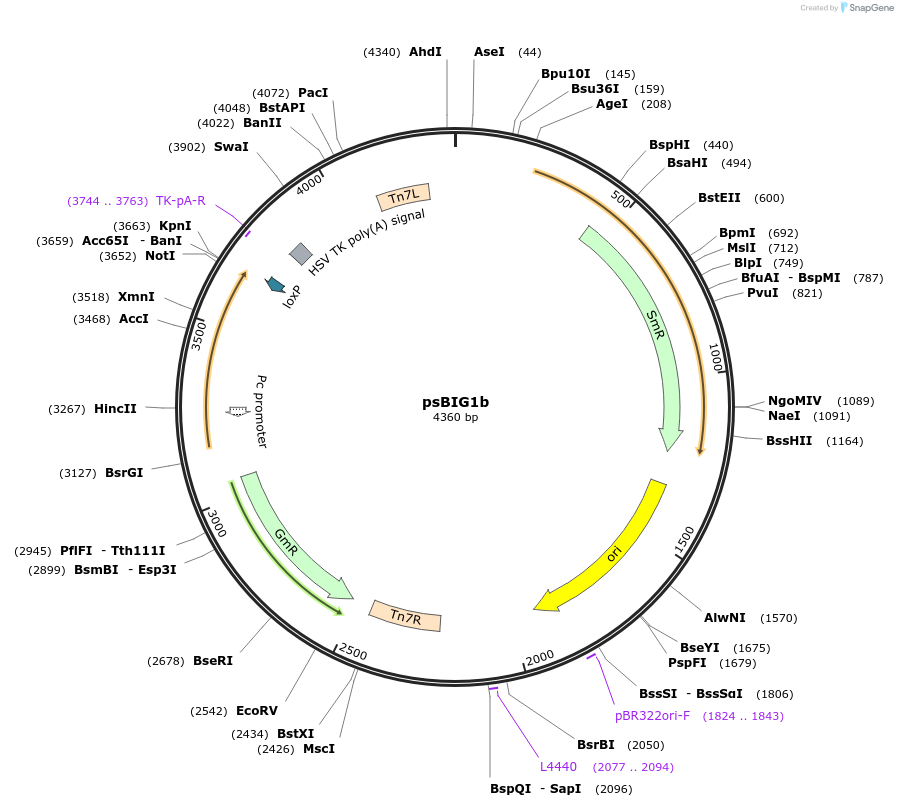
psBIG1b
Plasmid#229958PurposebigMamAct is evolution of biGBac for mammalian cells. psBIG plasmids are recipient of multi-assembly step by Gibson cloningDepositorTypeEmpty backboneUseCRISPRExpressionMammalianAvailable SinceFeb. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
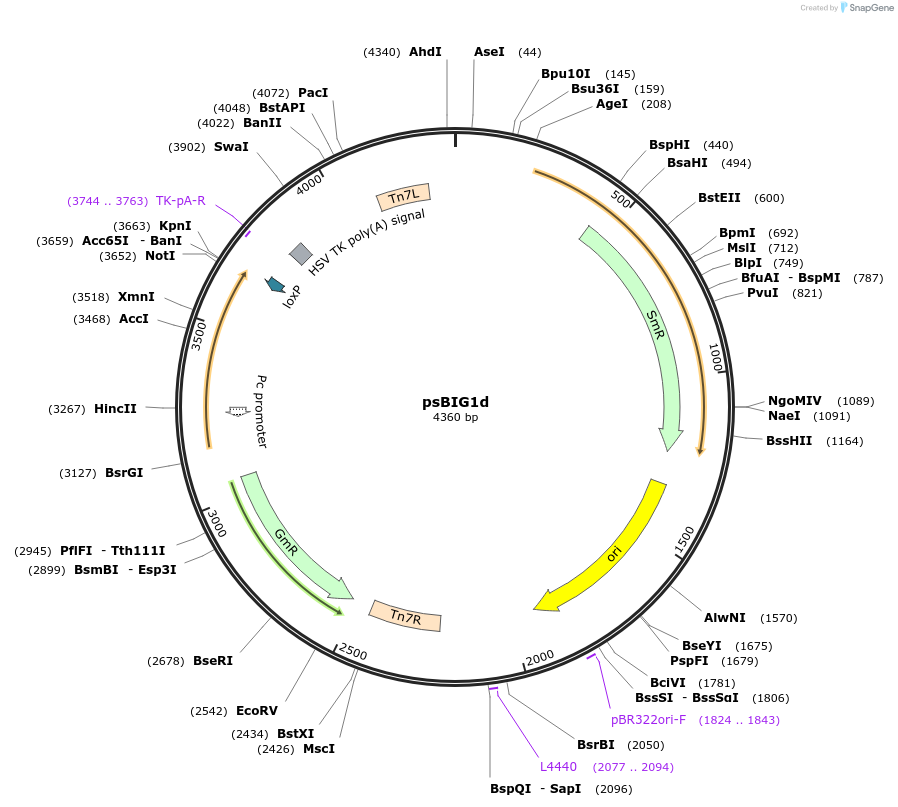
psBIG1d
Plasmid#229960PurposebigMamAct is evolution of biGBac for mammalian cells. psBIG plasmids are recipient of multi-assembly step by Gibson cloningDepositorTypeEmpty backboneUseCRISPRExpressionMammalianAvailable SinceFeb. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
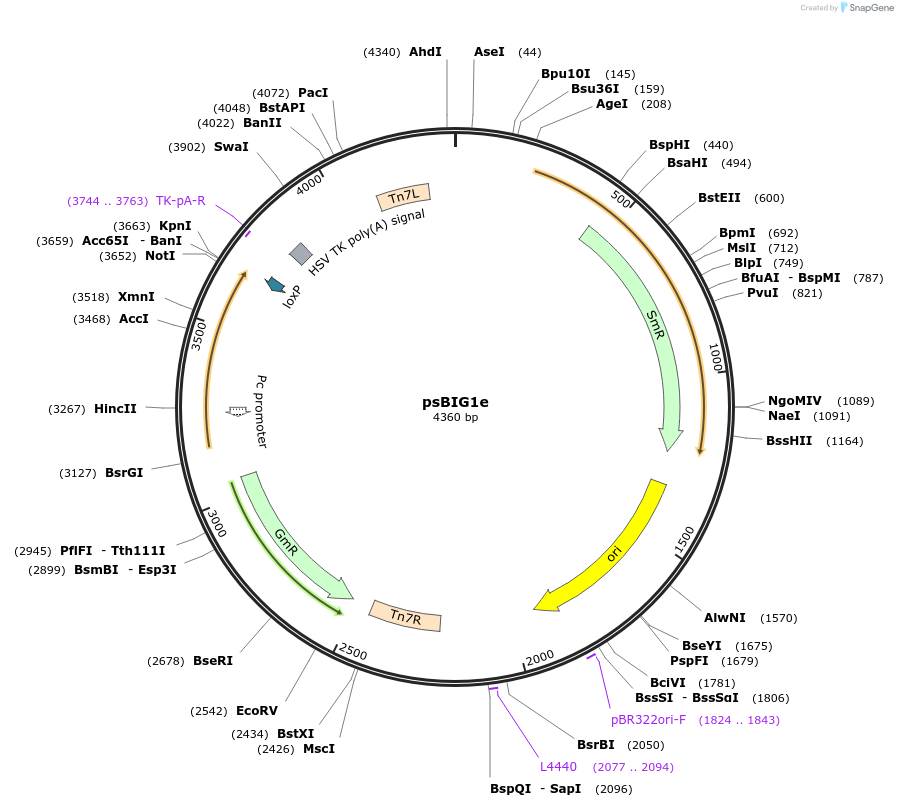
psBIG1e
