We narrowed to 5,790 results for: SPI;
-
Plasmid#224530PurposeA Gateway compatible 3' entry clone containing a V5 tagged mClover3 followed by a polyADepositorInsertV5-mClover3-pA
UseGateway cloningAvailable SinceSept. 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
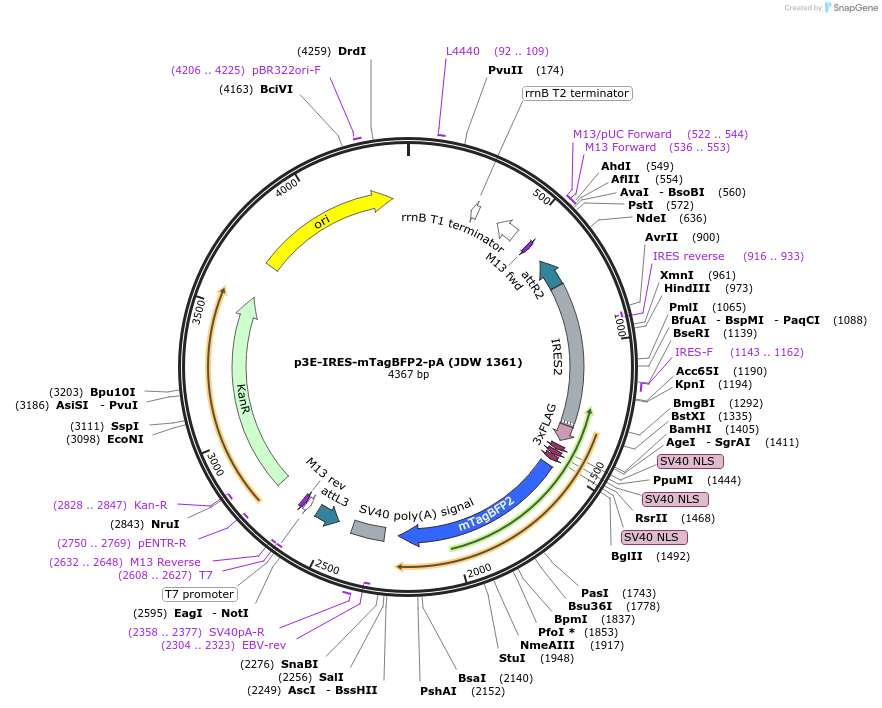
p3E-IRES-mTagBFP2-pA (JDW 1361)
Plasmid#224535PurposeA Gateway compatible 3' entry clone containing an IRES-FLAG-NLS-mTagBFP2-pADepositorInsertIRES-FLAG-NLS-mTagBFP2-pA
UseGateway cloningAvailable SinceSept. 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
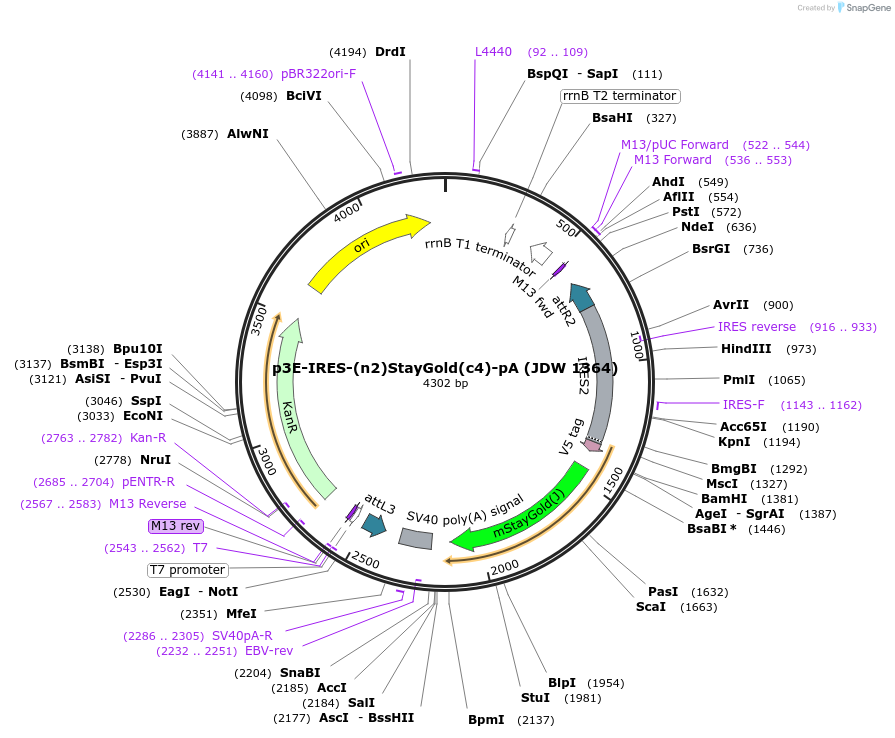
p3E-IRES-(n2)StayGold(c4)-pA (JDW 1364)
Plasmid#224537PurposeA Gateway compatible 3' entry clone containing IRES-(n2)StayGold(c4)-pADepositorInsertIRES-(n2)StayGold(c4)-pA
UseGateway cloningAvailable SinceSept. 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
