We narrowed to 2,376 results for: pet28
-
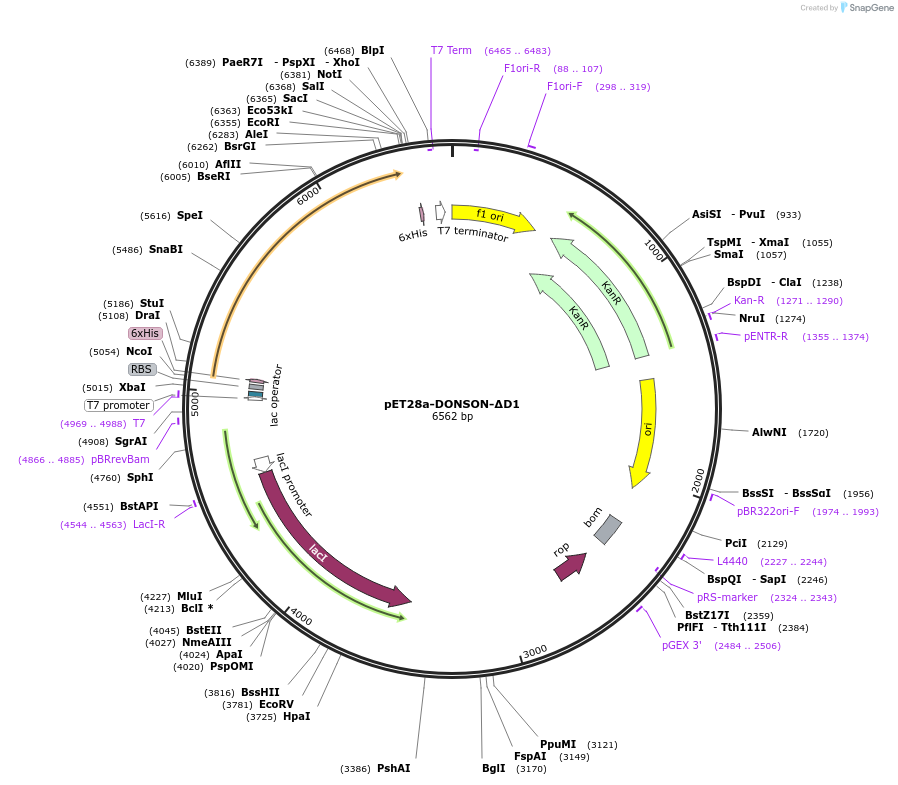
Plasmid#208804PurposeXenopus laevis His6-DONSON (1-155aa removed)DepositorInsertDONSON (donson.L Frog)
TagsHisExpressionBacterialMutationΔD1, deletion of 1-155aaPromoterT7Available SinceFeb. 8, 2024AvailabilityAcademic Institutions and Nonprofits only -
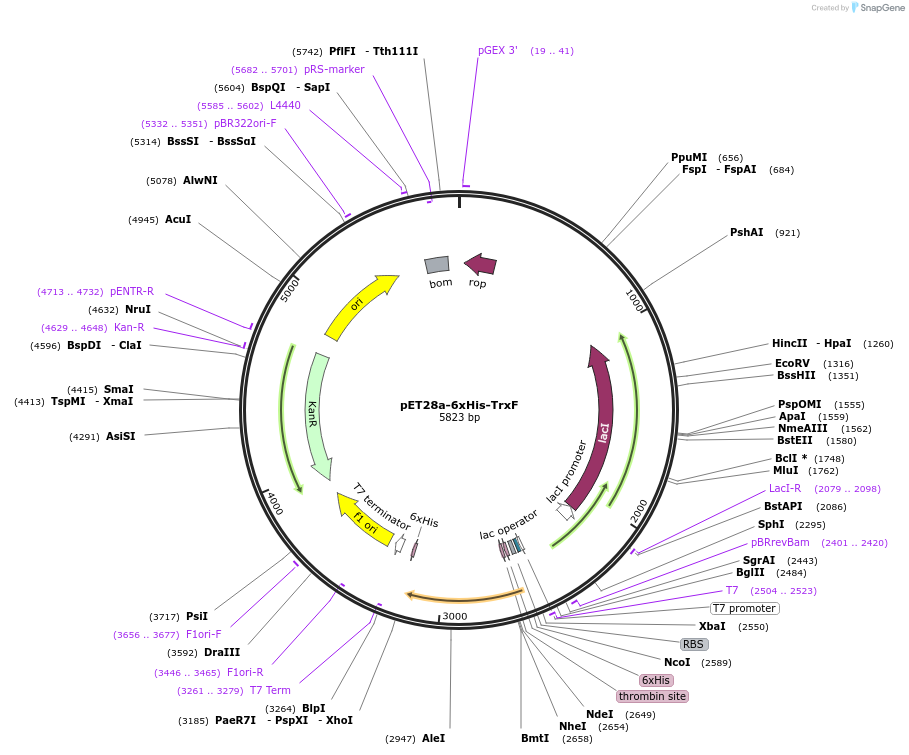
pET28a-6xHis-TrxF
Plasmid#207454PurposeE. coli expression of Mesembryanthemum crystallinum Thioredoxin F with thrombin-cleavable N-terminal 6xHis tagDepositorInsertThioredoxin F