Plasmid#229961PurposebigMamAct is evolution of biGBac for mammalian cells. psBIG plasmids are recipient of multi-assembly step by Gibson cloningDepositorTypeEmpty backboneUseCRISPRExpressionMammalianAvailable SinceFeb. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
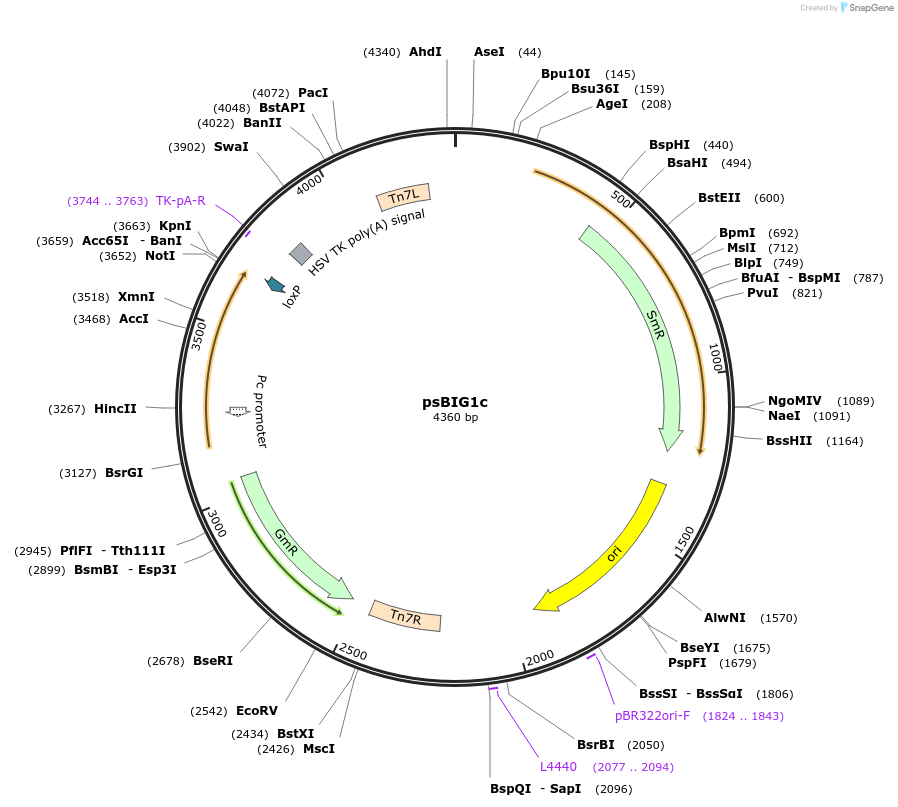
psBIG1c
Plasmid#229959PurposebigMamAct is evolution of biGBac for mammalian cells. psBIG plasmids are recipient of multi-assembly step by Gibson cloningDepositorTypeEmpty backboneUseCRISPRExpressionMammalianAvailable SinceFeb. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
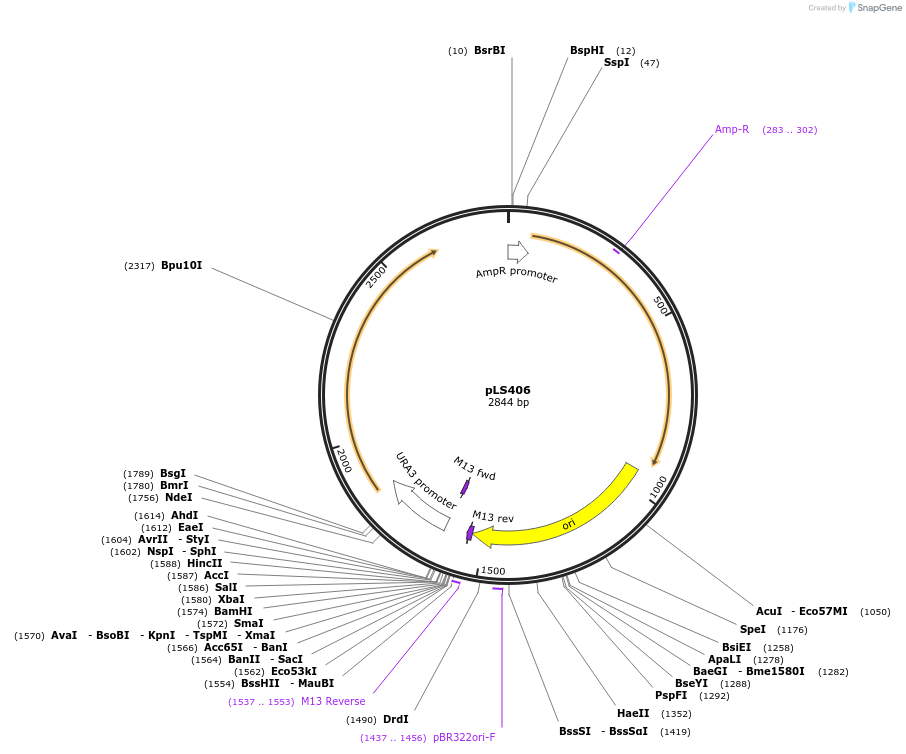
pLS406
