We narrowed to 41,926 results for: TRO
-
Plasmid#227551PurposeLevel 1 - Position 1 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CoACT3_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
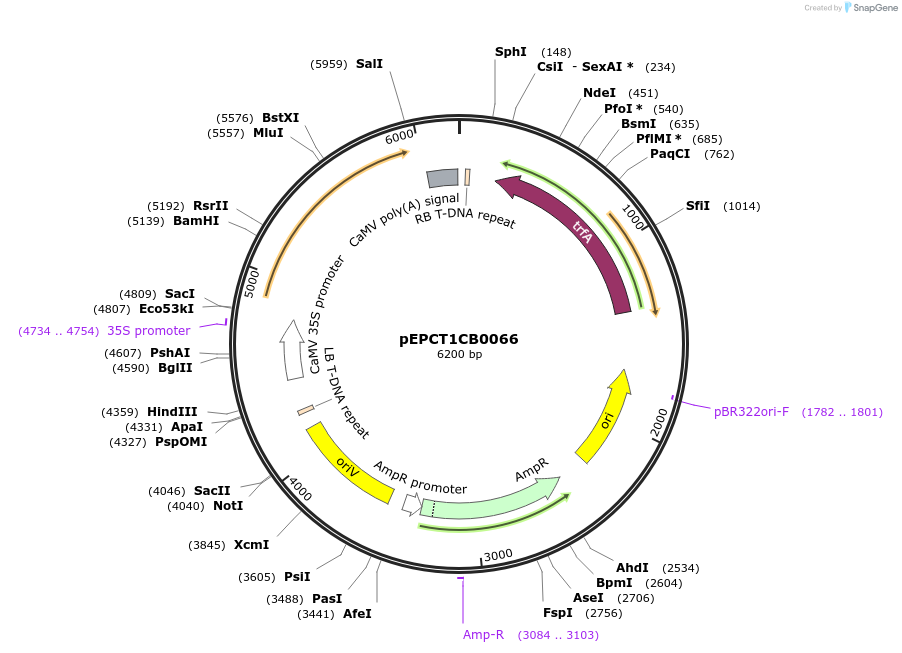
pEPCT1CB0066
Plasmid#227552PurposeLevel 1 - Position 3 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CoACT4_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
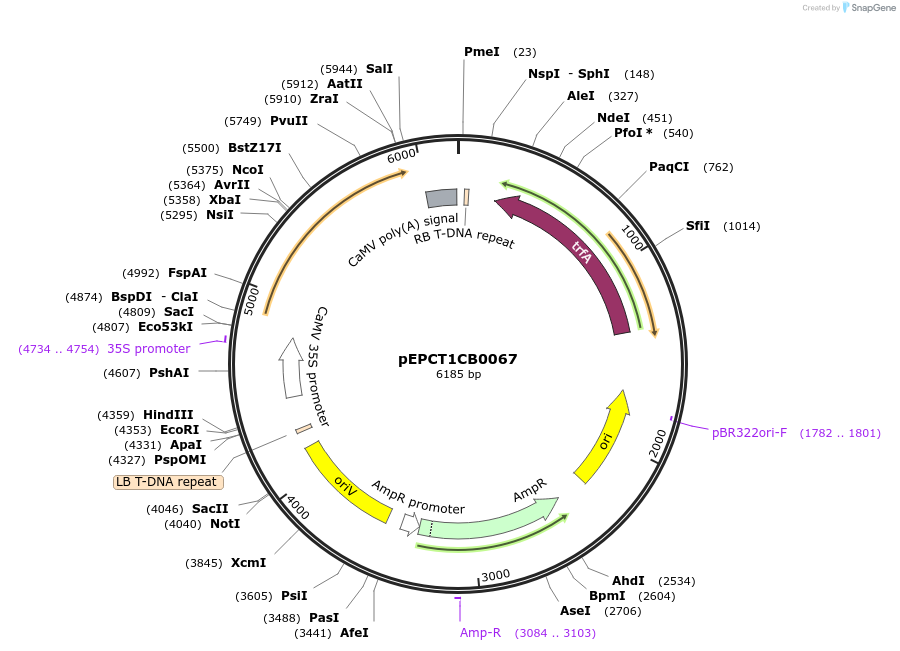
pEPCT1CB0067
Plasmid#227553PurposeLevel 1 - Position 3 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CoACT5_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
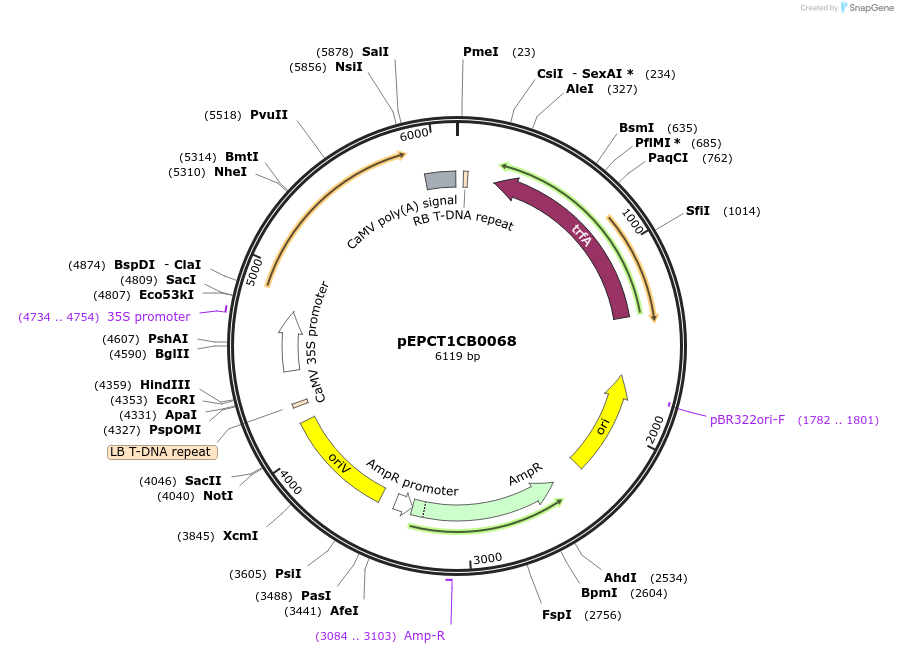
pEPCT1CB0068