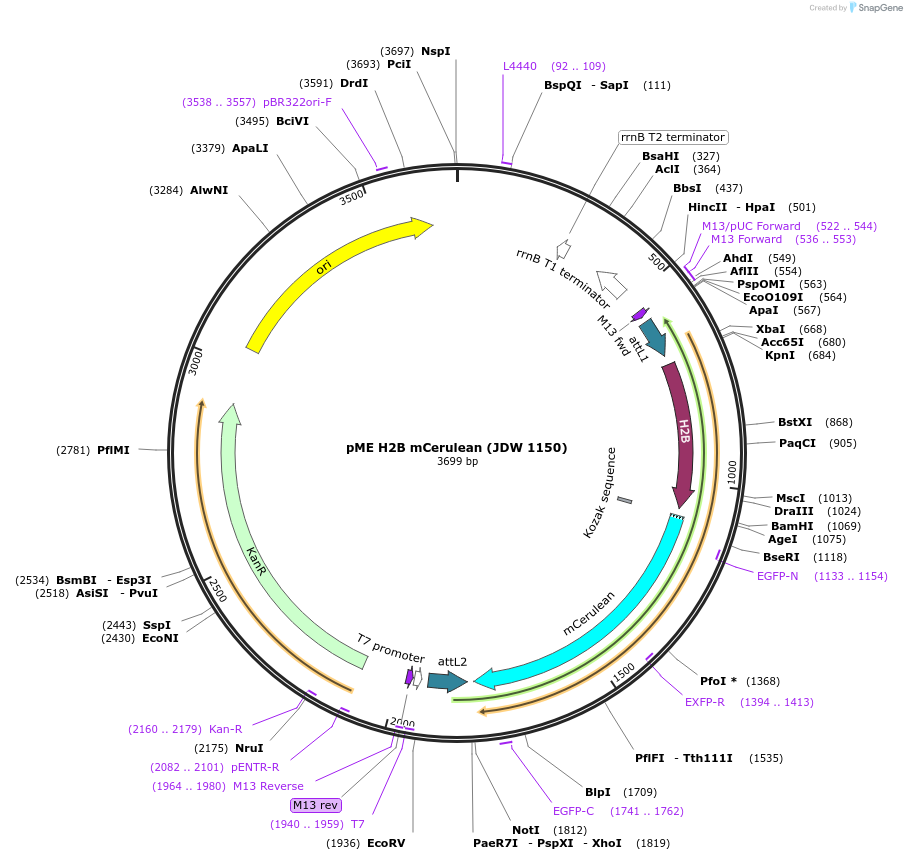
pME H2B mCerulean (JDW 1150)
Plasmid#224543PurposeA Gateway compatible middle entry clone containing a histone H2B fusion to mCerulean to label the nucleusDepositorInsertH2B-mCerulean
UseGateway cloningAvailable SinceSept. 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
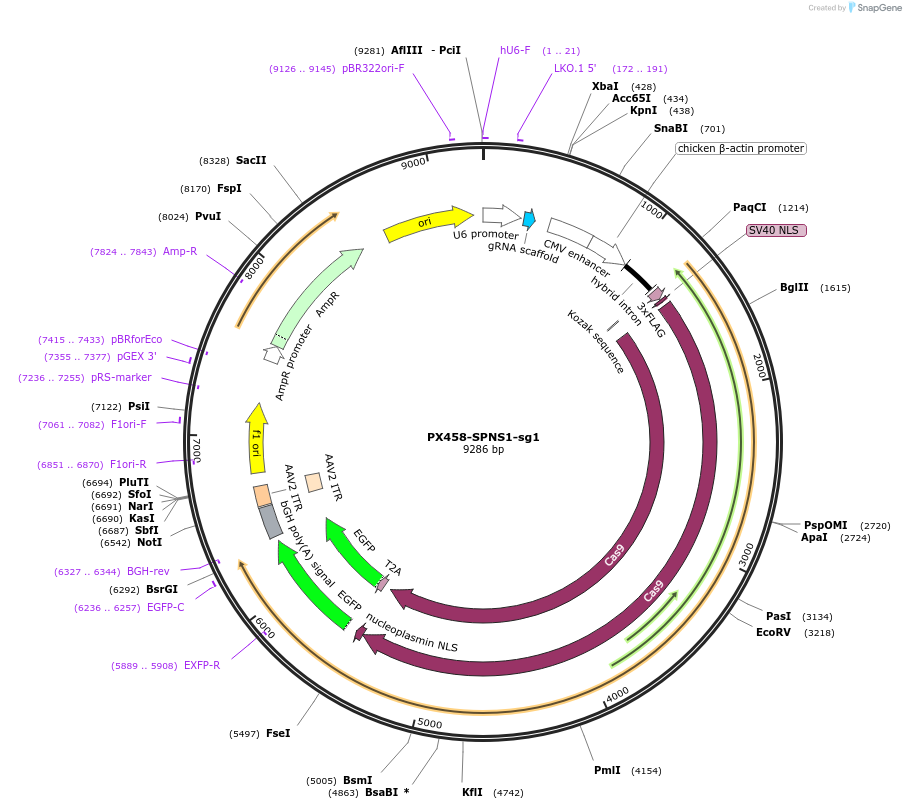
PX458-SPNS1-sg1
Plasmid#198566PurposeSPNS1-targeting sgRNA in PX458 vectorDepositorAvailable SinceJuly 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
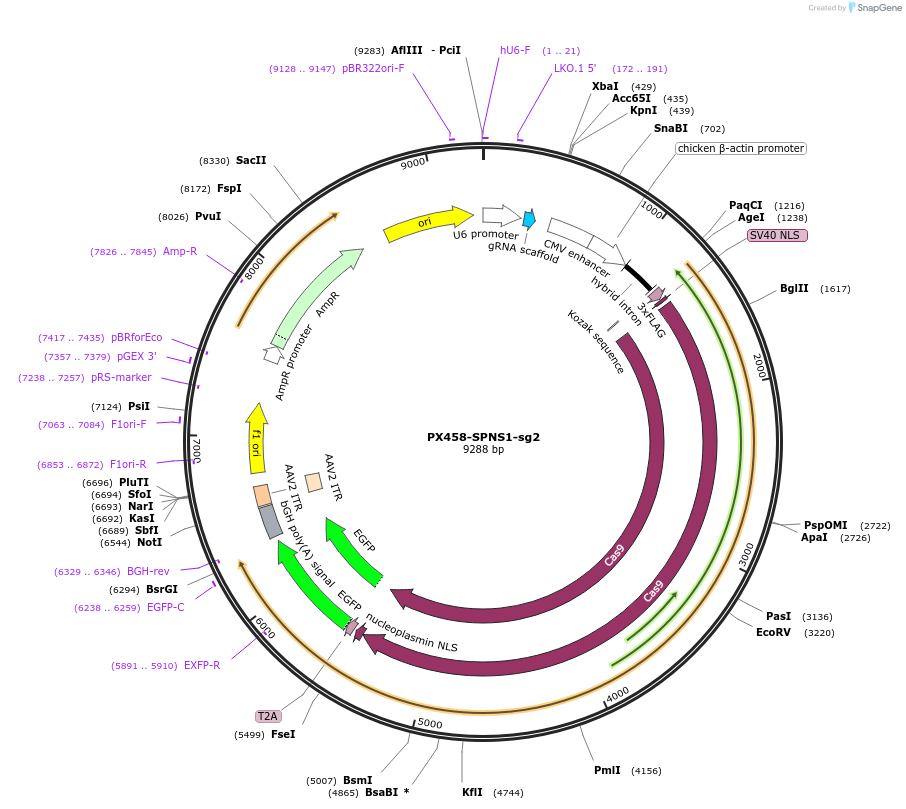
PX458-SPNS1-sg2
Plasmid#198567PurposeSPNS1-targeting sgRNA in PX458 vectorDepositorAvailable SinceJuly 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
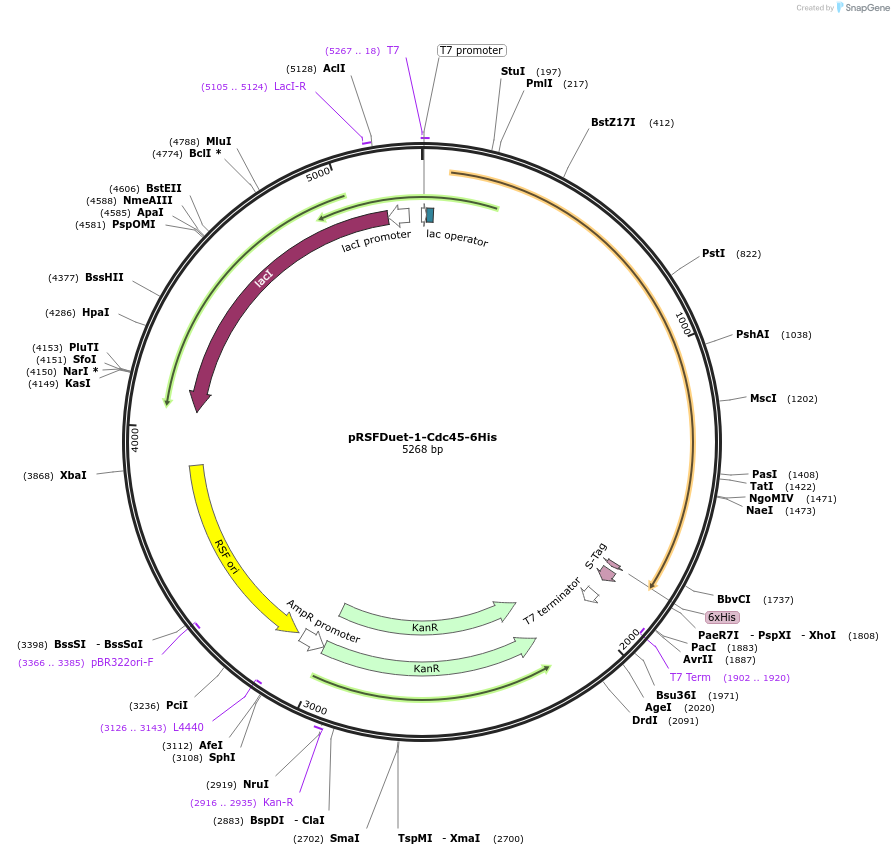
pRSFDuet-1-Cdc45-6His
Plasmid#211379Purposebacterial expression of human Cdc45 C-terminally fused to 6xHis-TagDepositorAvailable SinceJan. 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
