We narrowed to 10,180 results for: yeast
-
-
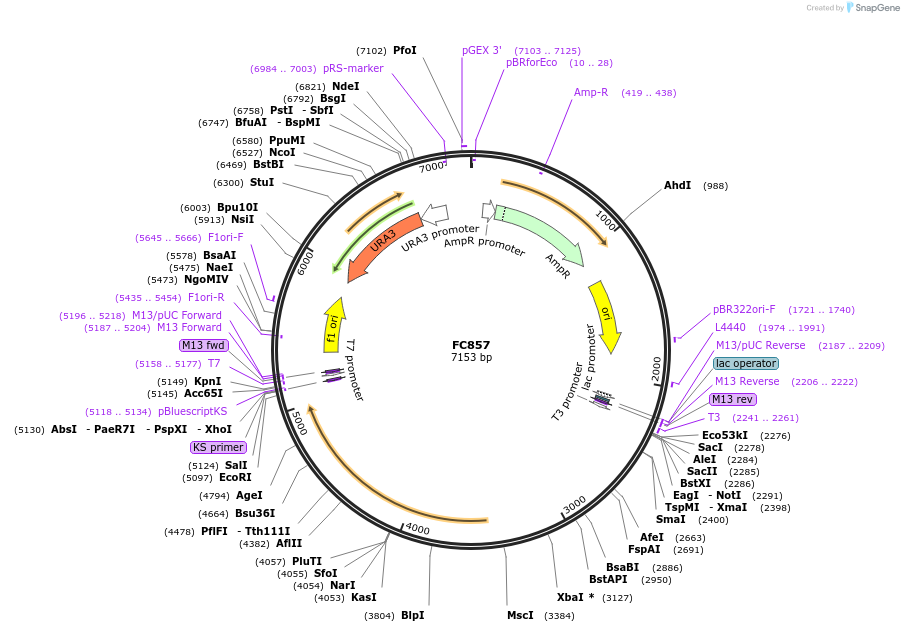
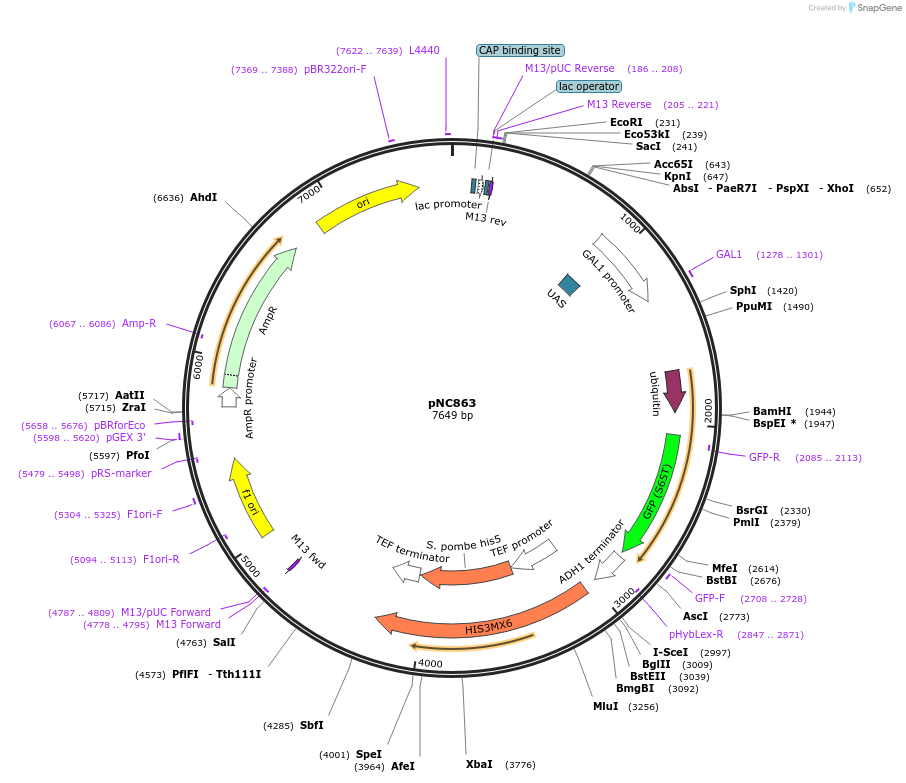
pNC820
Plasmid#20383DepositorInsertUAS cassette (FUS1)
ExpressionYeastMutationX=UbiM (see schematic)Available SinceMarch 6, 2009AvailabilityAcademic Institutions and Nonprofits only -
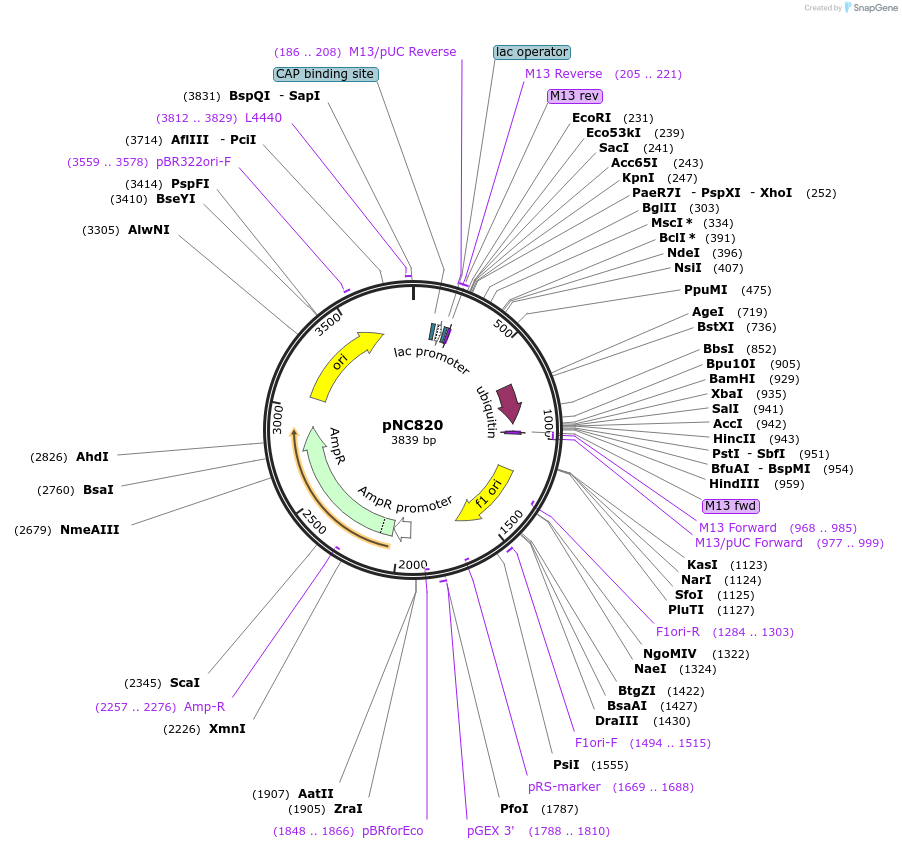
pNC951
Plasmid#20400DepositorInsertCFP-reporter
ExpressionYeastMutationX=UbiY (see schematic) linker=deltakAvailable SinceMarch 6, 2009AvailabilityAcademic Institutions and Nonprofits only -
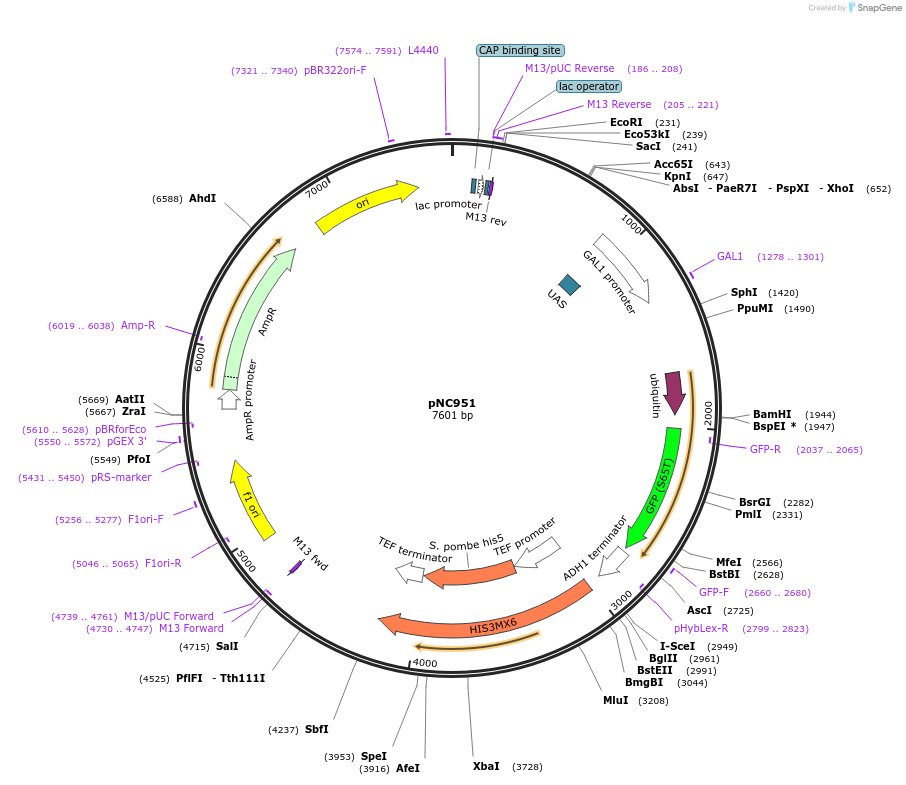
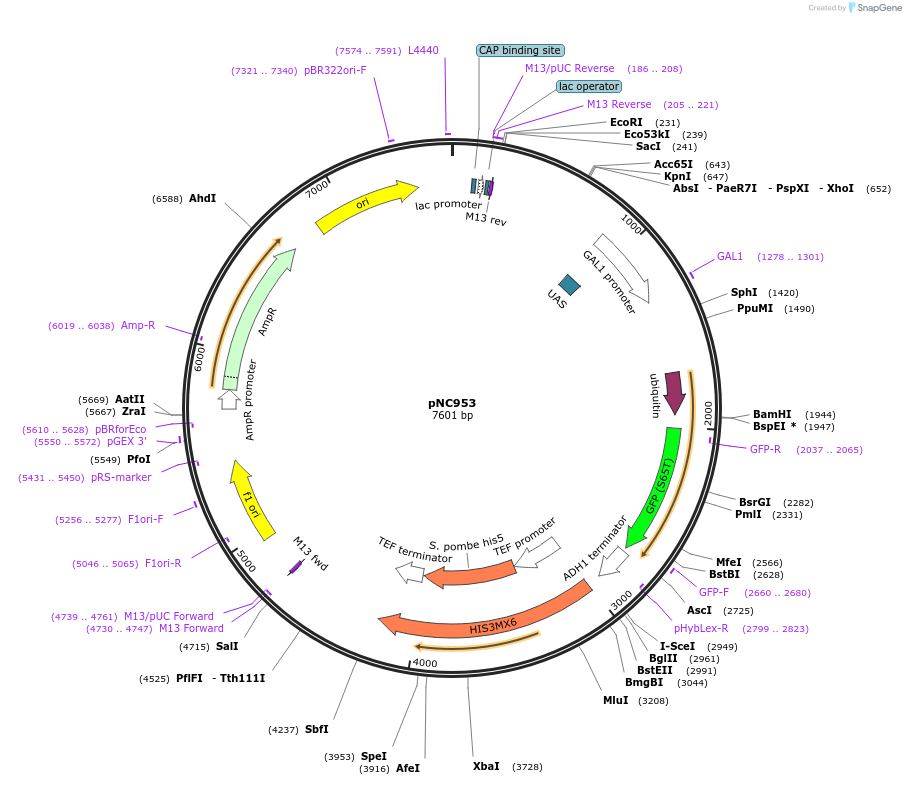
pNC953
Plasmid#20402DepositorInsertCFP-reporter
ExpressionYeastMutationX=UbiE (see schematic) linker=deltakAvailable SinceMarch 6, 2009AvailabilityAcademic Institutions and Nonprofits only -