Plasmid#227554PurposeLevel 1 - Position 3 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CoACT6_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
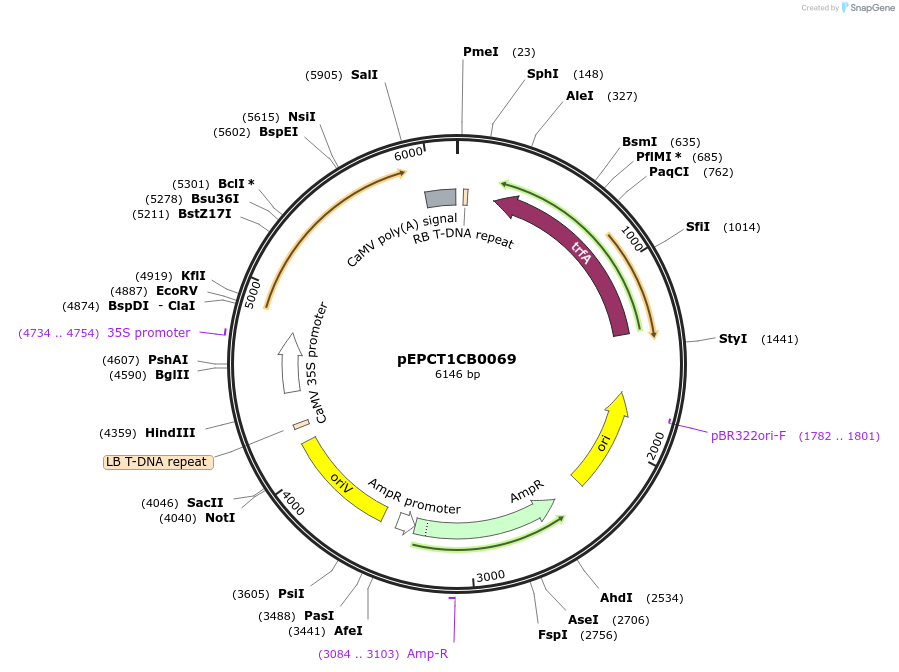
pEPCT1CB0069
Plasmid#227555PurposeLevel 1 - Position 3 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CoACT7_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
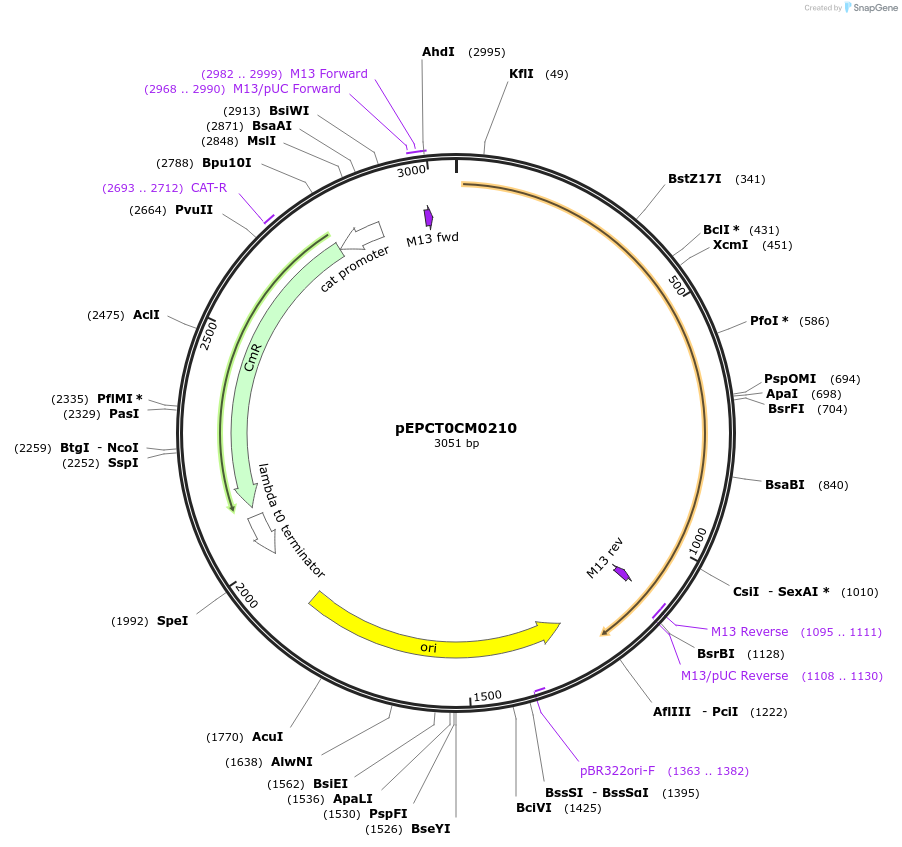
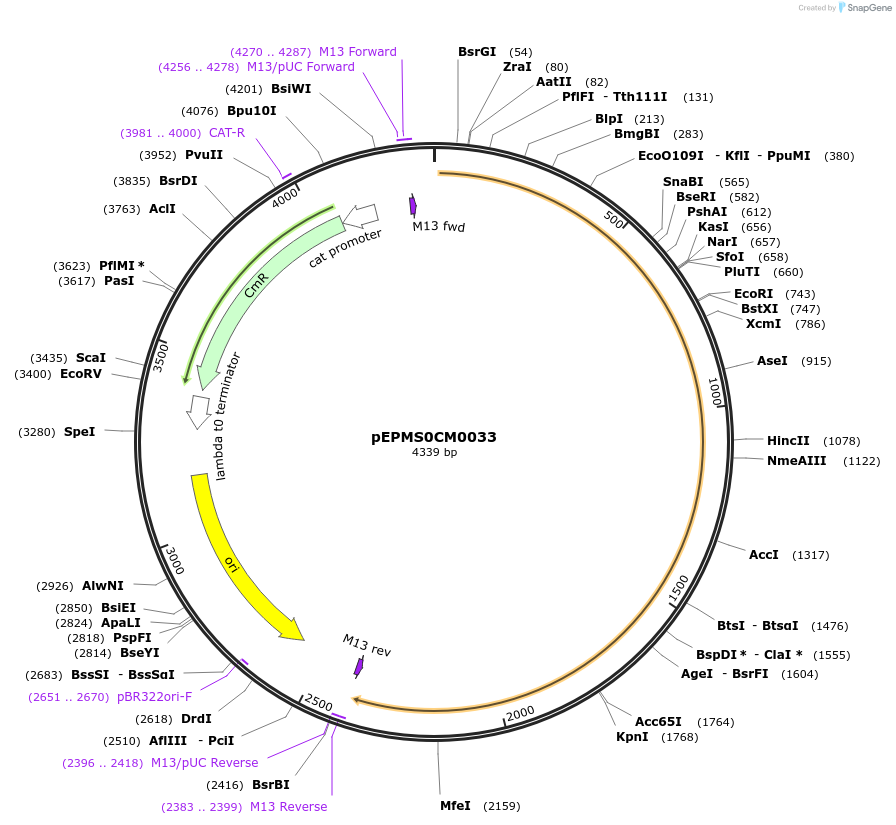
pEPCT0CM0210
Plasmid#227532PurposeL0 part - CDS (without STOP codon)DepositorInsertCoACT7 (Calendula officinalis acyltransferase 7)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
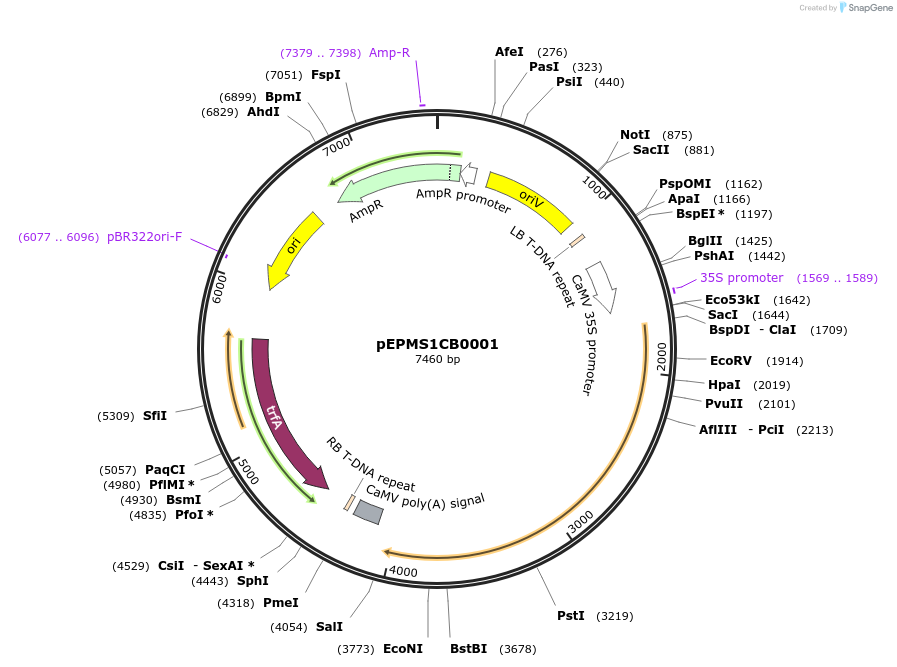
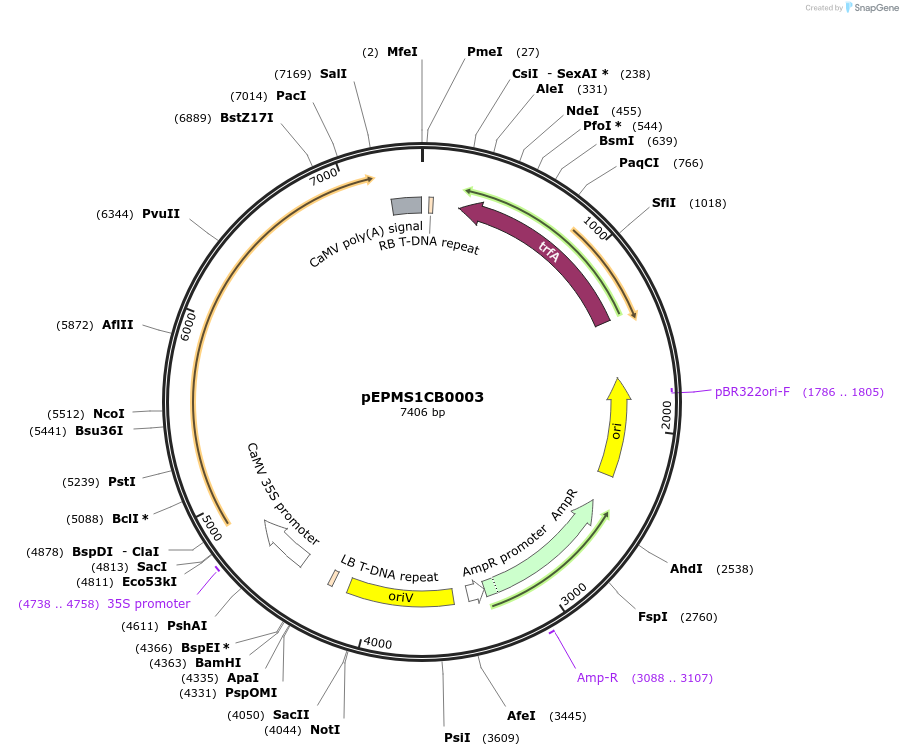
pEPMS1CB0001