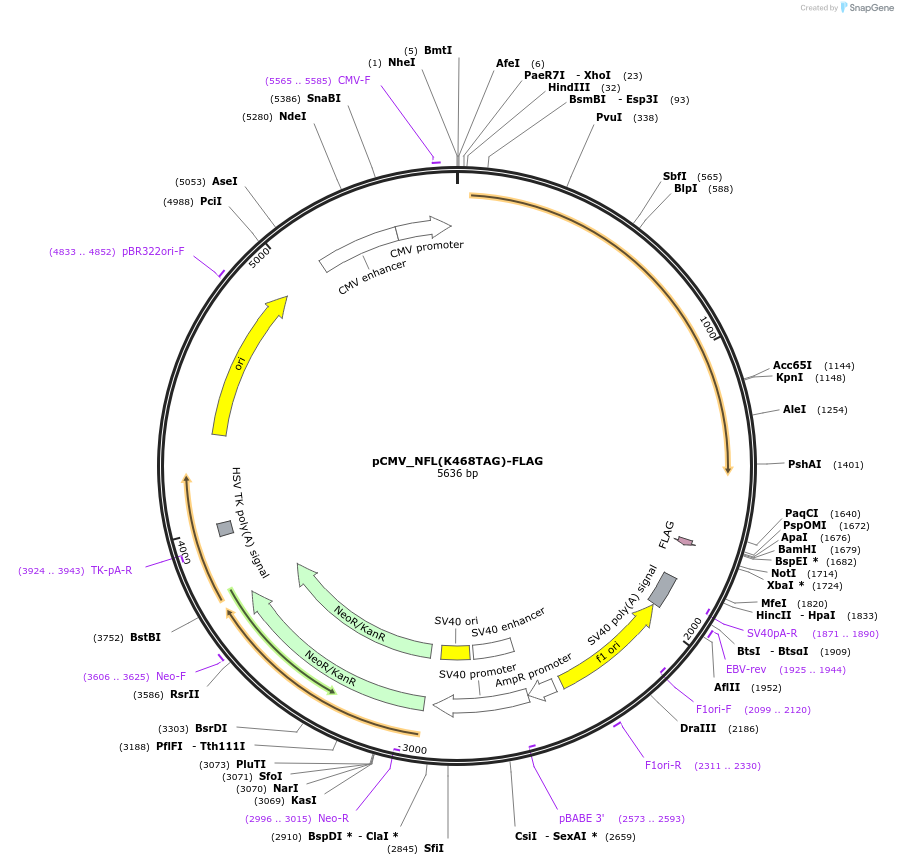
pCMV_NFL(K468TAG)-FLAG
Plasmid#182658PurposeExpresses mouse neurofilament light chain with a TAG codon at the position K468 and a C-terminal FLAG tag (DYKDDDDK) in mammalian cells.DepositorInsertmouse neurofilament light chain with a K468TAG mutation (Nefl Mouse)
TagsFLAG tag (DYKDDDDK)ExpressionMammalianMutationK468TAGPromoterCMVAvailable SinceMay 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
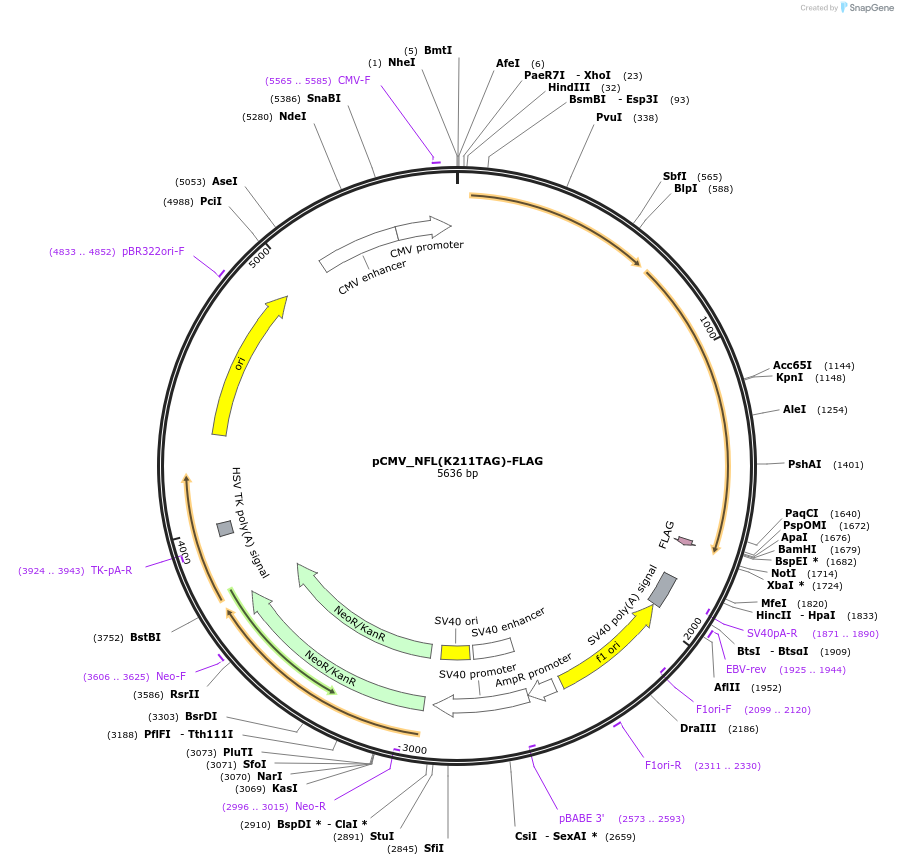
pCMV_NFL(K211TAG)-FLAG
Plasmid#182655PurposeExpresses mouse neurofilament light chain with a TAG codon at the position K211 and a C-terminal FLAG tag (DYKDDDDK) in mammalian cells.DepositorInsertmouse neurofilament light chain with a K211TAG mutation (Nefl Mouse)