pCAB42
Plasmid#17268DepositorTypeEmpty backboneTagsB42•HAExpressionYeastAvailable SinceJan. 22, 2010AvailabilityAcademic Institutions and Nonprofits only -
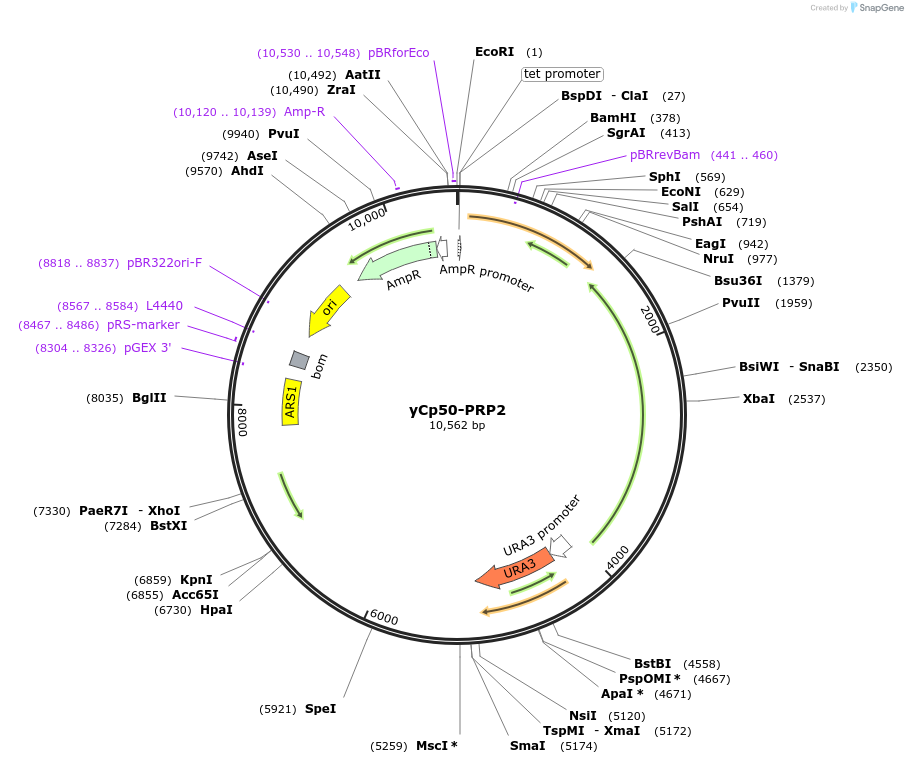
yCp50-PRP2
Plasmid#111304PurposePrp2 in yCp50 plasmidDepositorInsertPRP2
ExpressionYeastAvailable SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
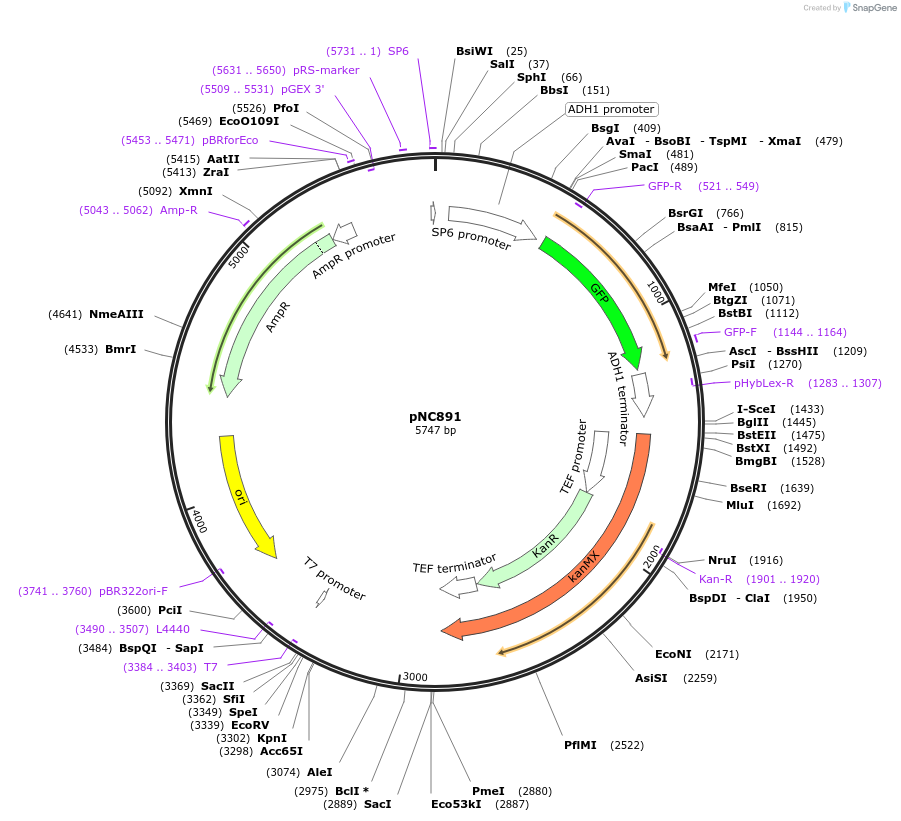
pNC891
Plasmid#20397DepositorInsertYFP-reporter
ExpressionYeastMutationX=M (see schematic)Available SinceMarch 6, 2009AvailabilityAcademic Institutions and Nonprofits only -
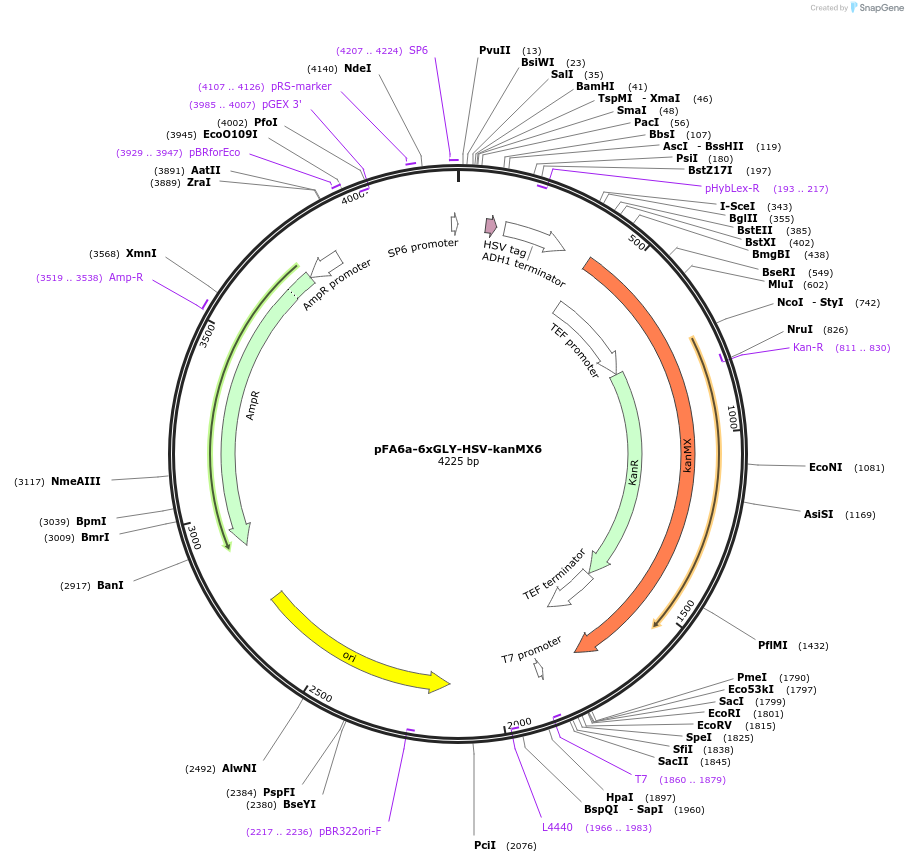
pFA6a-6xGLY-HSV-kanMX6
Plasmid#20775DepositorTypeEmpty backboneUsePcr-based c-terminal taggingExpressionYeastAvailable SinceMarch 23, 2009AvailabilityAcademic Institutions and Nonprofits only -