Plasmid#227533PurposeLevel 1 - Position 1 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CoTXSS_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
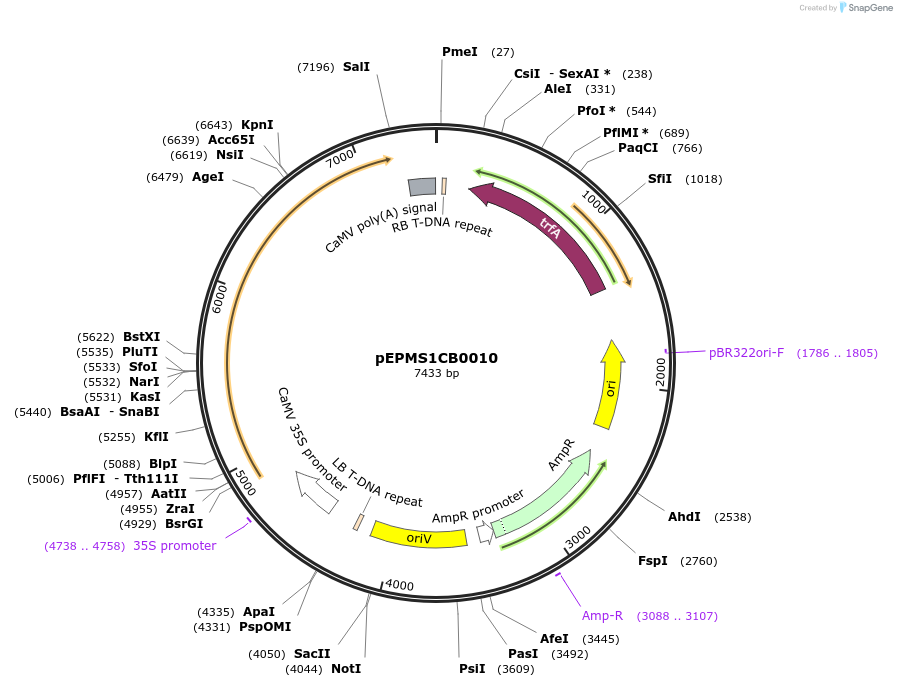
pEPMS1CB0010
Plasmid#227534PurposeLevel 1 - Position 1 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CcTXSS_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
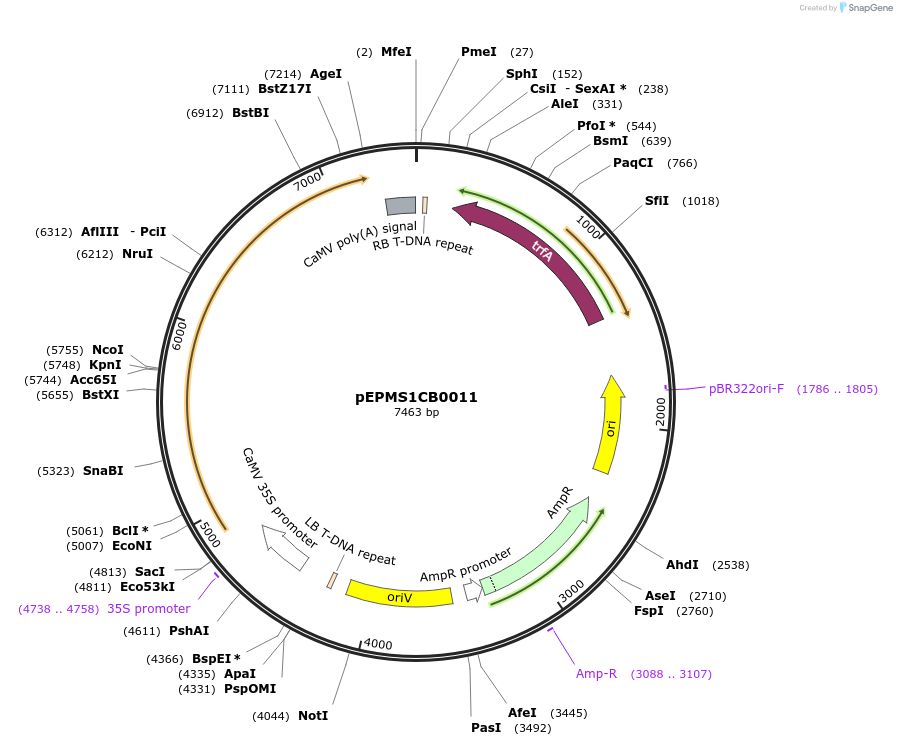
pEPMS1CB0011
Plasmid#227536PurposeLevel 1 - Position 1 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CeTXSS_35S
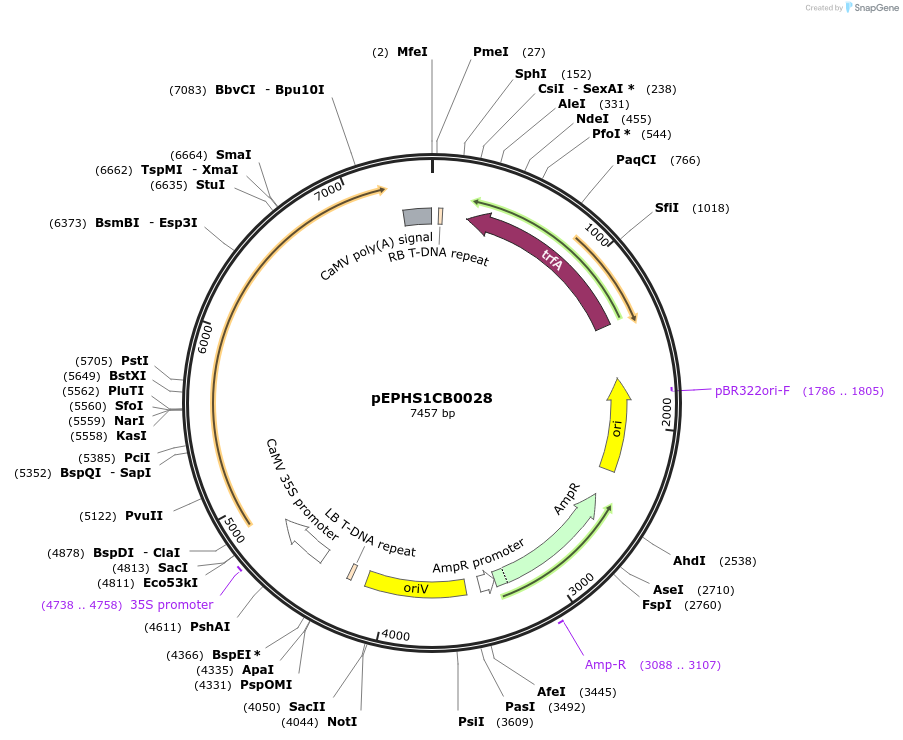
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
pEPHS1CB0028