Plasmid#231078PurposeMinimal integrating shuttle vector depleted of restriction sites outside the polylinker region, yeast selection marker URA3DepositorTypeEmpty backboneUseIntegratingExpressionBacterial and YeastAvailable SinceFeb. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
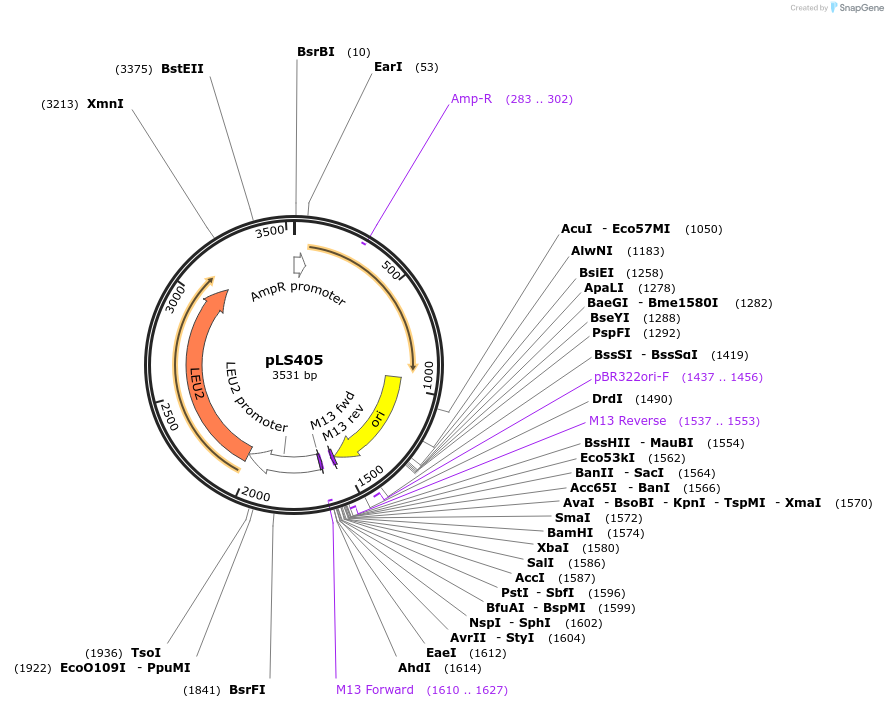
pLS405
Plasmid#231077PurposeMinimal integrating shuttle vector depleted of restriction sites outside the polylinker region, yeast selection marker LEU2DepositorTypeEmpty backboneUseIntegratingExpressionBacterial and YeastAvailable SinceJan. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
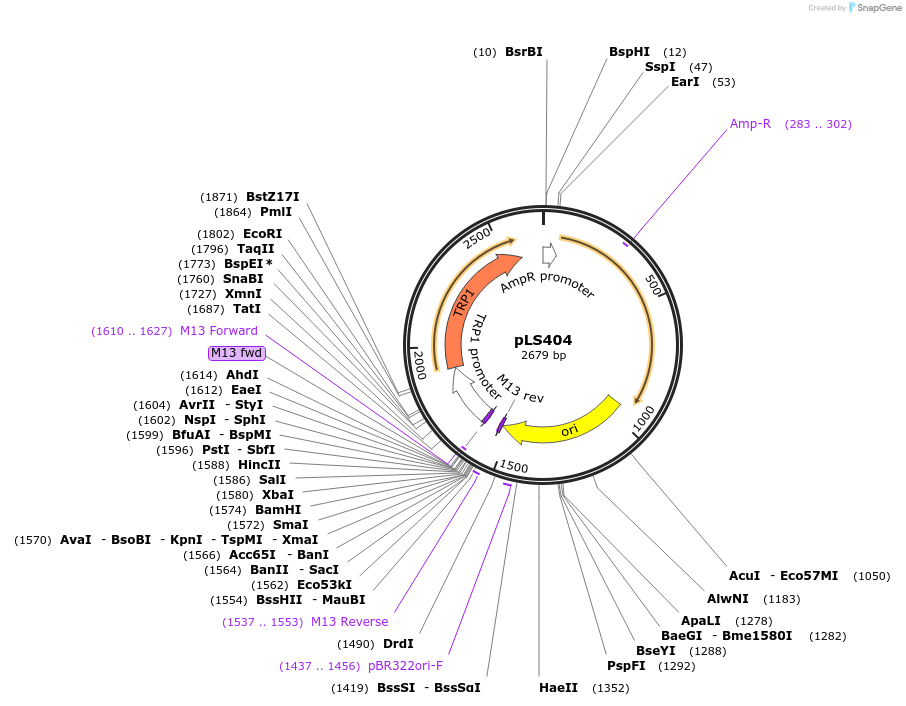
pLS404
Plasmid#231076PurposeMinimal integrating shuttle vector depleted of restriction sites outside the polylinker region, yeast selection marker TRP1DepositorTypeEmpty backboneUseIntegratingExpressionBacterial and YeastAvailable SinceJan. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
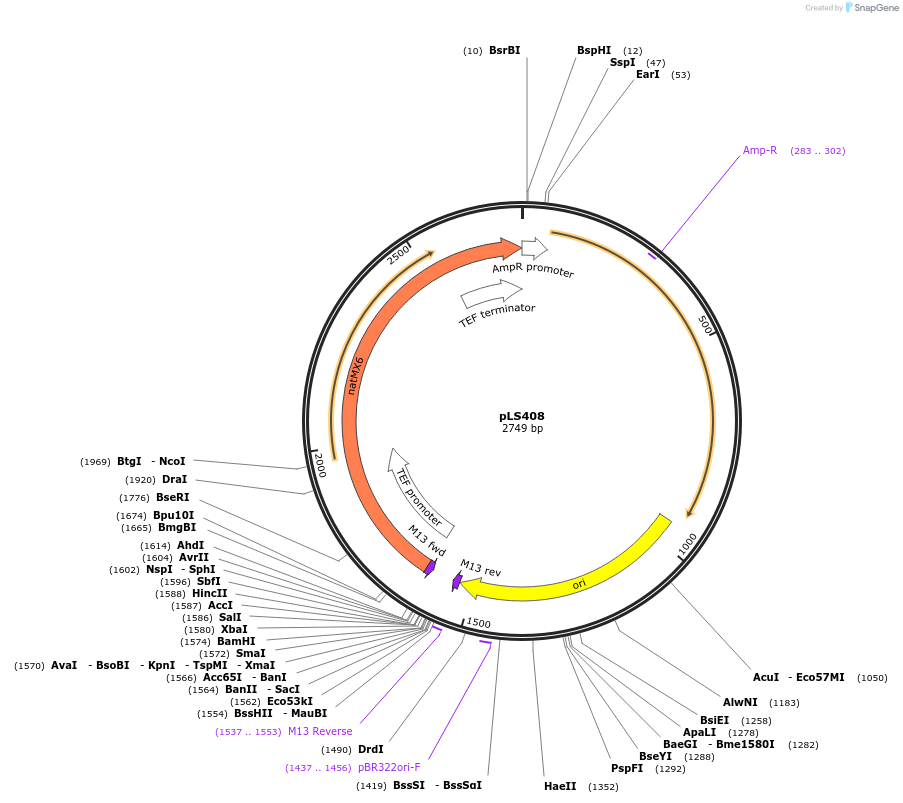
pLS408
Plasmid#231079PurposeMinimal integrating shuttle vector depleted of restriction sites outside the polylinker region, yeast selection marker natMX6DepositorTypeEmpty backboneUseIntegratingExpressionBacterial and YeastAvailable SinceJan. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
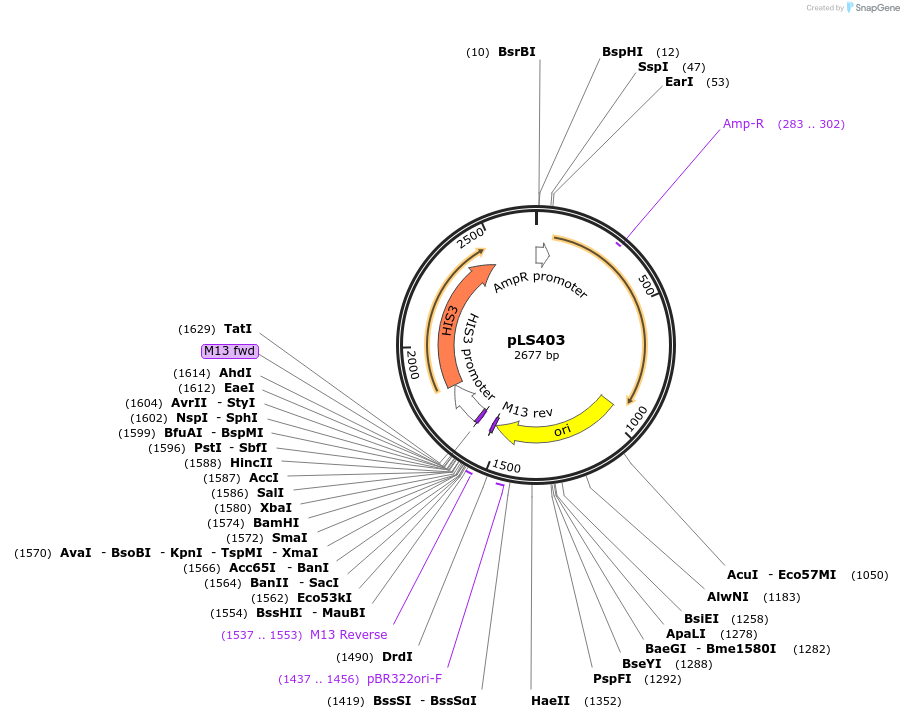
pLS403
Plasmid#231075PurposeMinimal integrating shuttle vector depleted of restriction sites outside the polylinker region, yeast selection marker HIS3DepositorTypeEmpty backboneUseIntegratingExpressionBacterial and YeastAvailable SinceJan. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