Tags6xHisExpressionBacterialMutationCodon optimized for E coli expressionPromoterT7Available SinceJan. 17, 2024AvailabilityAcademic Institutions and Nonprofits only -
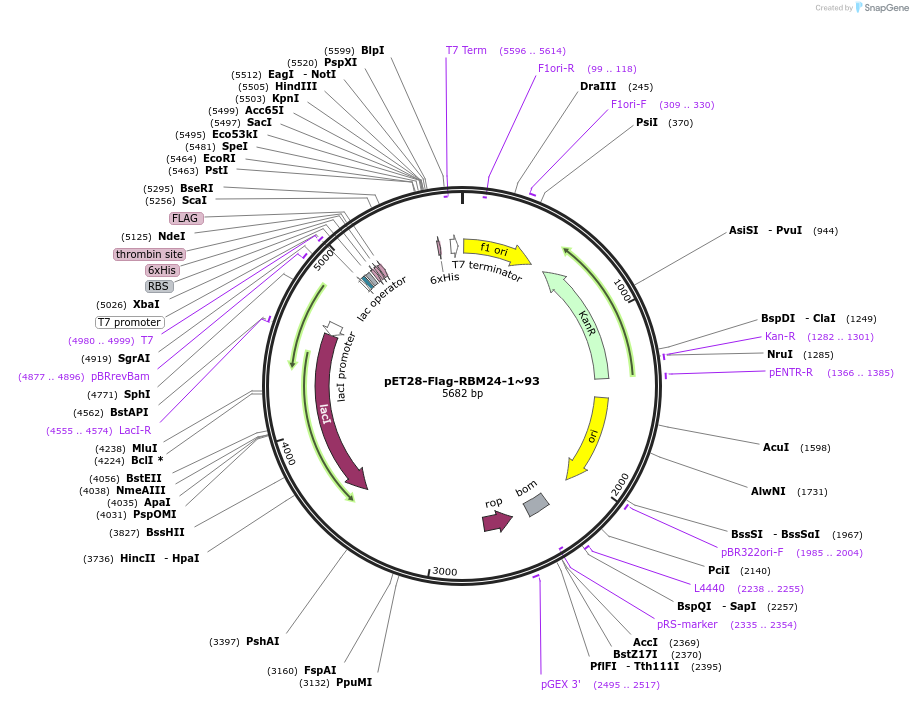
pET28-Flag-RBM24-1~93
Plasmid#208633Purposefor E. coli expression of truncated Flag tagged human RBM24 (ENST00000379052), aa1~93DepositorInserthuman RBM24 (ENST00000216146) aa 1~93 (RBM24 Human)
Tags6xHis and FlagExpressionBacterialMutationonly aa 1~93, other residues not includedPromoterT7Available SinceJan. 9, 2024AvailabilityAcademic Institutions and Nonprofits only -
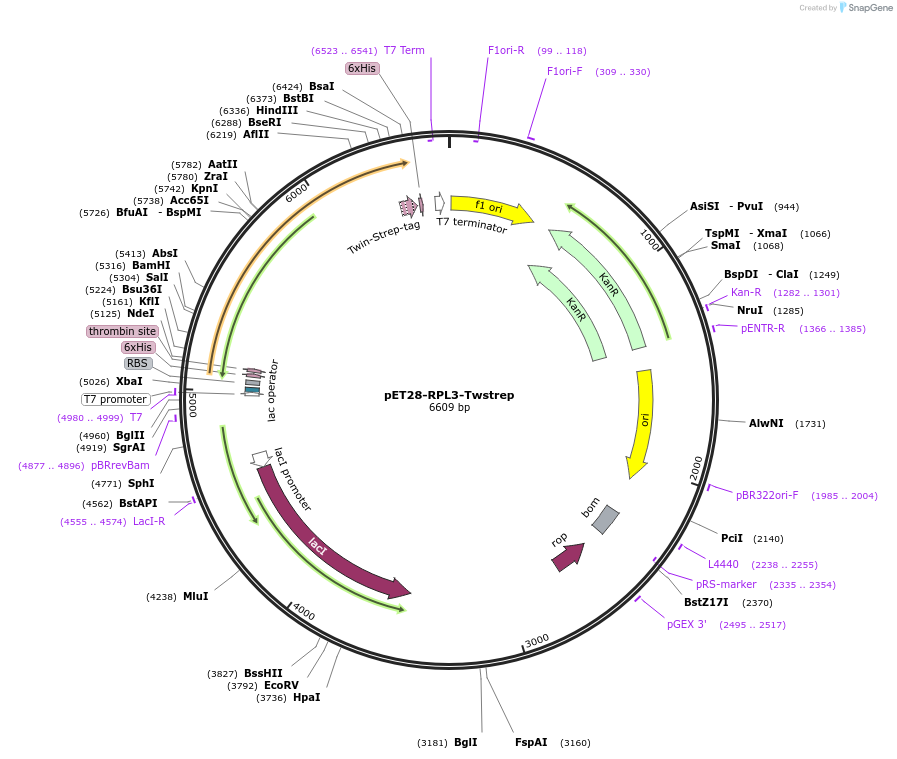