TagsFLAG tag (DYKDDDDK)ExpressionMammalianMutationK211TAGPromoterCMVAvailable SinceMay 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
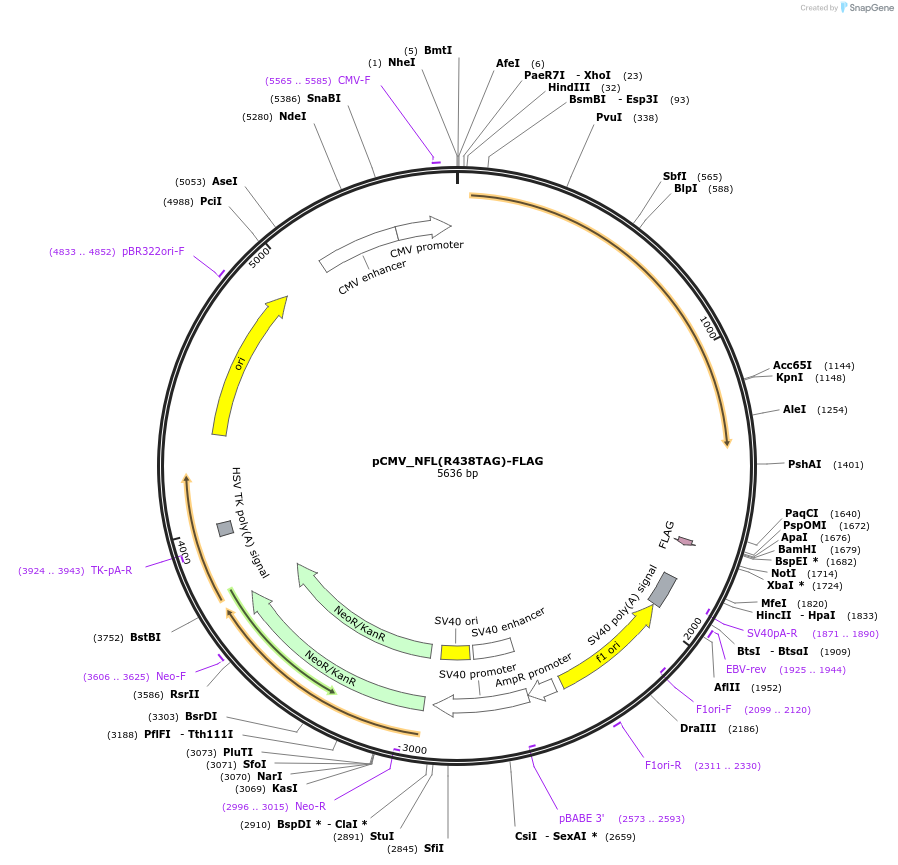
pCMV_NFL(R438TAG)-FLAG
Plasmid#182657PurposeExpresses mouse neurofilament light chain with a TAG codon at the position R438 and a C-terminal FLAG tag (DYKDDDDK) in mammalian cells.DepositorInsertmouse neurofilament light chain with a R438TAG mutation (Nefl Mouse)
TagsFLAG tag (DYKDDDDK)ExpressionMammalianMutationR438TAGPromoterCMVAvailable SinceMay 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
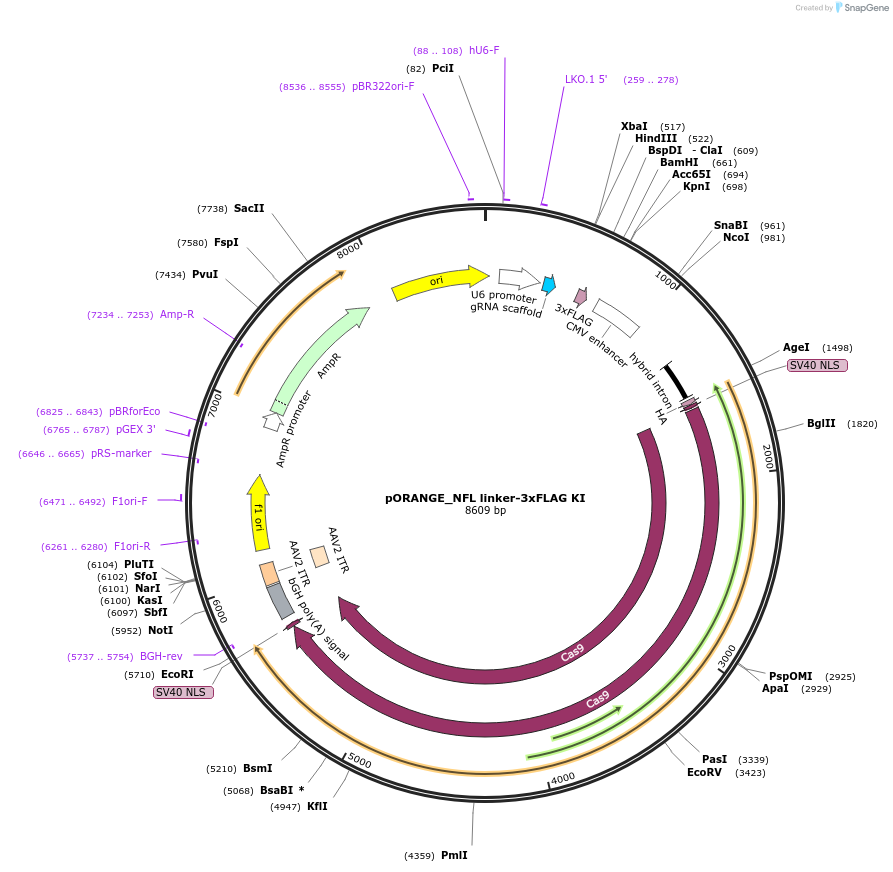
pORANGE_NFL linker-3xFLAG KI
Plasmid#182679PurposeExpression of spCas9, gRNA targeting the end of mouse Nefl gene and donor linker-3xFLAG. Can be used for C-terminal tagging of endogenous NFL with a linker-3xFLAG tag.DepositorInsertNefl-targeting gRNA and linker-3xFLAG donor sequence (Nefl Mouse)