Plasmid#227537PurposeLevel 1 - Position 1 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_TdTXSS_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
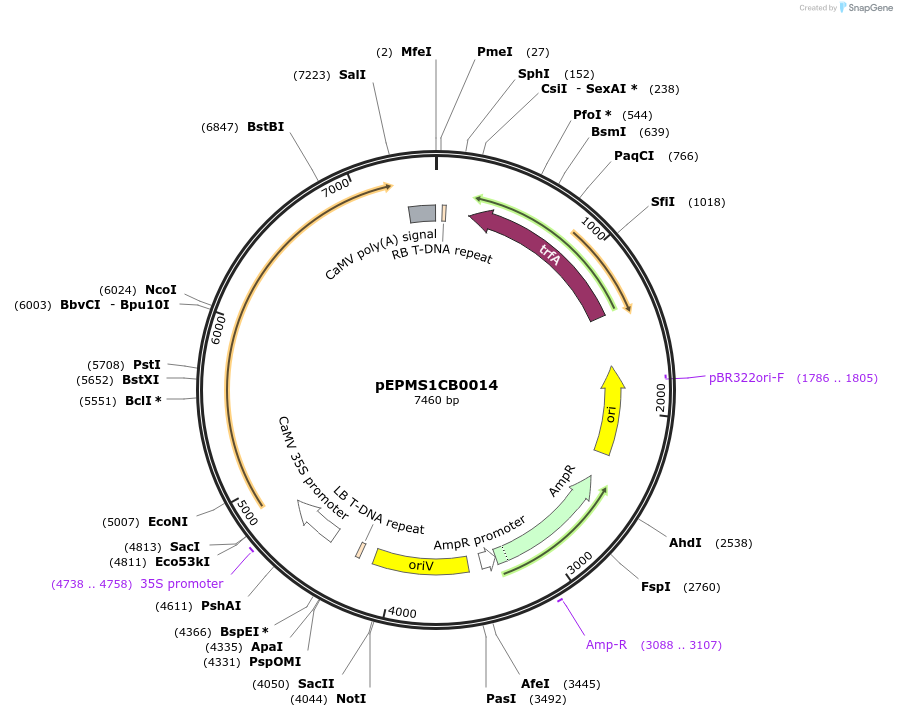
pEPMS1CB0014
Plasmid#227538PurposeLevel 1 - Position 1 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_LsTXSS_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
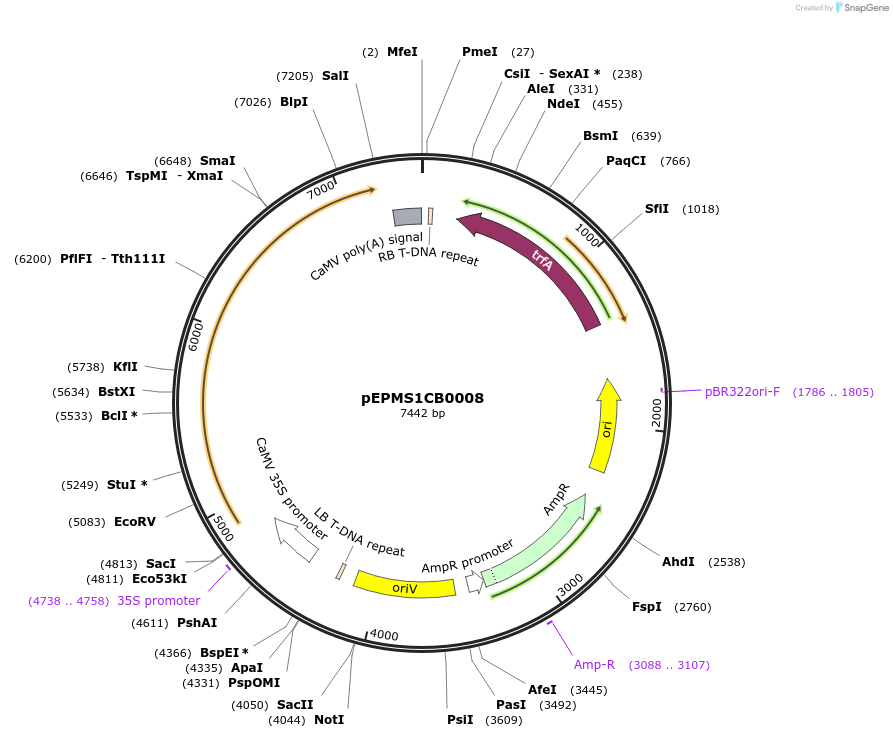
pEPMS1CB0008
Plasmid#227539PurposeLevel 1 - Position 1 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_TkTXSS _35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
pEPMS1CB0009
Plasmid#227540PurposeLevel 1 - Position 1 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_TcTXSS_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