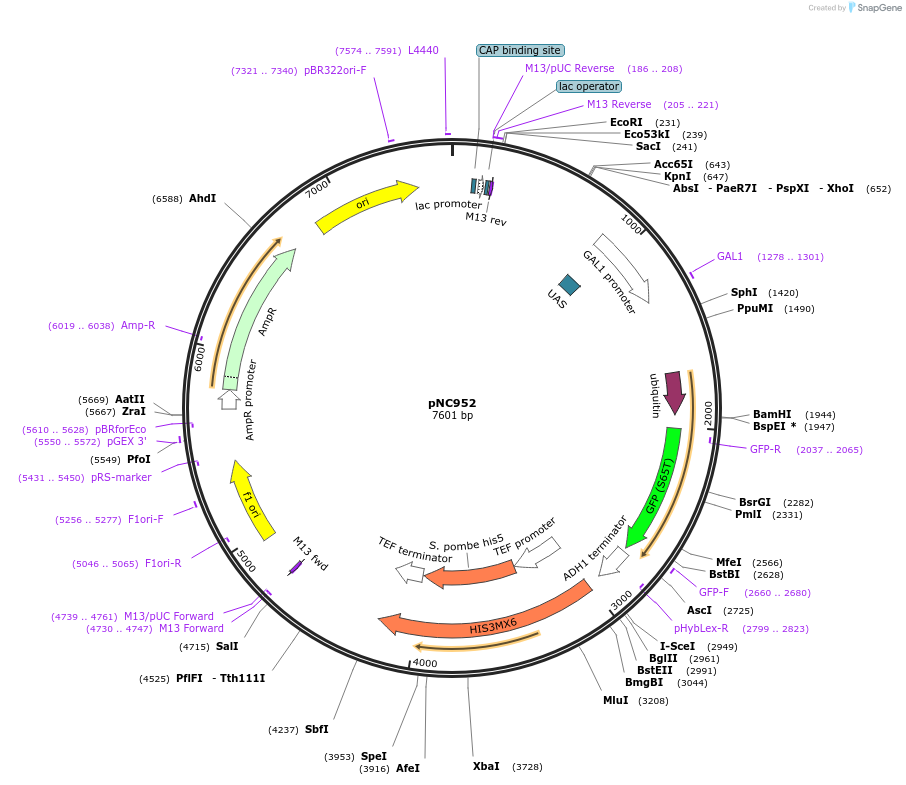
pNC952
Plasmid#20401DepositorInsertCFP-reporter
ExpressionYeastMutationX=UbiM (see schematic) linker=deltakAvailable SinceMarch 6, 2009AvailabilityAcademic Institutions and Nonprofits only -
pCGLex/Zeo
Plasmid#17259DepositorTypeEmpty backboneTagsLexAExpressionYeastAvailable SinceJan. 22, 2010AvailabilityAcademic Institutions and Nonprofits only -
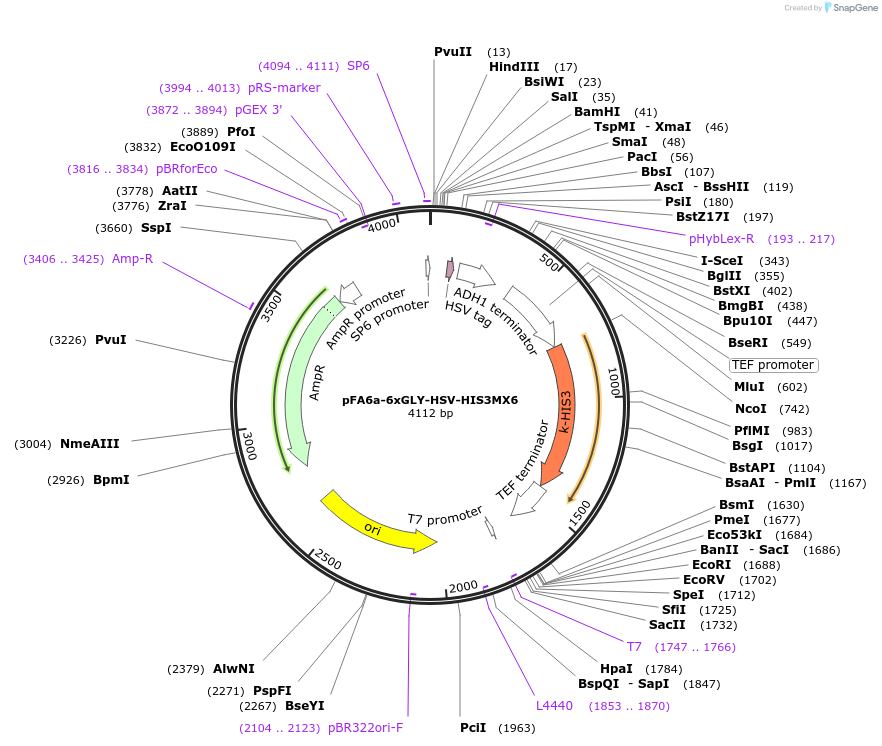
pFA6a-6xGLY-HSV-HIS3MX6
Plasmid#20774DepositorTypeEmpty backboneUsePcr-based c-terminal taggingExpressionYeastAvailable SinceMarch 27, 2009AvailabilityAcademic Institutions and Nonprofits only -
pCETT2
Plasmid#19784DepositorTypeEmpty backboneTagsHA and c-MycExpressionYeastAvailable SinceNov. 21, 2008AvailabilityAcademic Institutions and Nonprofits only -