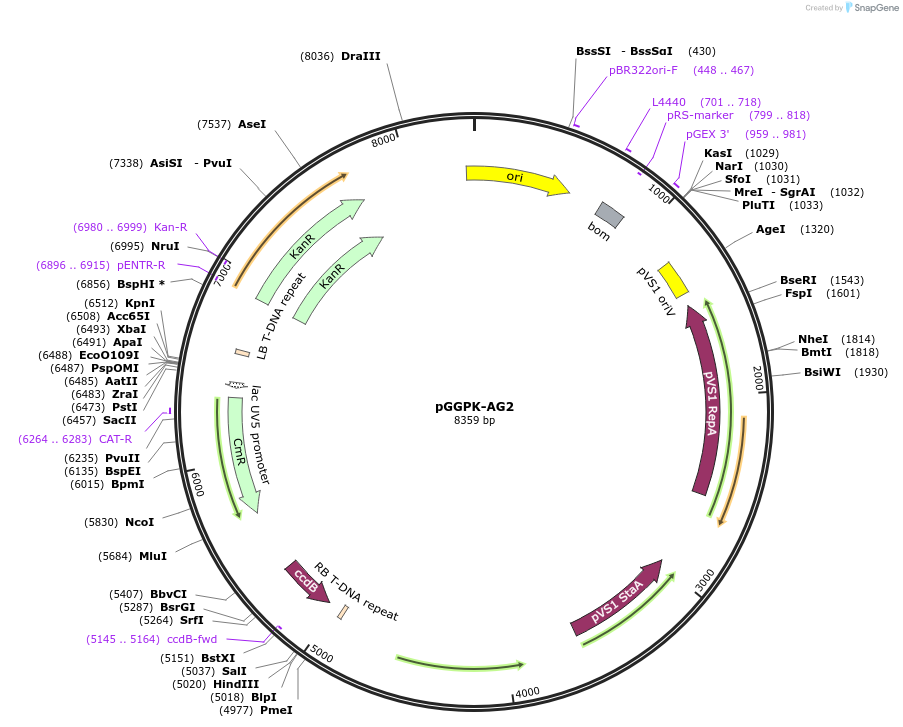
pGGPK-AG2
Plasmid#222027PurposeEmpty Kanamycin resistant GreenGate destination vector based on pGGP-AG.DepositorTypeEmpty backboneUseSynthetic Biology; Greengate compatible cloning v…Available SinceDec. 16, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
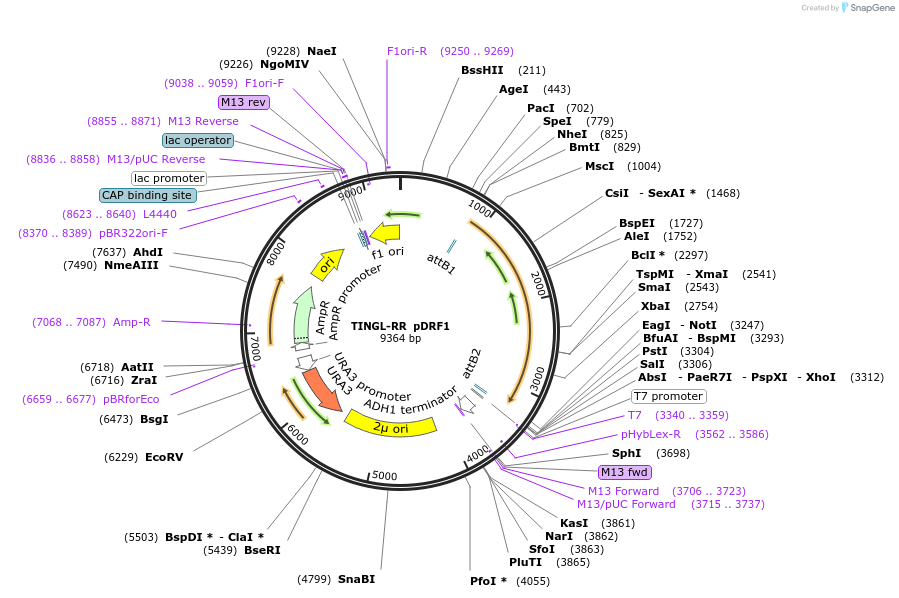
TINGL-RR pDRF1
Plasmid#226435PurposeExpresses TINGL-RR (mTq2-based glucose sensor - O2A - ymScarletI) in yeastDepositorInsertTINGL-O2A-ymScarletI
ExpressionYeastPromoterPMA1Available SinceDec. 2, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
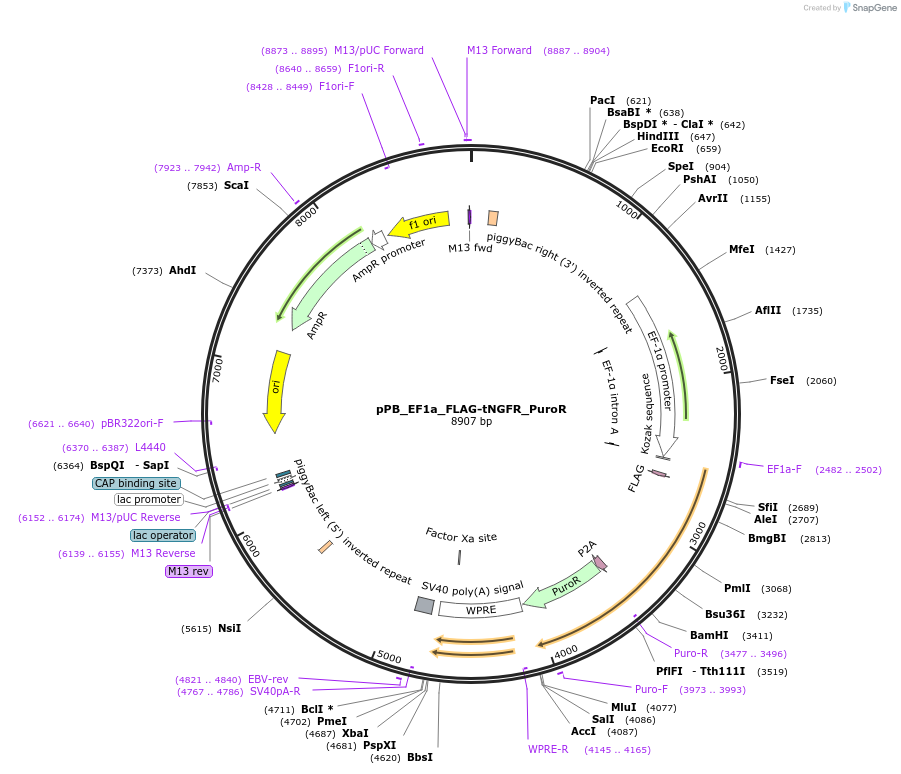