pET28-RPL3-Twstrep
Plasmid#208623Purposefor E. coli expression of C-ter Twin Strep tagged human RPL3 (ENST00000216146)DepositorInserthuman RPL3 (ENST00000216146) (RPL3 Human)
Tags6xHis and Twin StrepExpressionBacterialPromoterT7Available SinceJan. 9, 2024AvailabilityAcademic Institutions and Nonprofits only -
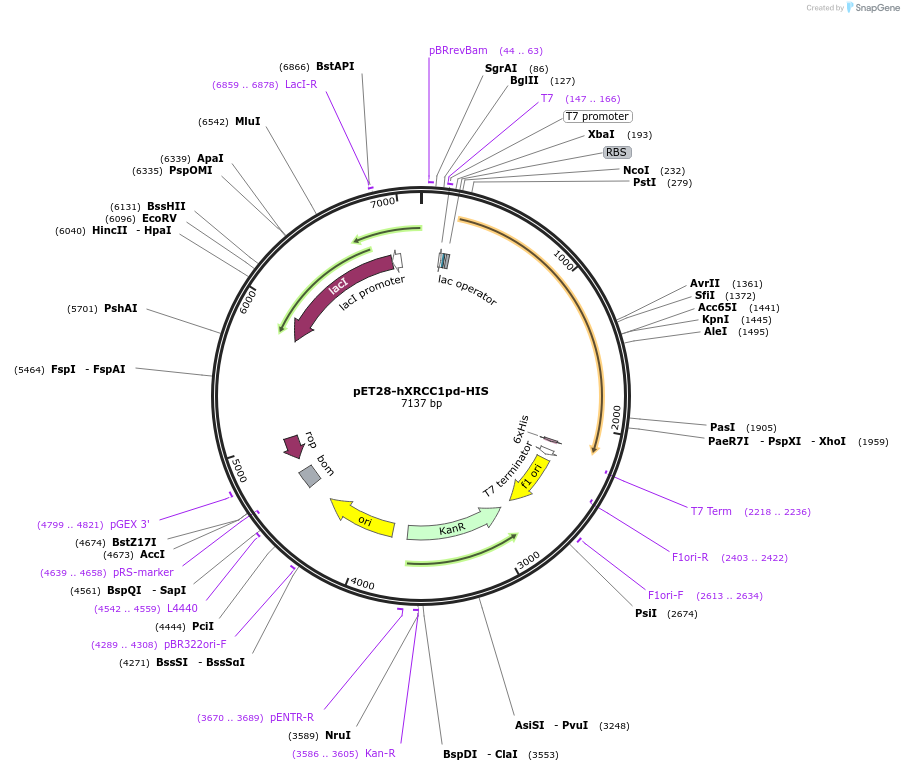
pET28-hXRCC1pd-HIS
Plasmid#206036PurposeBacterial expression of PAR-deficient hXRCC1pd with 6xHis tag, under the control of T7 promoterDepositorInserthXRCC1