ExpressionMammalianPromoterU6 and chicken beta-actin promoterAvailable SinceMay 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
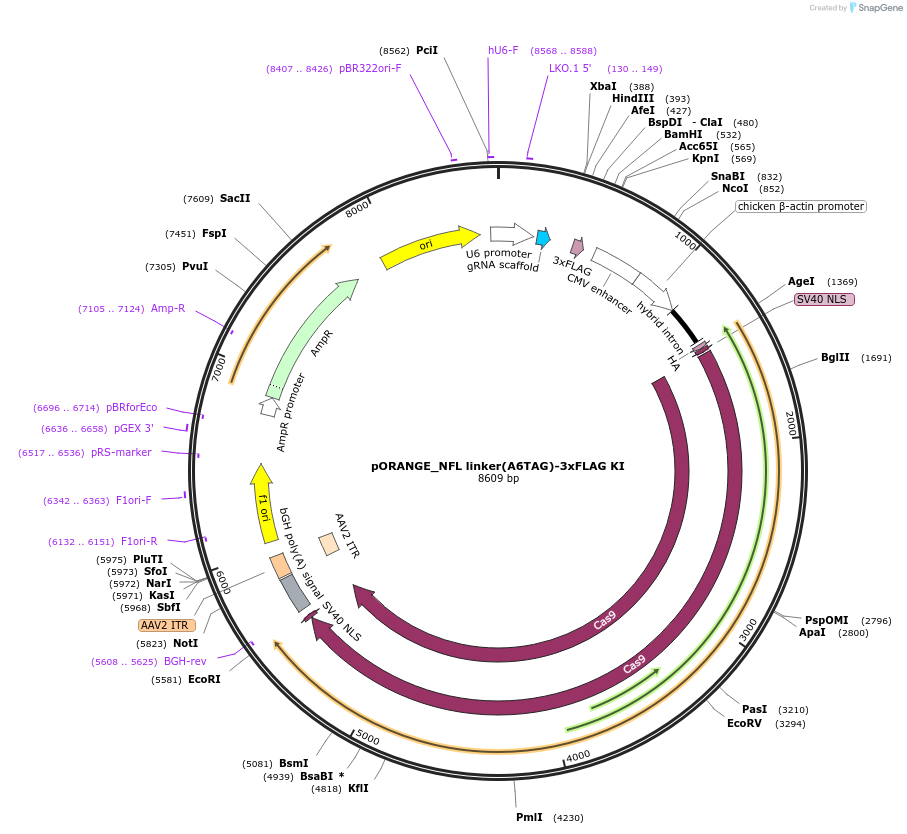
pORANGE_NFL linker(A6TAG)-3xFLAG KI
Plasmid#182680PurposeExpression of spCas9, gRNA targeting the end of mouse Nefl gene and donor linker(A6TAG)-3xFLAG. Can be used for amber codon suppression and click chemistry labeling of endogenous NFL.DepositorInsertNefl-targeting gRNA and linker(A6TAG)-3xFLAG donor sequence (Nefl Mouse)
ExpressionMammalianPromoterU6 and chicken beta-actin promoterAvailable SinceMay 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
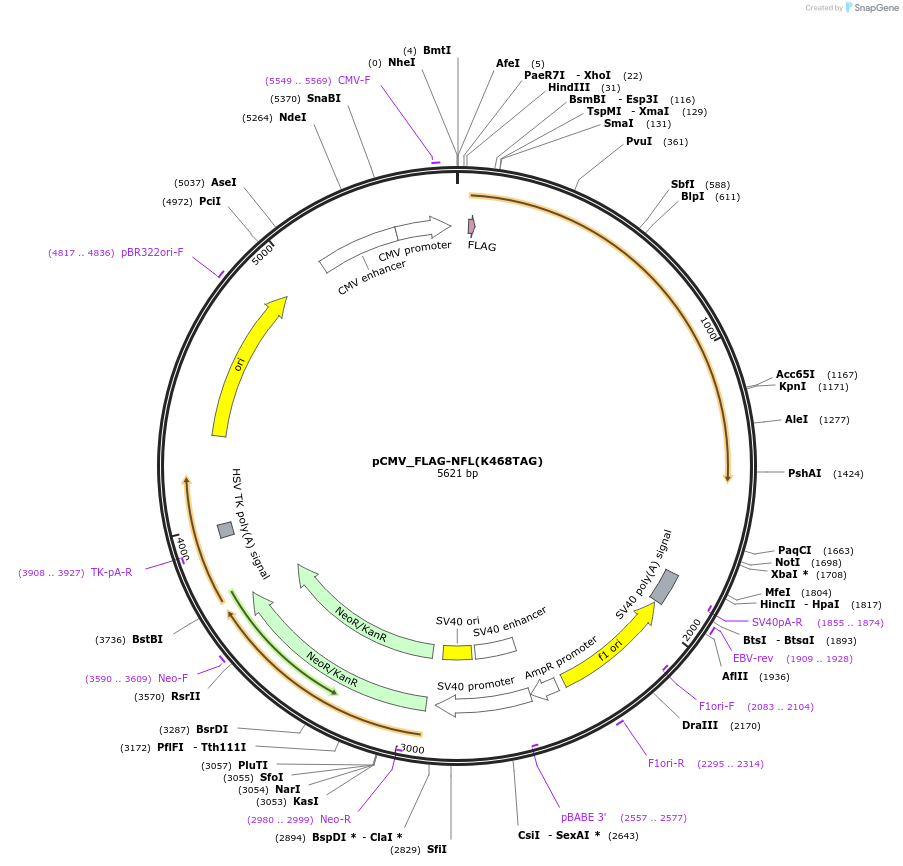
pCMV_FLAG-NFL(K468TAG)
Plasmid#182663PurposeExpresses mouse neurofilament light chain with a TAG codon at the position K468 and an N-terminal FLAG tag (DYKDDDDK) in mammalian cells.DepositorInsertmouse neurofilament light chain with a K468TAG mutation (Nefl Mouse)