p2AB42
Plasmid#17267DepositorTypeEmpty backboneTagsB42•HAExpressionYeastAvailable SinceJan. 22, 2010AvailabilityAcademic Institutions and Nonprofits only -
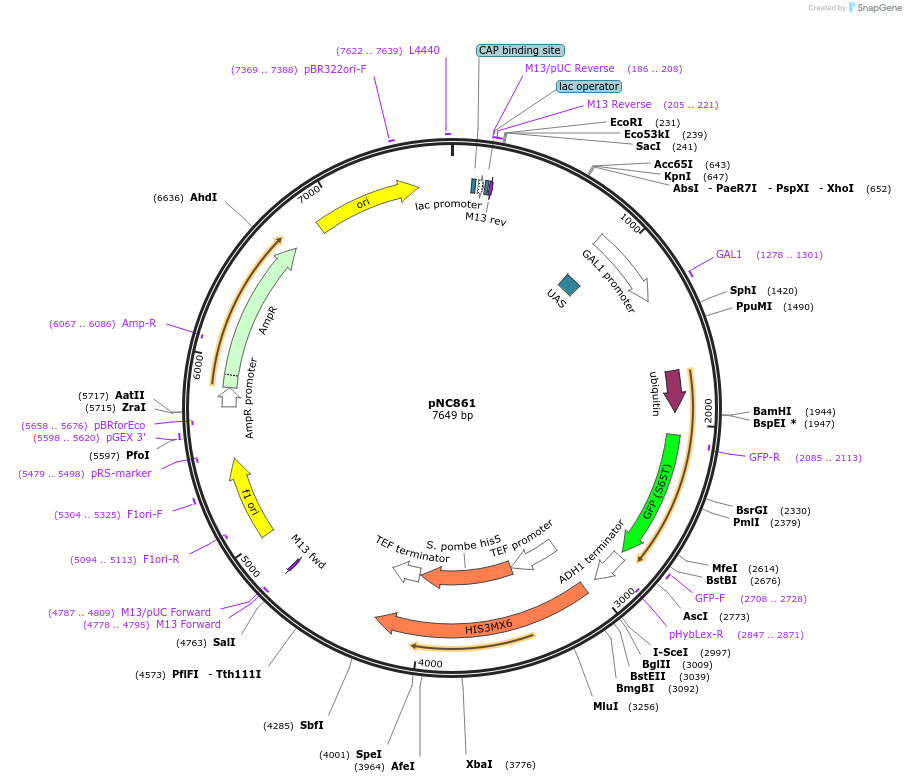
pNC861
Plasmid#20391DepositorInsertCFP-reporter
ExpressionYeastMutationX=UbiY (see schematic) linker: ekAvailable SinceMarch 6, 2009AvailabilityAcademic Institutions and Nonprofits only -
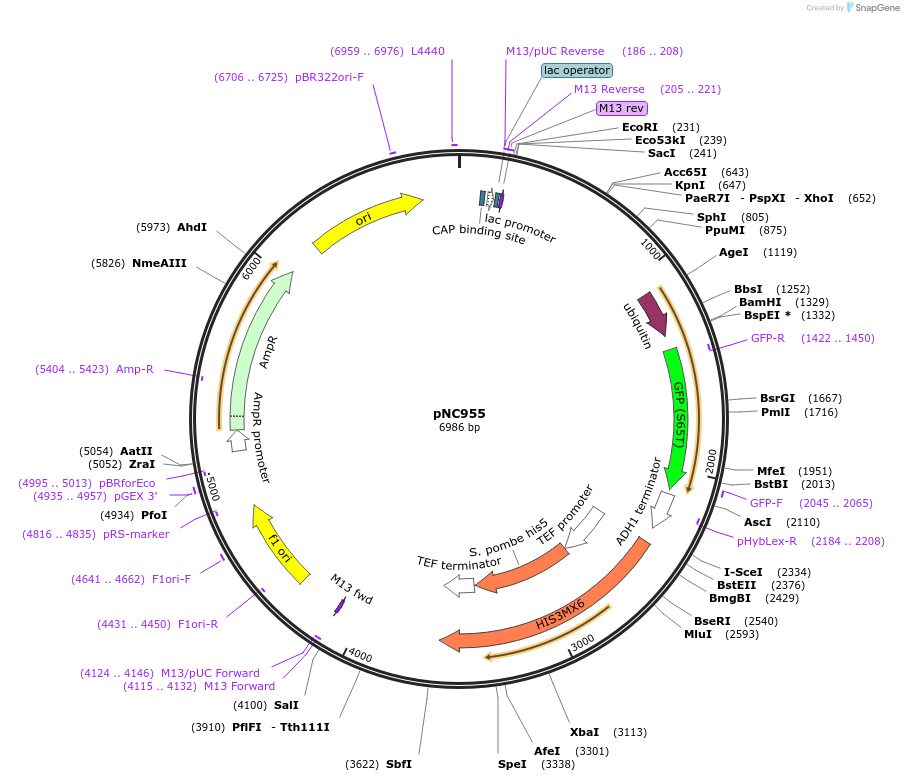
pNC955
Plasmid#20405DepositorInsertCFP-reporter
ExpressionYeastMutationX=UbiY (see schematic) linker=deltakAvailable SinceMarch 6, 2009AvailabilityAcademic Institutions and Nonprofits only -
pYES2.1-ScDcr1deltaN
Plasmid#32038DepositorInsertDrc1(111-610)
ExpressionYeastPromoterGAL1Available SinceOct. 5, 2011AvailabilityAcademic Institutions and Nonprofits only -