TagsHISExpressionBacterialMutationS103A,R186A,S184A,S193A,S219A,S220A,S236A,S268APromoterT7Available SinceJan. 3, 2024AvailabilityAcademic Institutions and Nonprofits only -
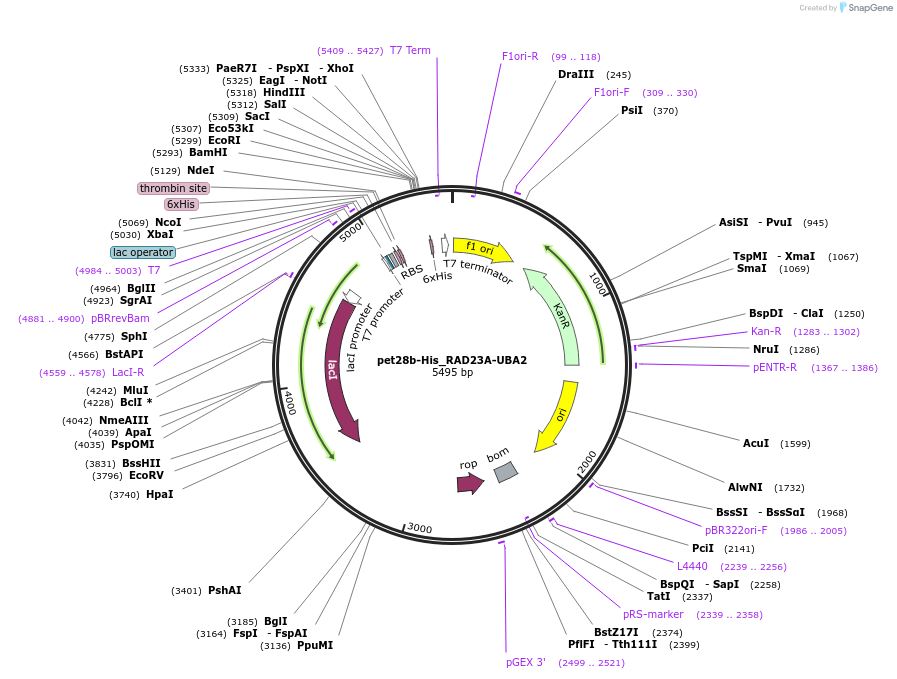
pet28b-His_RAD23A-UBA2
Plasmid#201550PurposeExpresses UBA2 of human RAD23A in E. coliDepositorInsertUV excision repair protein RAD23 homolog A (RAD23A Human)
Tags6x HisExpressionBacterialMutationExpresses amino acids 311-363 of DNA polymerase i…PromoterT7Available SinceNov. 8, 2023AvailabilityAcademic Institutions and Nonprofits only -
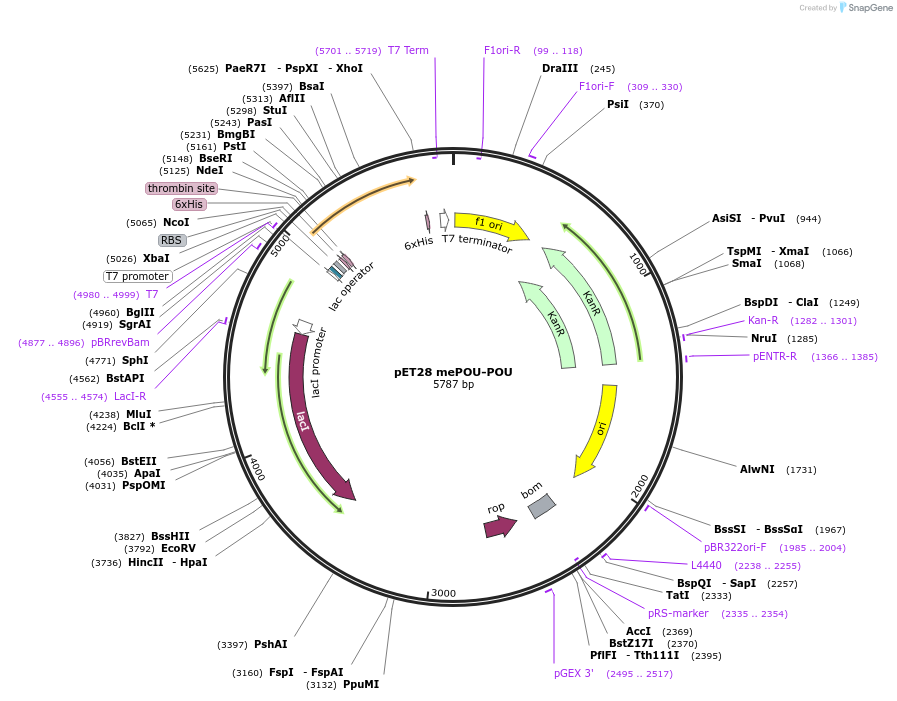
pET28 mePOU-POU