TagsFLAG tag (DYKDDDDK)ExpressionMammalianMutationK468TAGPromoterCMVAvailable SinceMay 27, 2022AvailabilityAcademic Institutions and Nonprofits only -
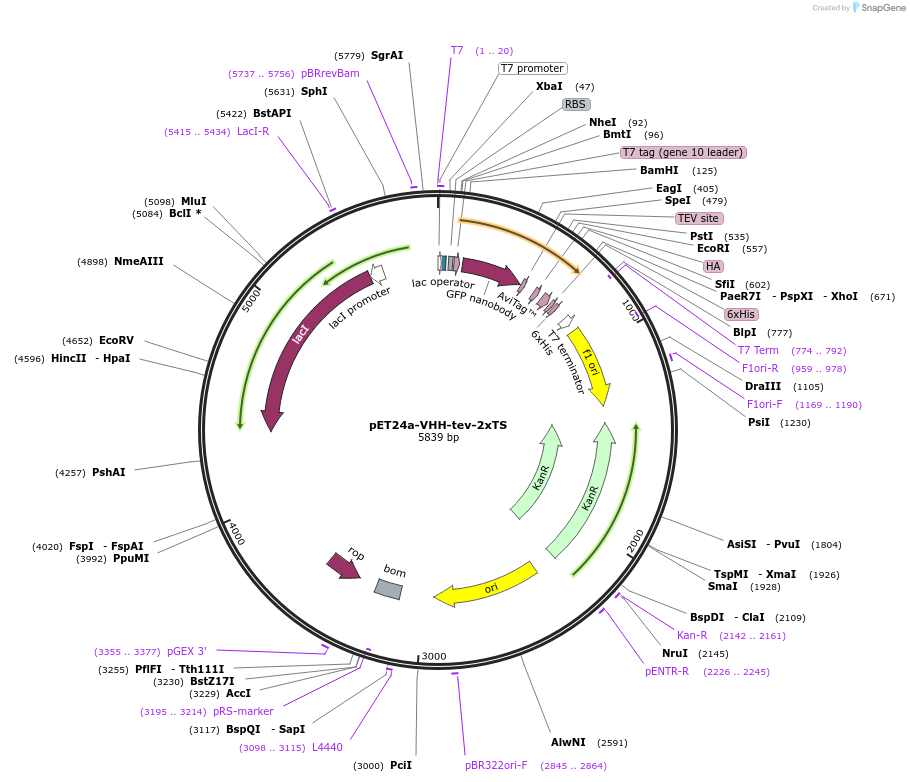
pET24a-VHH-tev-2xTS
Plasmid#117750PurposeBacterial expression of a functionalized anti-GFP (VHH) nanobody fused to two tyrosine sulfation (TS) motifs. VHH-2xTS also contains a T7, HA, BAP, tev cleavage site and His6 epitopeDepositorInsertanti-GFP nanobody fused to a T7, TS, HA, BAP and His6 epitope
TagsBAP, HA, His6, T7, and Tyrosine sulfation (TS) se…ExpressionBacterialPromoterT7Available SinceFeb. 27, 2019AvailabilityAcademic Institutions and Nonprofits only -
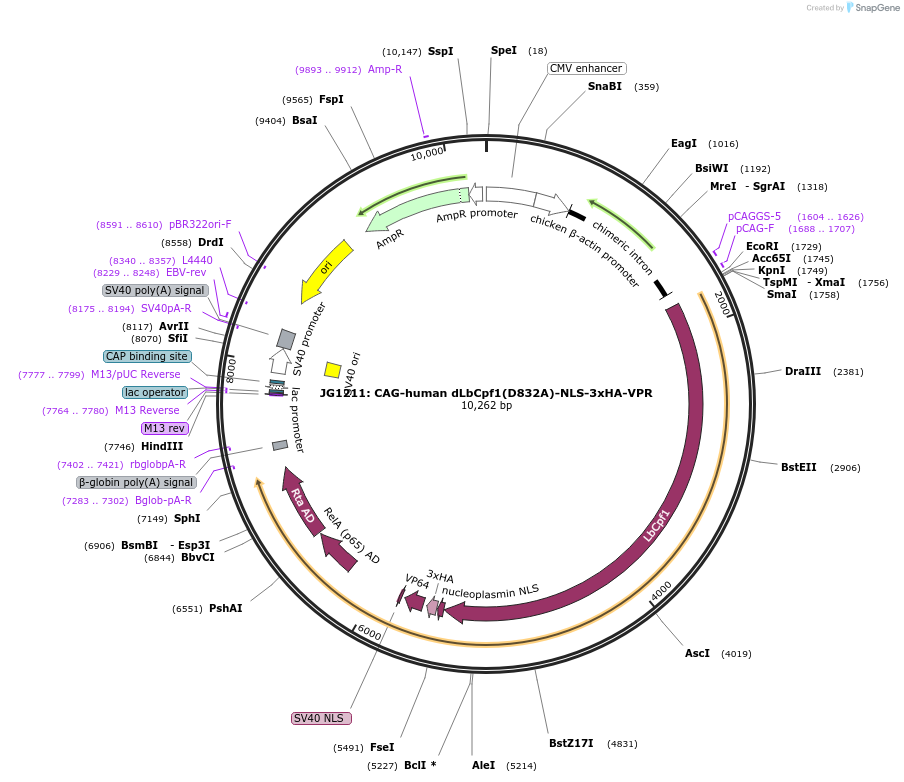
JG1211: CAG-human dLbCpf1(D832A)-NLS-3xHA-VPR