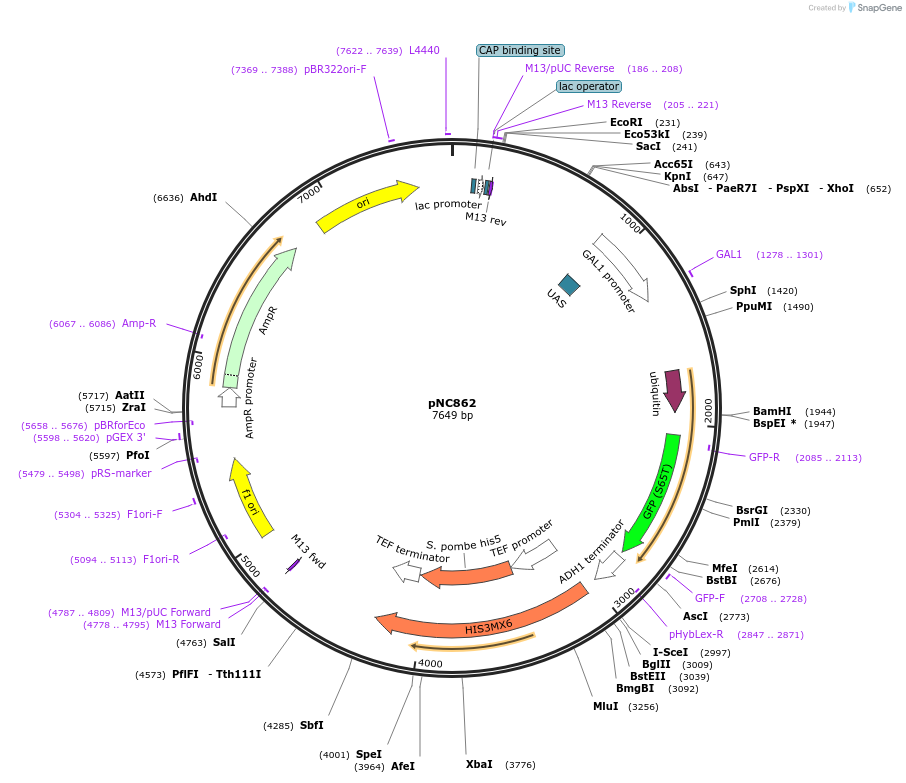
pNC862
Plasmid#20392DepositorInsertCFP-reporter
ExpressionYeastMutationX=UbiM (see schematic) linker: ekAvailable SinceMarch 6, 2009AvailabilityAcademic Institutions and Nonprofits only -
pNC863
Plasmid#20393DepositorInsertCFP-reporter
ExpressionYeastMutationX=UbiE (see schematic) linker: ekAvailable SinceMarch 6, 2009AvailabilityAcademic Institutions and Nonprofits only -
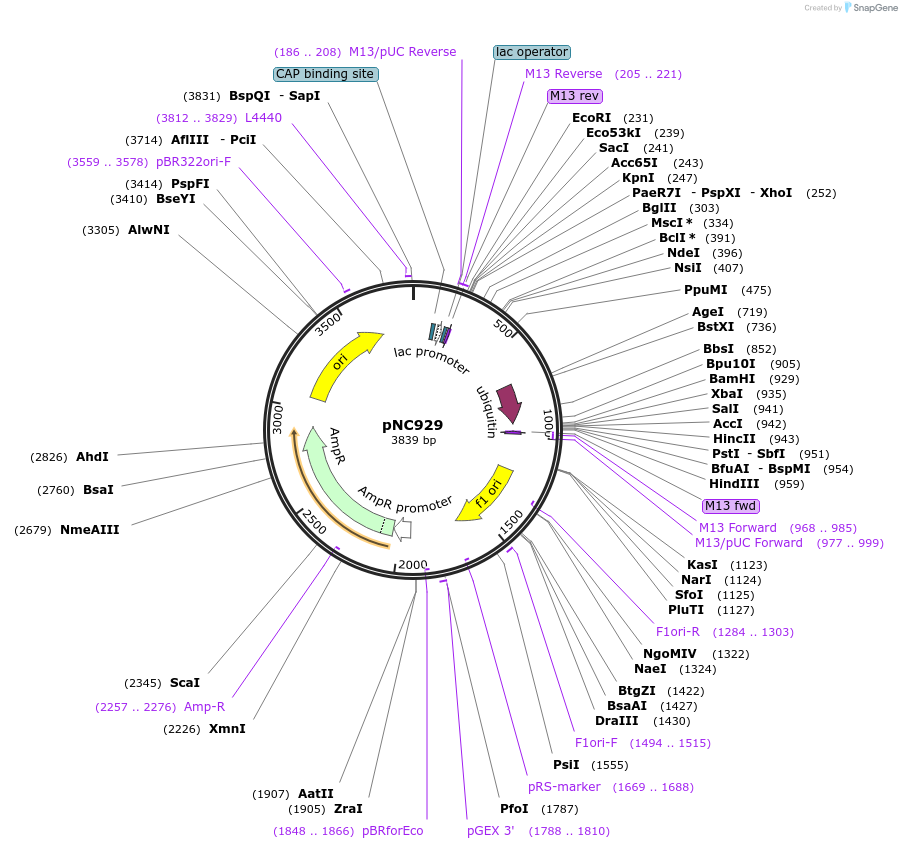
pNC929
Plasmid#20398DepositorInsertUAS cassette (FUS1)
ExpressionYeastMutationX=UbiE (see schematic)Available SinceMarch 6, 2009AvailabilityAcademic Institutions and Nonprofits only -
pCGB42
Plasmid#17266DepositorTypeEmpty backboneTagsB42•HAExpressionYeastAvailable SinceJan. 22, 2010AvailabilityAcademic Institutions and Nonprofits only -