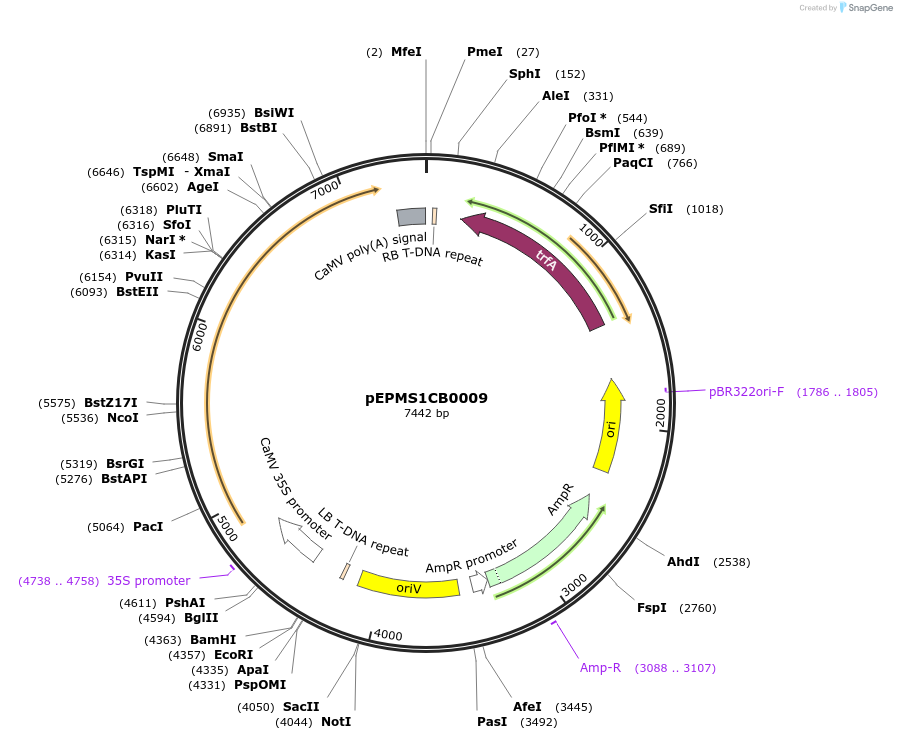
pEPMS1CB0003
Plasmid#227541PurposeLevel 1 - Position 1 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CoMAS_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
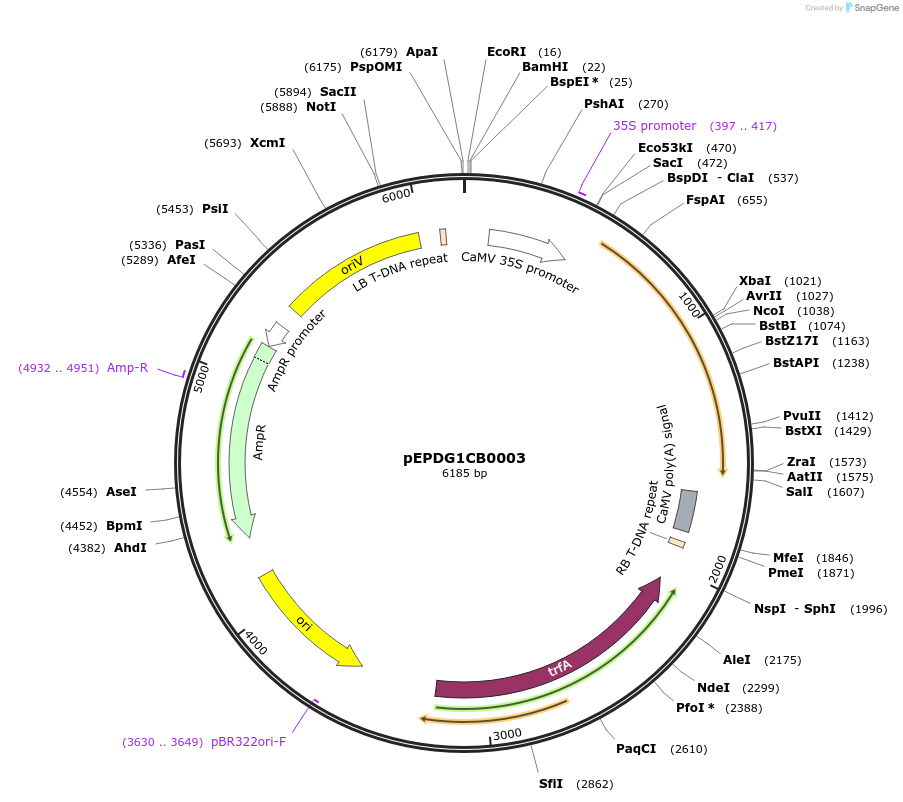
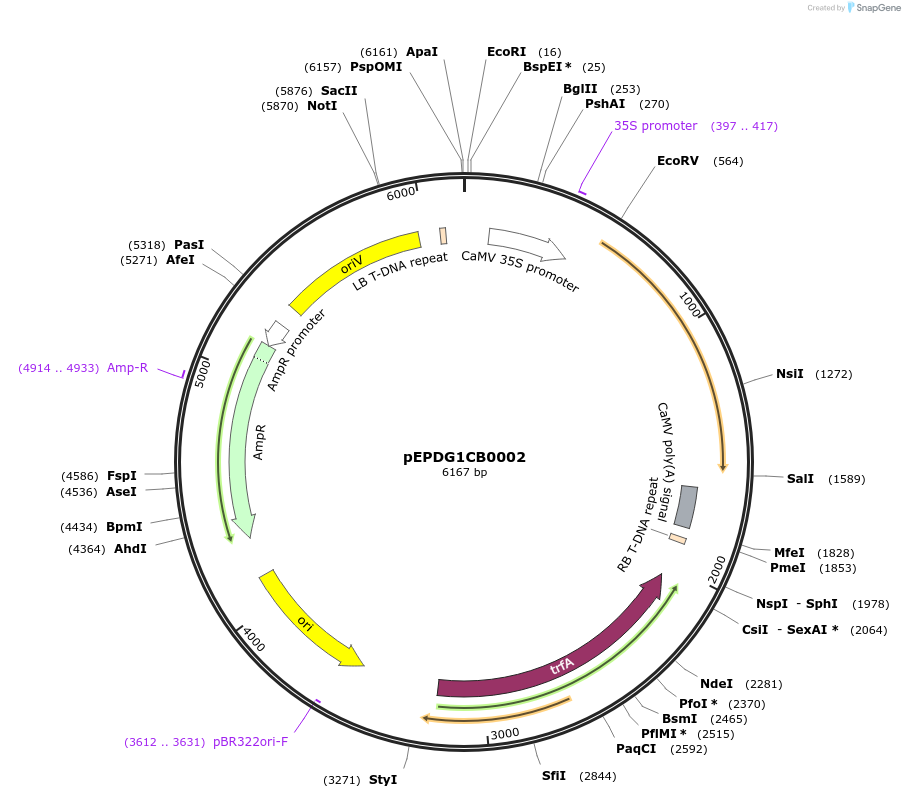
pEPDG1CB0004
Plasmid#227542PurposeLevel 1 - Position 1 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CaTXSS_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
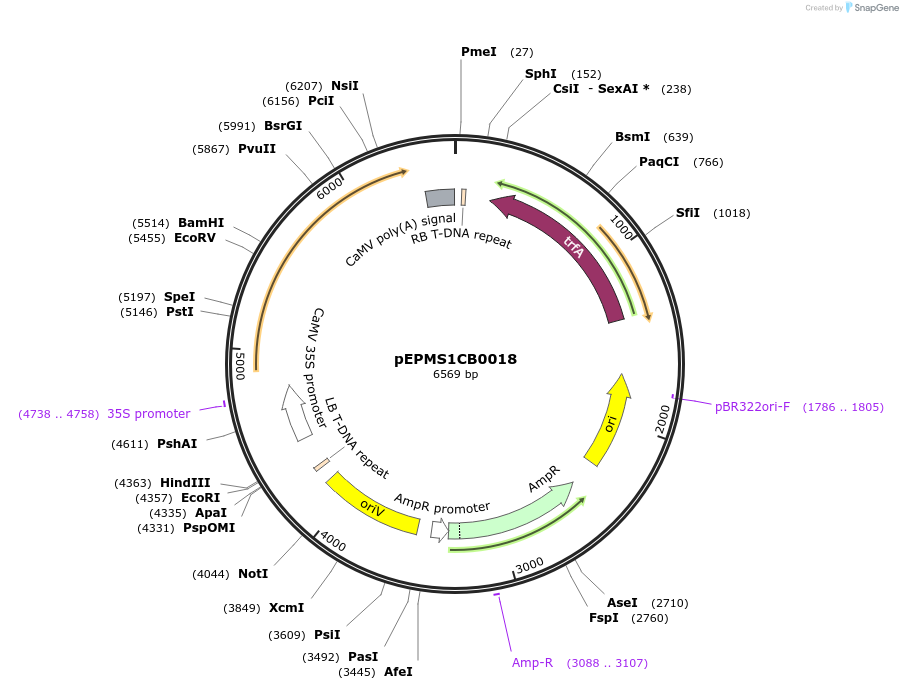
pEPMS1CB0018
Plasmid#227543PurposeLevel 1 - Position 2 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CYP716A392_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
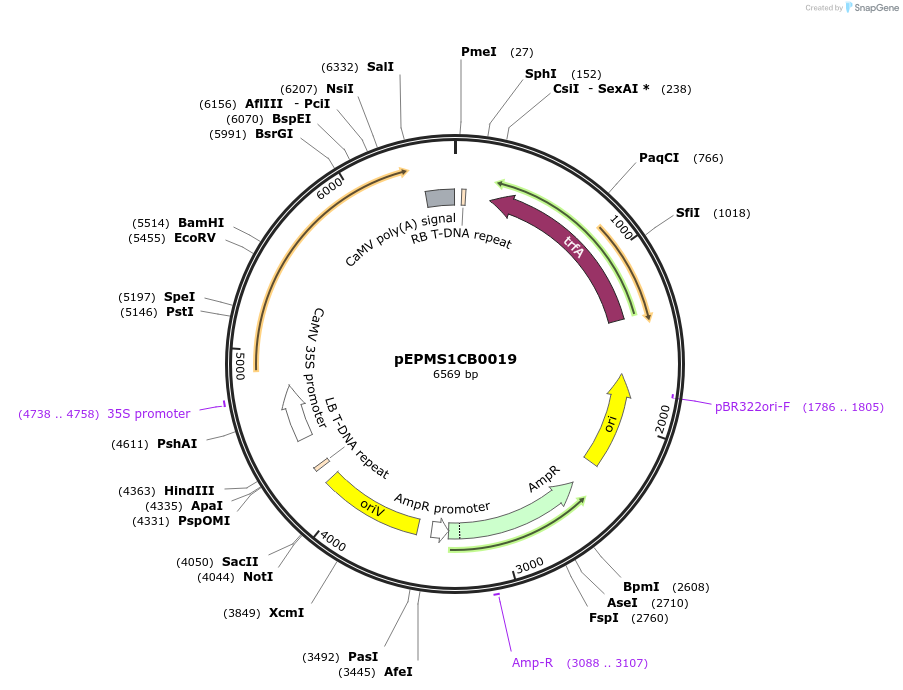