Plasmid#104567PurposeMammalian expression vector for catalytically inactive Cpf1 from Lachnospiraceae bacterium (dLbCpf1) fused to VPR activatorDepositorInserthuman codon optimized ‘dead’ Cpf1 fused to HSV VPR activation domain
UseCRISPRTagsNLS-3xHA-VPRExpressionMammalianMutationD832APromoterCAGAvailable SinceJan. 17, 2018AvailabilityAcademic Institutions and Nonprofits only -
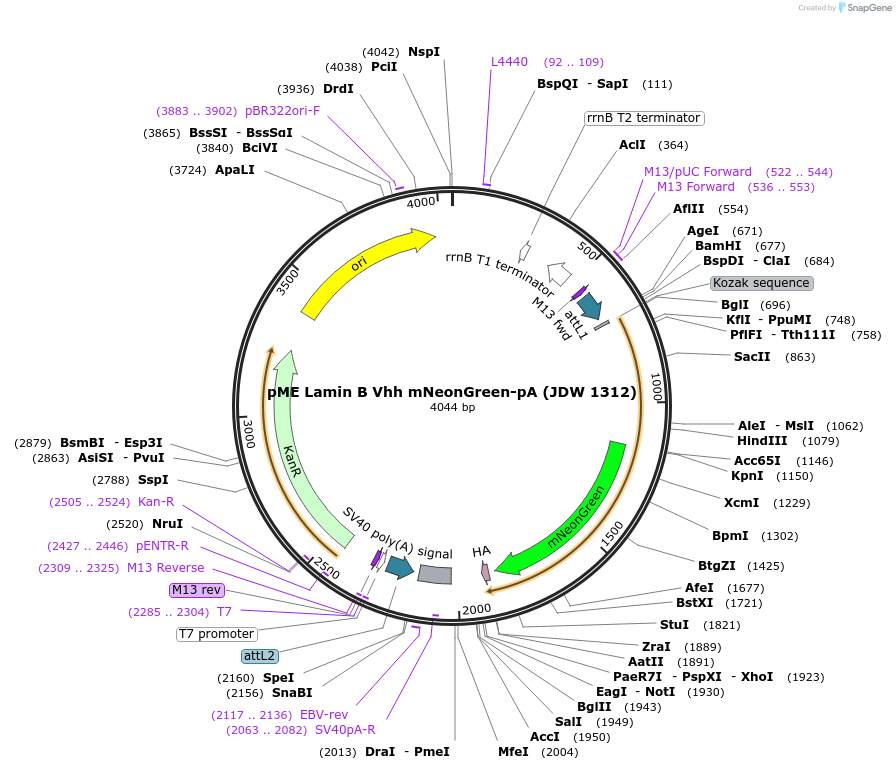
pME Lamin B Vhh mNeonGreen-pA (JDW 1312)
Plasmid#224501PurposeGateway compatible middle entry clone containing Lamin B nanobody fused to mNeonGreen with HA Tag (For visualizing the nuclear lamina, lamin chromobody)DepositorInsertLamin B Vhh mNeonGreen-pA
UseGateway cloningAvailable SinceSept. 27, 2024AvailabilityAcademic Institutions and Nonprofits only -
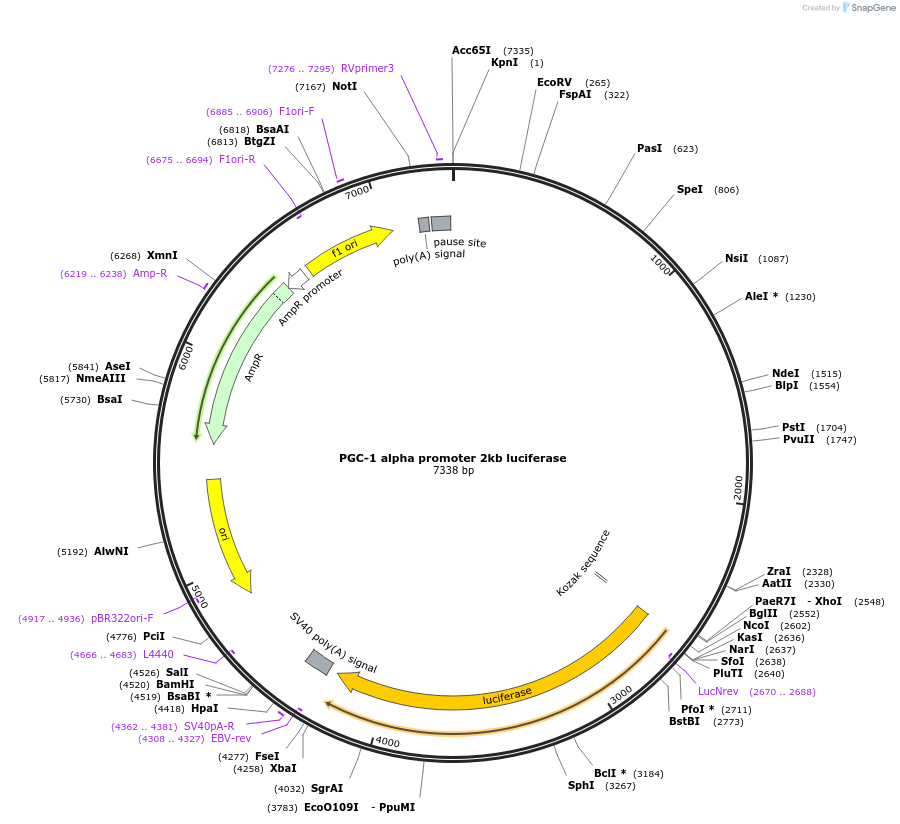
PGC-1 alpha promoter 2kb luciferase
Plasmid#8887DepositorAvailable SinceJan. 5, 2006AvailabilityAcademic Institutions and Nonprofits only -