pCGB42/Kan
Plasmid#17261DepositorTypeEmpty backboneTagsB42•HAExpressionYeastAvailable SinceJan. 22, 2010AvailabilityAcademic Institutions and Nonprofits only -
FC512
Plasmid#27331DepositorAvailable SinceFeb. 28, 2011AvailabilityAcademic Institutions and Nonprofits only -
pNC843
Plasmid#20388DepositorInsertCFP-reporter
ExpressionYeastMutationX=UbiM (see schematic)Available SinceMarch 6, 2009AvailabilityAcademic Institutions and Nonprofits only -
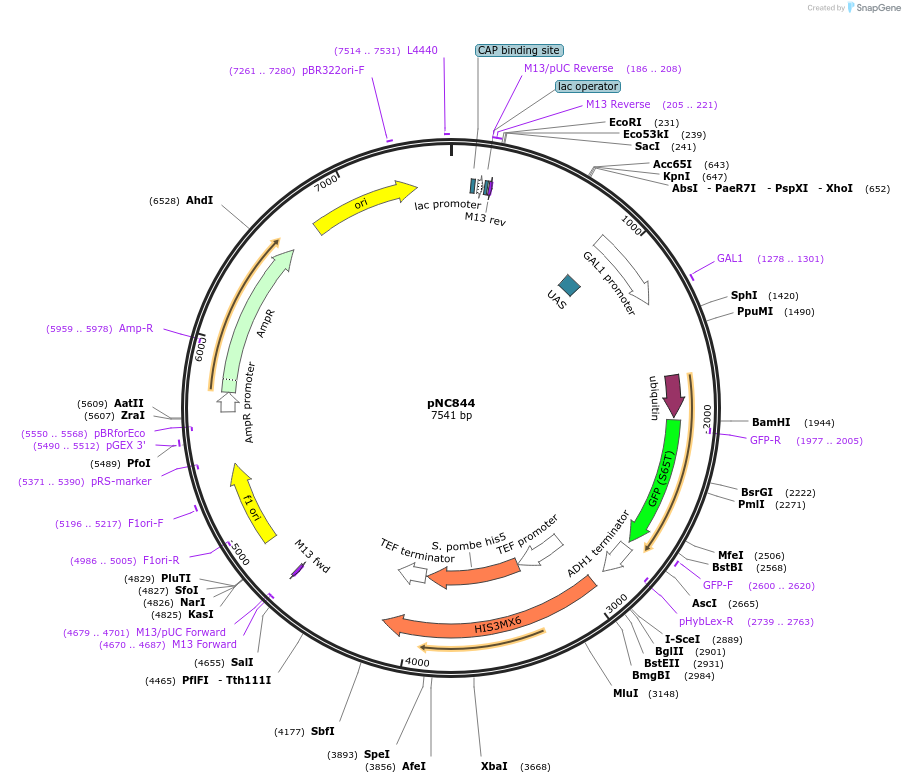
pNC844
Plasmid#20389DepositorInsertCFP-reporter
ExpressionYeastMutationX=UbiY (see schematic)Available SinceMarch 6, 2009AvailabilityAcademic Institutions and Nonprofits only -
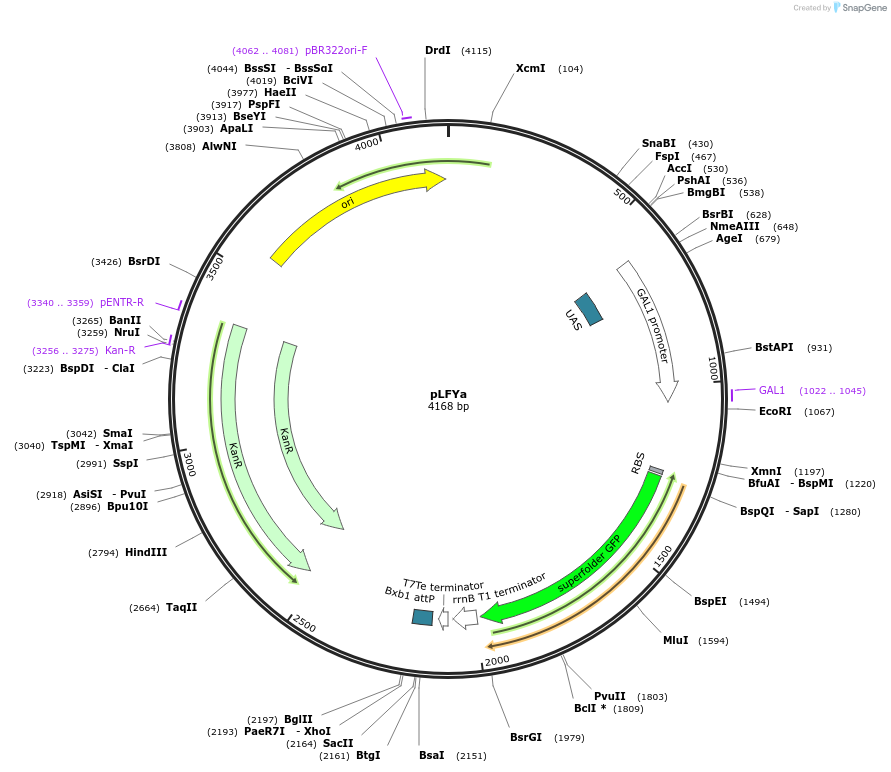
pLFYa