pEPMS1CB0019
Plasmid#227544PurposeLevel 1 - Position 2 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CYP716A393_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
pEPMS1CB0020
Plasmid#227545PurposeLevel 1 - Position 2 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CYP716A429_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
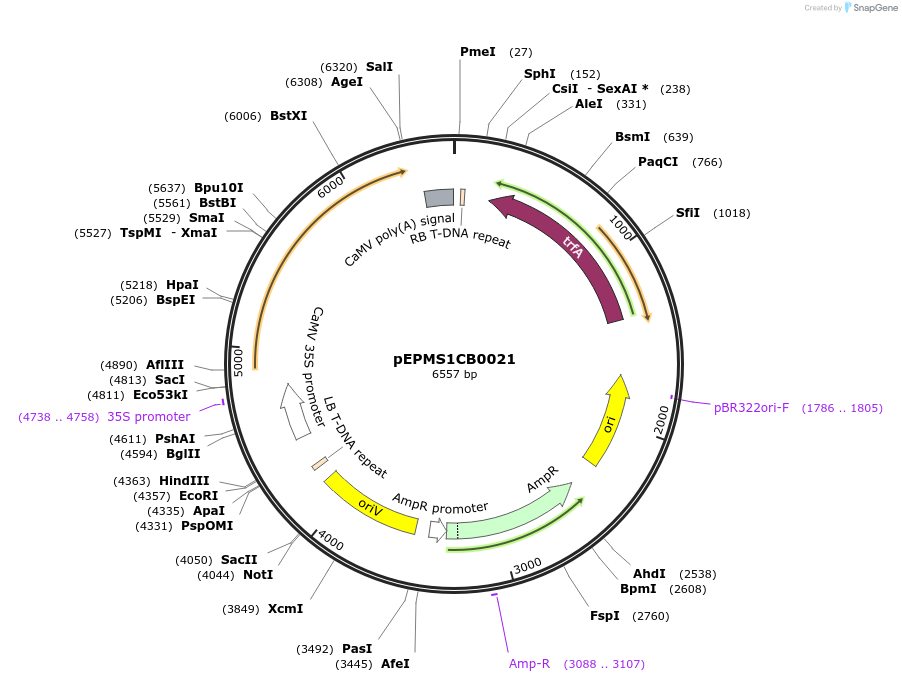
pEPMS1CB0021
Plasmid#227546PurposeLevel 1 - Position 2 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CYP716A430_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
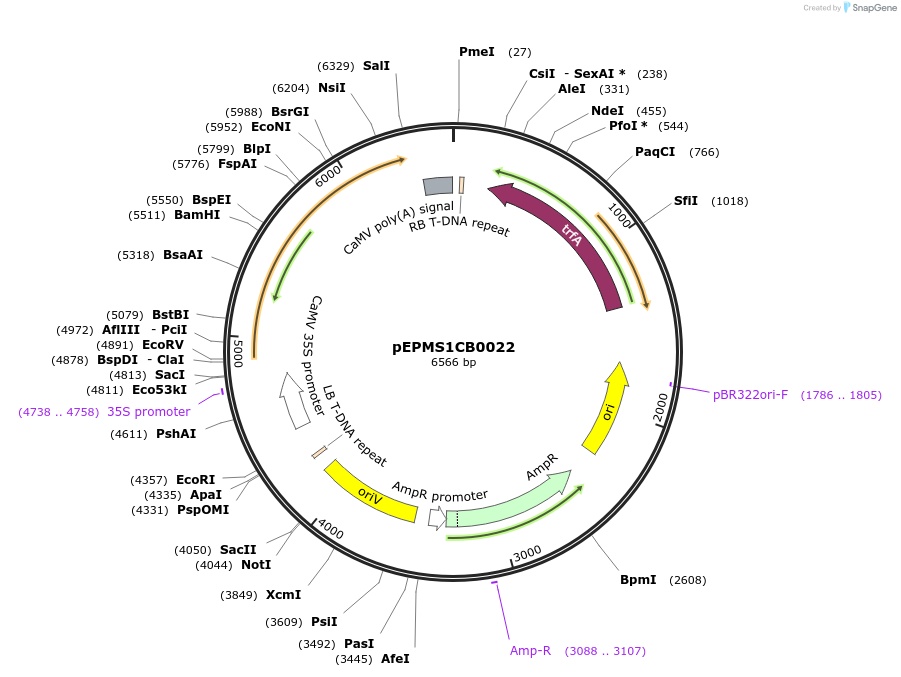
pEPMS1CB0022