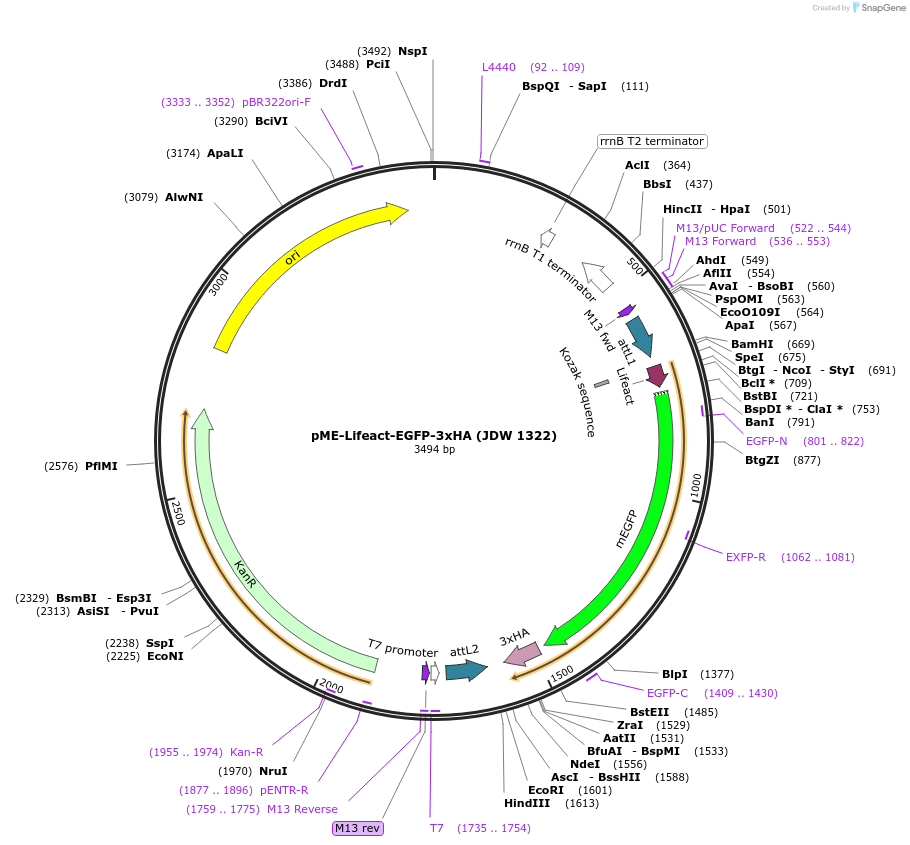
pME-Lifeact-EGFP-3xHA (JDW 1322)
Plasmid#224494PurposeGateway compatible middle entry clone containing Lifeact-EGFP (EGFP F-actin reporter)DepositorInsertLiefact-EGFP-3xHA
UseGateway cloningAvailable SinceSept. 27, 2024AvailabilityAcademic Institutions and Nonprofits only -
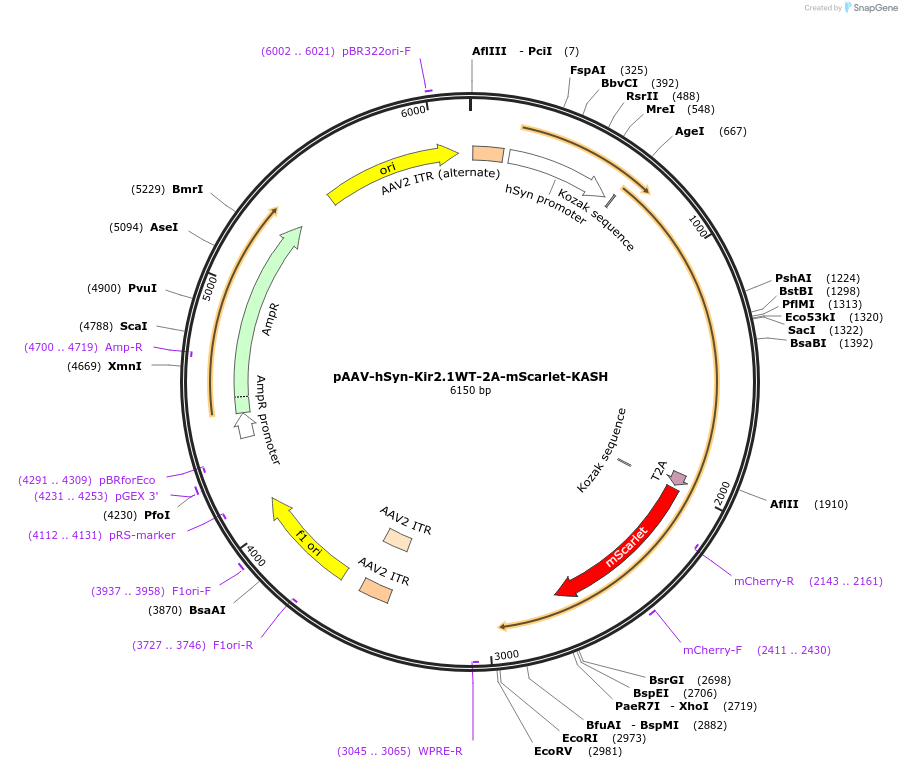
pAAV-hSyn-Kir2.1WT-2A-mScarlet-KASH
Plasmid#203840PurposeKir2.1 expression for silencing neurons and mScarlet-KASH expressionDepositorAvailable SinceAug. 9, 2023AvailabilityAcademic Institutions and Nonprofits only -
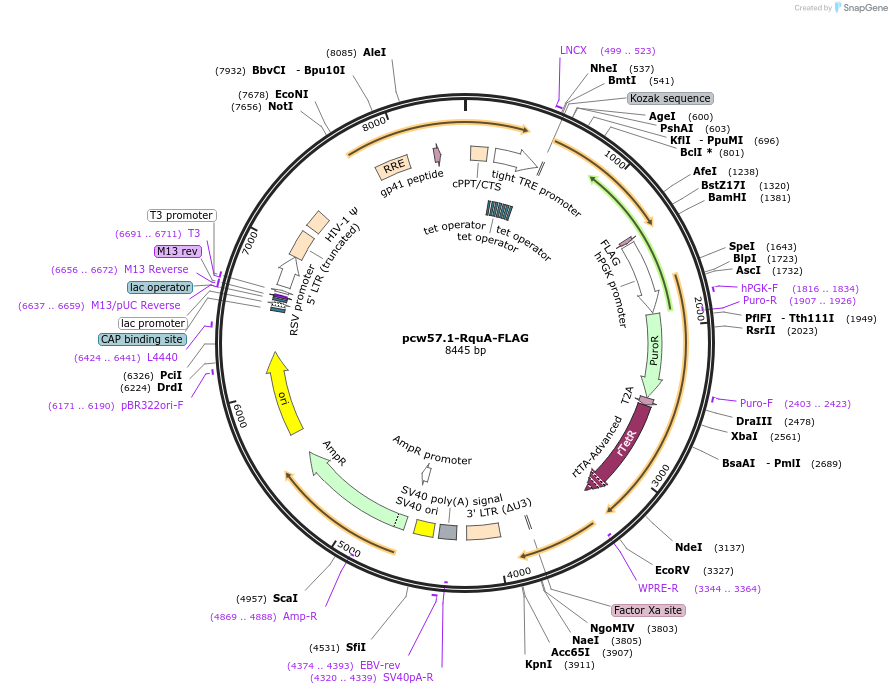
pcw57.1-RquA-FLAG
Plasmid#229685PurposeDoxycycline-inducible expression of RquA-FLAGDepositorInsertRquA-FLAG
UseLentiviralExpressionMammalianPromoterTREAvailable SinceFeb. 6, 2025AvailabilityAcademic Institutions and Nonprofits only