Plasmid#243803PurposeDropout vector for genomic integration into yeast strain LFYa. GFP is flanked by BamHI sites for easy cloning using Gibson assembly. Construct contains the attP site for the serine recombinase BxB1.DepositorArticleTypeEmpty backboneUseSynthetic BiologyAvailable SinceOct. 8, 2025AvailabilityAcademic Institutions and Nonprofits only -
pLFYalpha
Plasmid#243804PurposeDropout vector for genomic integration into yeast strain LFYalpha. GFP is flanked by BamHI sites for easy cloning using Gibson assembly. Construct contains attB site for the serine recombinase BxB1.DepositorArticleTypeEmpty backboneUseSynthetic BiologyAvailable SinceOct. 8, 2025AvailabilityAcademic Institutions and Nonprofits only -
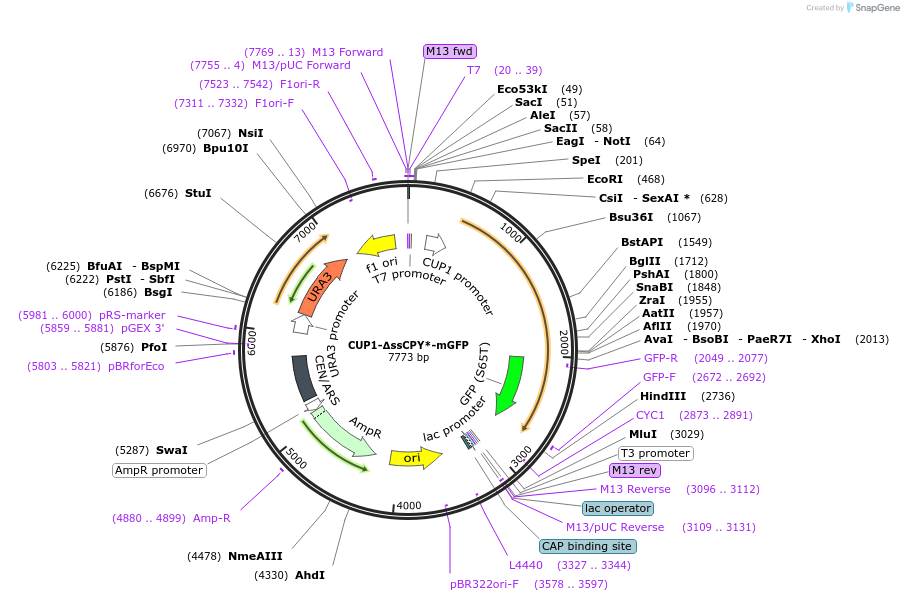
CUP1-∆ssCPY*-mGFP