Plasmid#227547PurposeLevel 1 - Position 2 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CYP716A431_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
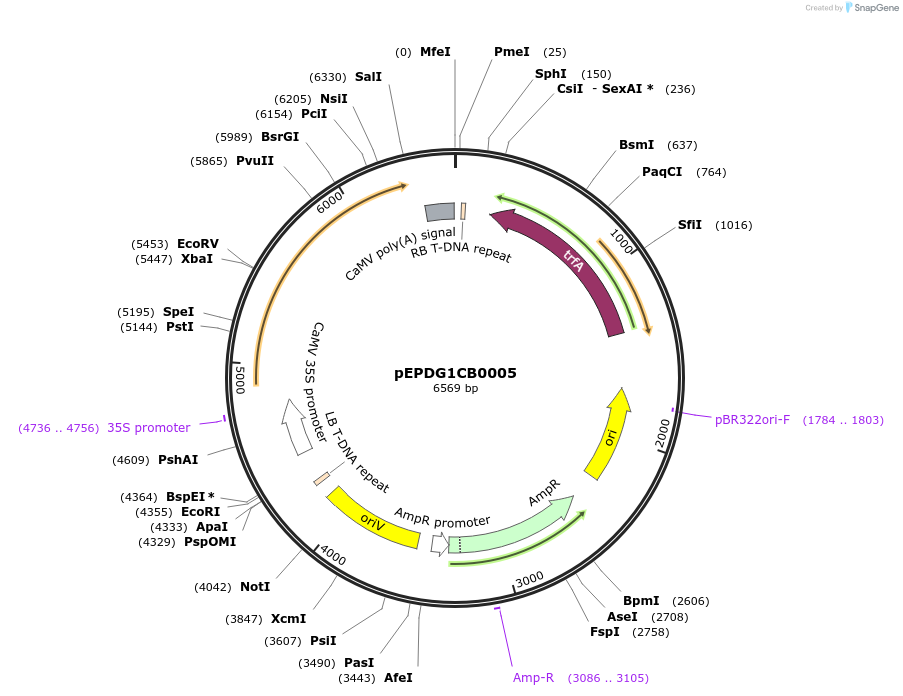
pEPDG1CB0005
Plasmid#227548PurposeLevel 1 - Position 1 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CYP716A392a_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
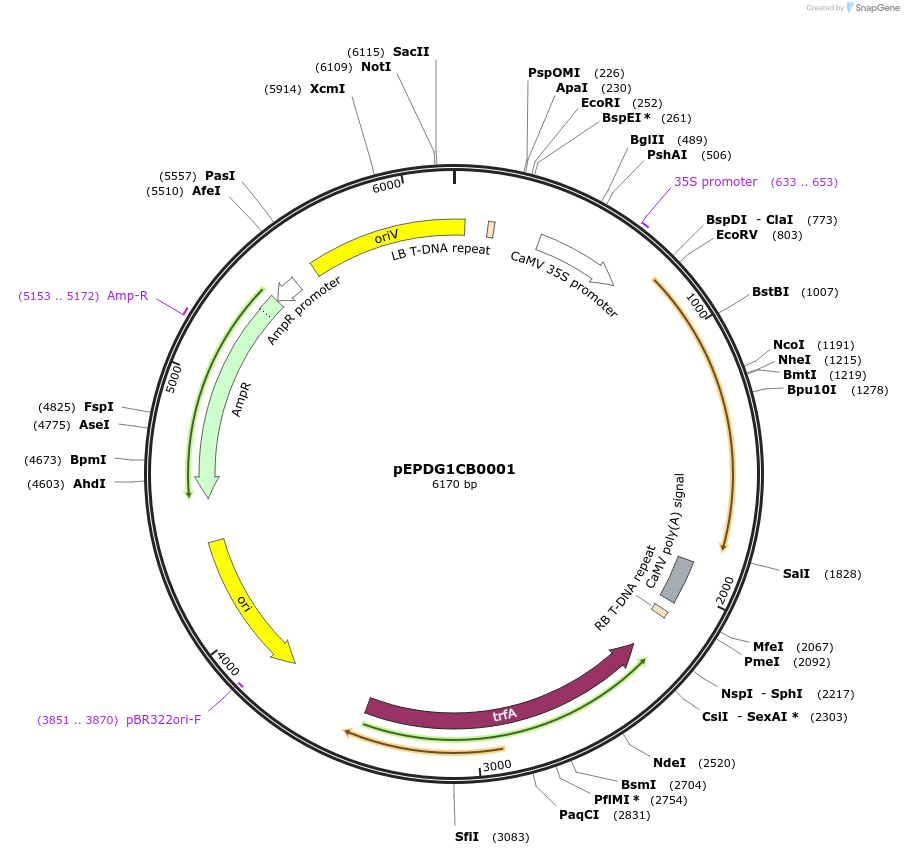
pEPDG1CB0001
Plasmid#227549PurposeLevel 1 - Position 1 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CoACT1_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
pEPDG1CB0002
Plasmid#227550PurposeLevel 1 - Position 1 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CoACT2_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
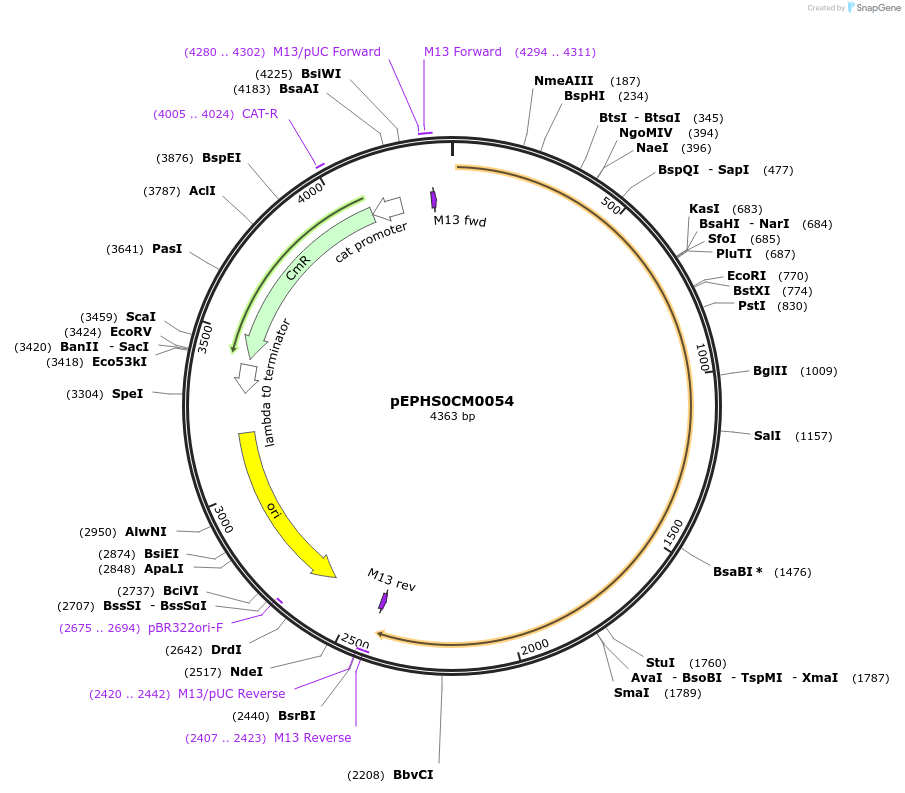