pPB_EF1a_FLAG-tNGFR_PuroR
Plasmid#212651PurposepiggyBac vector to express FLAG-tNGFR with puromycin resistanceDepositorInserttNGFR
UsePiggybacTagsFLAGExpressionMammalianPromoterEF1aAvailable SinceNov. 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
CPH3747_GST-TEV-Spt6p-tSH2-1223-1452
Plasmid#227086PurposeBacterial expression of R1282H single-mutant GST-tagged Spt6 tSH2 domain (1223-1452)DepositorInsertspt6-tsh2 (SPT6 Budding Yeast)
TagsGST-TEV_cleavage_siteExpressionBacterialMutationR1282HPromoterT7Available SinceNov. 4, 2024AvailabilityAcademic Institutions and Nonprofits only -
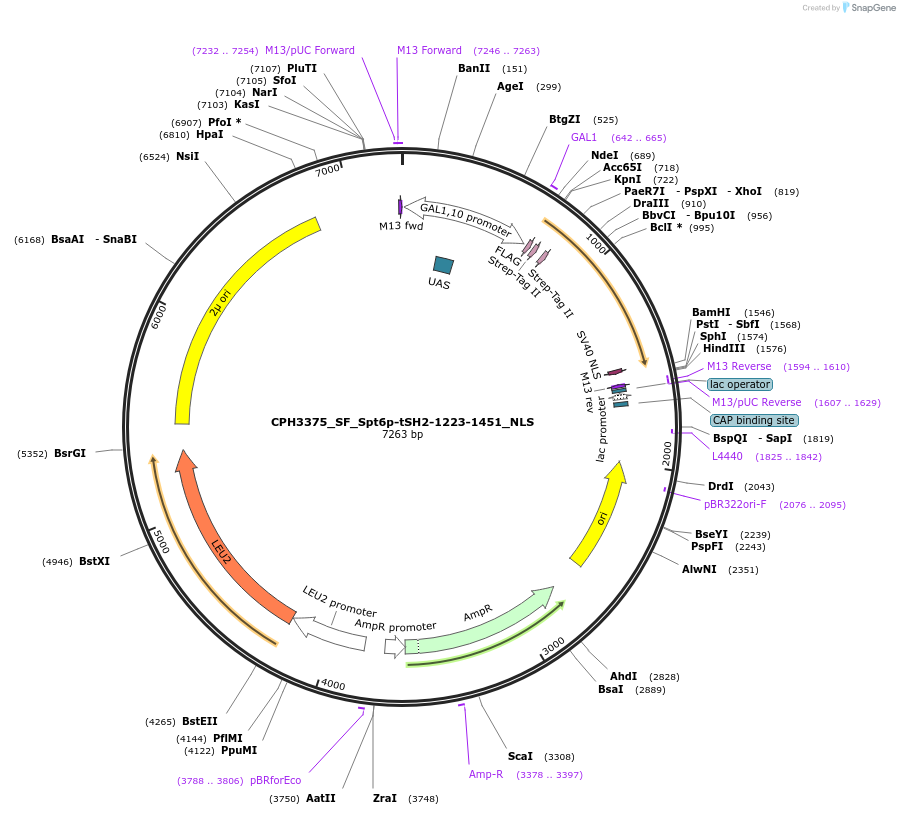
CPH3375_SF_Spt6p-tSH2-1223-1451_NLS