Plasmid#212197PurposeFluorescent reporter for cytoplasmic aggregatesDepositorInsertCarboxypeptidase Y variant (PRC1 Budding Yeast)
TagsmEGFP (monomeric version EGFP, A206K mutation)ExpressionYeastMutationDeletion signal peptide, G255A mutationPromoterCUP1Available SinceNov. 1, 2024AvailabilityAcademic Institutions and Nonprofits only -
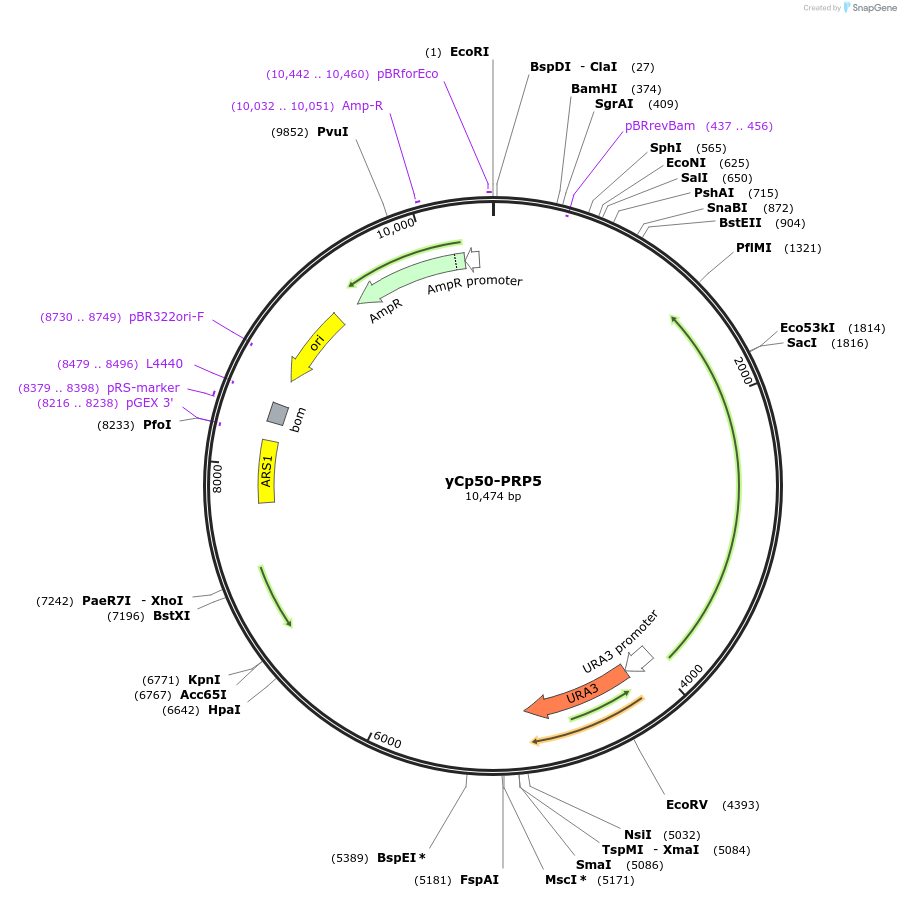
yCp50-PRP5
Plasmid#111303PurposePrp5 in yCp50 plasmidDepositorInsertPRP5
ExpressionYeastAvailable SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
pRM430
Plasmid#44530DepositorInsertHHT2
ExpressionYeastMutationhht2 deletion 4-30Available SinceJune 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pGF28
Plasmid#44960DepositorInserthhf2
ExpressionYeastMutationdel (4-14)Available SinceMay 21, 2013AvailabilityAcademic Institutions and Nonprofits only