Plasmid#206392PurposeExpresses the DNA binding domain of the ePOU with a 6xHis tag for protein purificationDepositorInsertePOU
Tags6xHisExpressionBacterialMutationPOU DNA binding domain onlyAvailable SinceOct. 6, 2023AvailabilityAcademic Institutions and Nonprofits only -
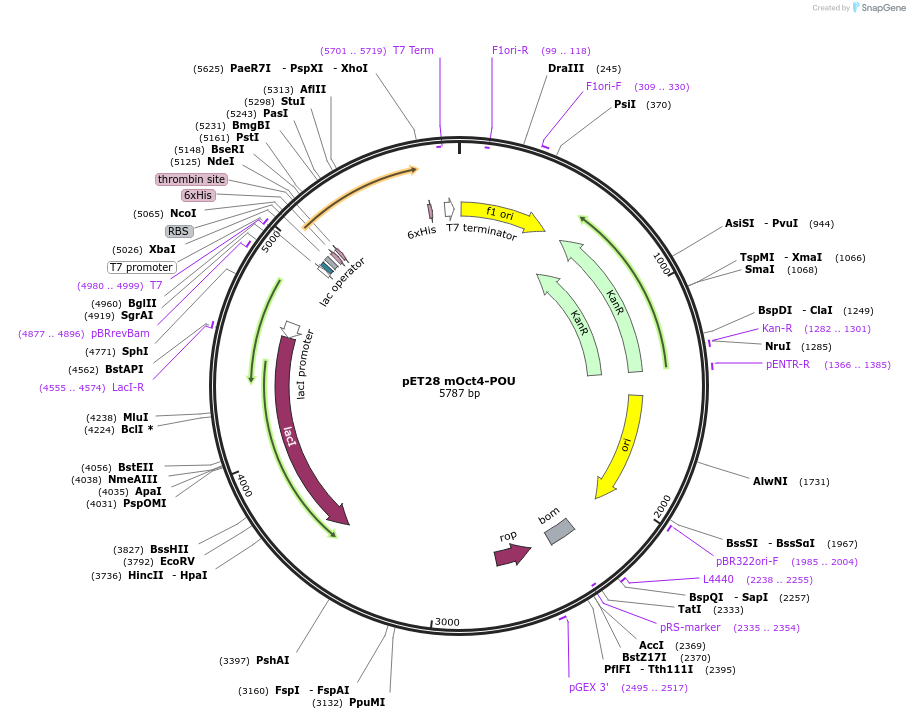
pET28 mOct4-POU
Plasmid#206393PurposeExpresses the DNA binding domain of the Oct4 with a 6xHis tag for protein purificationDepositorAvailable SinceSept. 20, 2023AvailabilityAcademic Institutions and Nonprofits only -
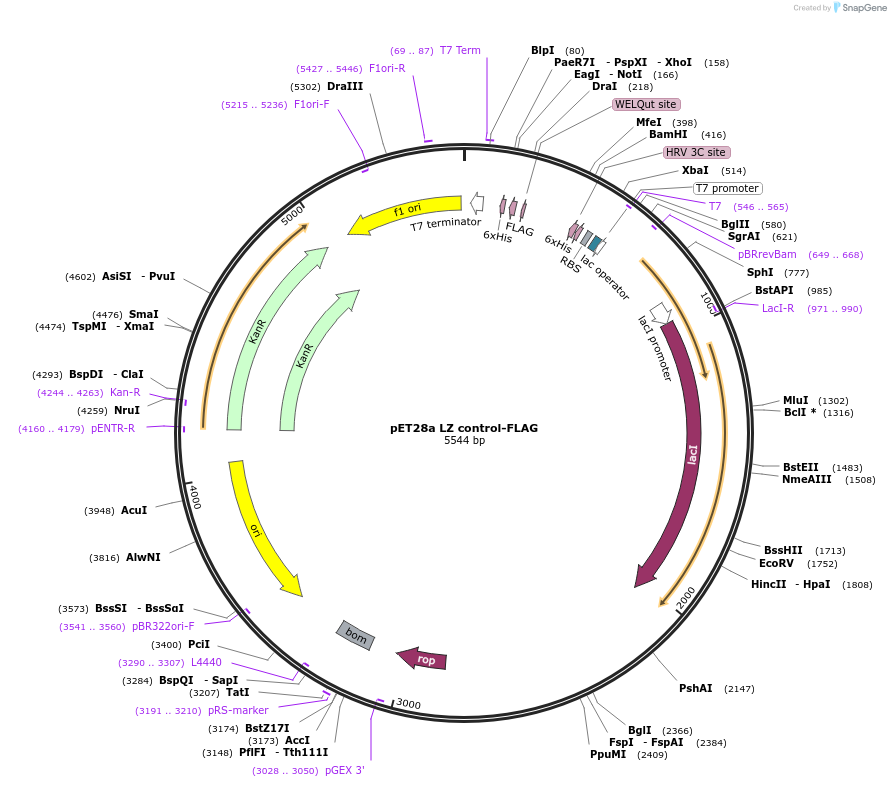
pET28a LZ control-FLAG