Plasmid#227084PurposeGal1p dependent budding yeast expression of tandem-affinity tagged spt6p tSH2 domain (1123-1451) localized to nucleusDepositorInsertspt6-tsh2 (SPT6 Budding Yeast)
TagsSTREPii-FLAG and SV40 NLS 1ExpressionYeastPromoterGal1-10Available SinceNov. 4, 2024AvailabilityAcademic Institutions and Nonprofits only -
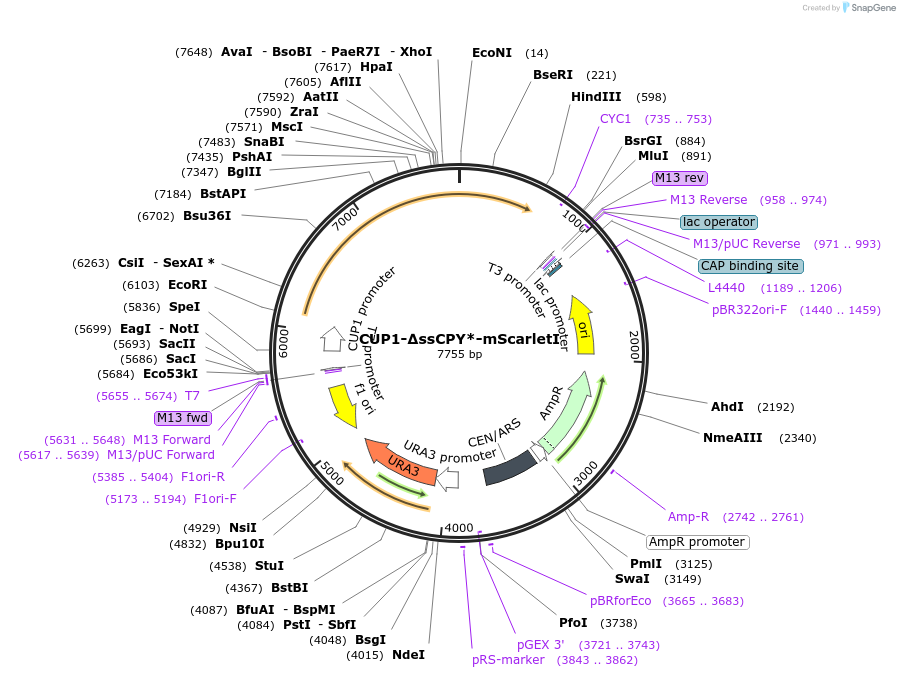
CUP1-∆ssCPY*-mScarletI
Plasmid#221157PurposeFluorescent reporter for cytoplasmic aggregatesDepositorInsertCarboxypeptidase Y (PRC1 Budding Yeast)
TagsmScarletIExpressionYeastMutationDeletion signal peptide, G255A mutationAvailable SinceNov. 1, 2024AvailabilityAcademic Institutions and Nonprofits only -
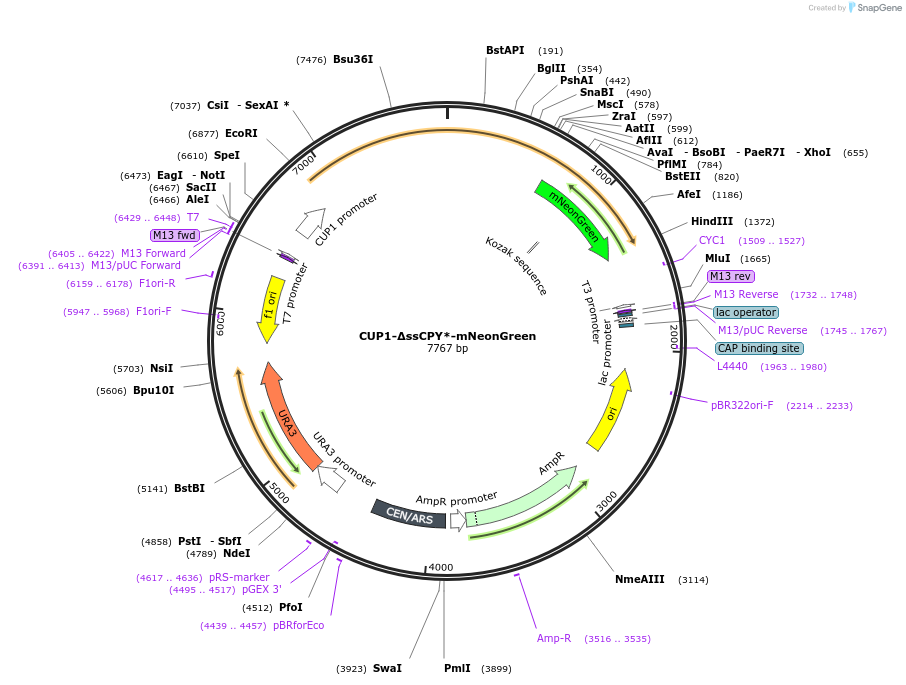
CUP1-∆ssCPY*-mNeonGreen
Plasmid#221156PurposeFluorescent reporter for cytoplasmic aggregatesDepositorInsertCarboxypeptidase Y (PRC1 Budding Yeast)
TagsmNeonGreenExpressionYeastMutationDeletion signal peptide, G255A mutationAvailable SinceNov. 1, 2024AvailabilityAcademic Institutions and Nonprofits only -
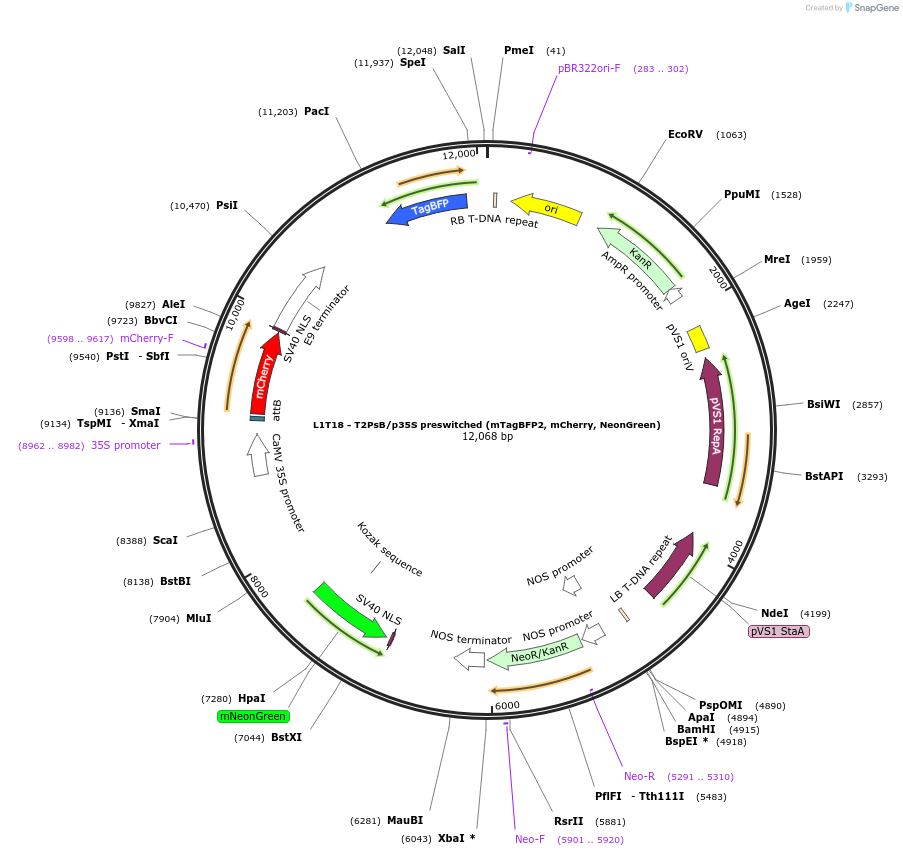
L1T18 - T2PsB/p35S preswitched (mTagBFP2, mCherry, NeonGreen)
Plasmid#219690PurposeLevel 1 preswitched History-dependent target, switch with Bxb1, p35S promoter, and output: mCherry then NeonGreen (for transformation, Kan plant resistance, Kan bacteria resistance).DepositorInsertT2PsB/p35S (mTagBFP2, mCherry, NeonGreen)
ExpressionPlantAvailable SinceJuly 16, 2024AvailabilityAcademic Institutions and Nonprofits only