Plasmid#199678PurposeBacterial expression of Leucine Zipper (LZ) control-FlagDepositorInsertLeucine zipper control
Tags6xHis and FlagExpressionBacterialPromoterT7 promoterAvailable SinceMay 23, 2023AvailabilityAcademic Institutions and Nonprofits only -
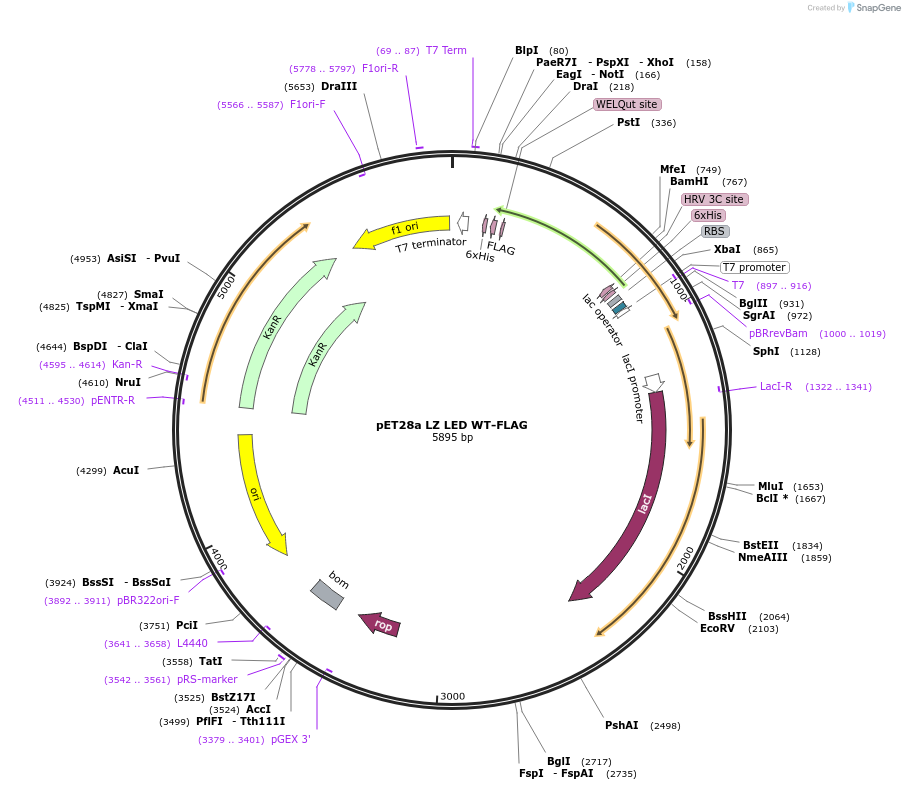
pET28a LZ LED WT-FLAG
Plasmid#199679PurposeBacterial expression of Leucine Zipper (LZ) LYCHOS LED-FlagDepositorInsertLeucine zipper LYCHOS LED WT (GPR155 Human)
Tags6xHis and FlagExpressionBacterialPromoterT7 promoterAvailable SinceMay 16, 2023AvailabilityAcademic Institutions and Nonprofits only