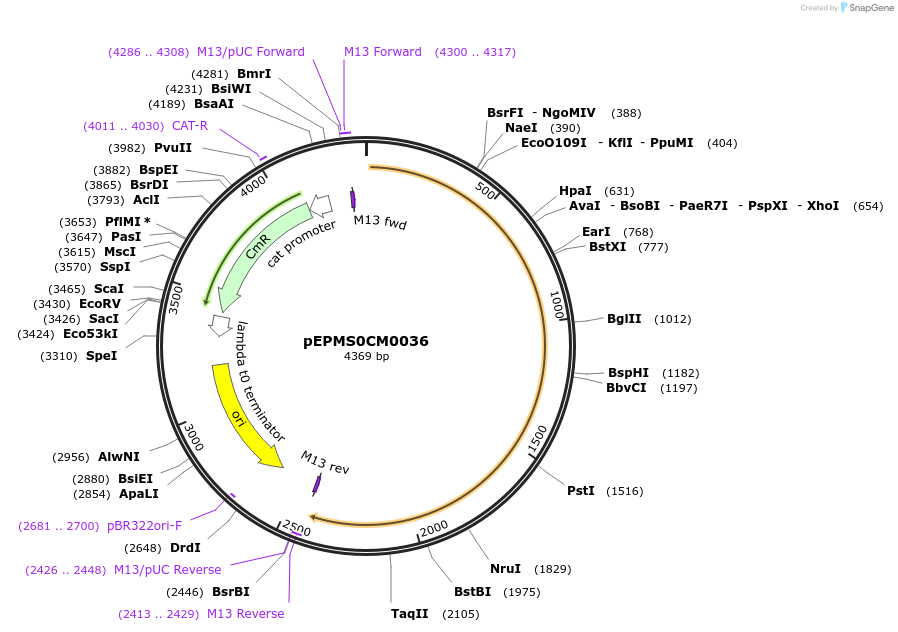
pEPHS0CM0054
Plasmid#227515PurposeL0 part - CDS (without STOP codon)DepositorInsertTdTXSS (Tragopogon dubius taraxasterol synthase)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
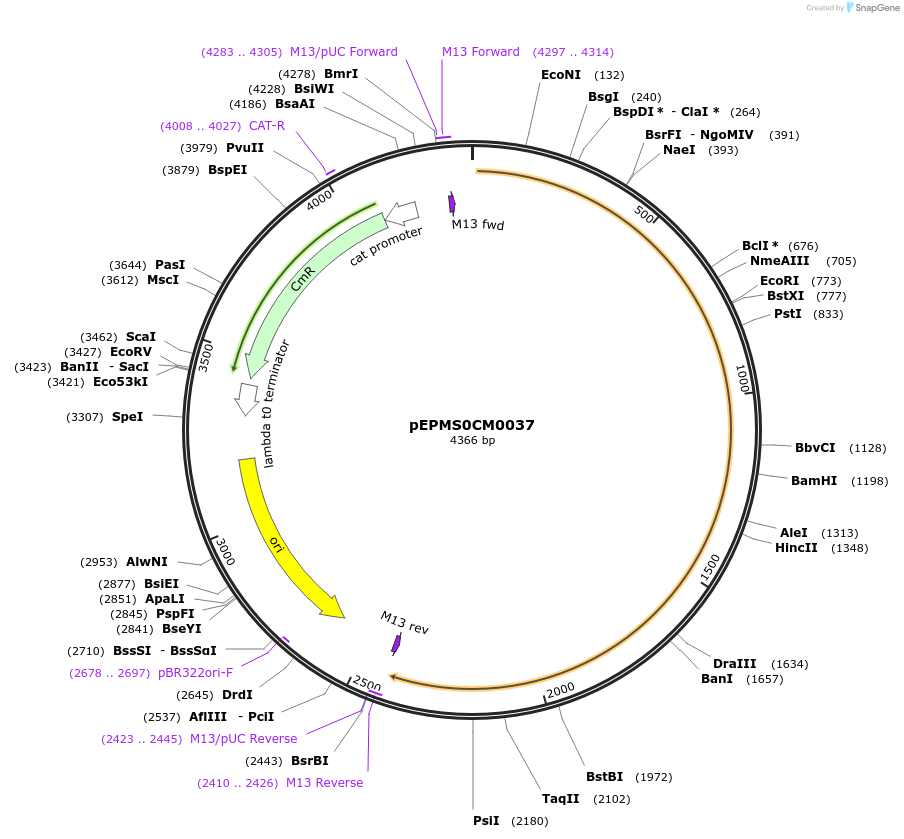
pEPMS0CM0037
Plasmid#227516PurposeL0 part - CDS (without STOP codon)DepositorInsertLsTXSS (Lactuca sativa taraxasterol synthase)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
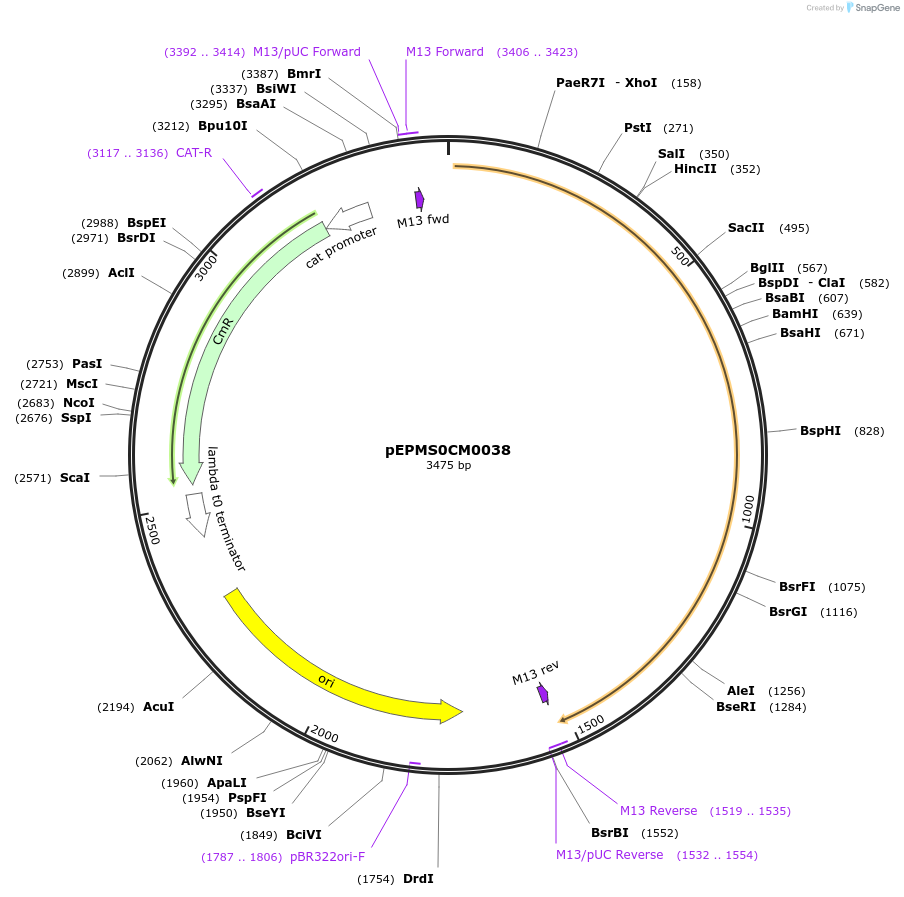
pEPMS0CM0038
Plasmid#227520PurposeL0 part - CDS (without STOP codon)DepositorInsertCYP716A392 (Calendula officinalis cytochrome P450 1)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
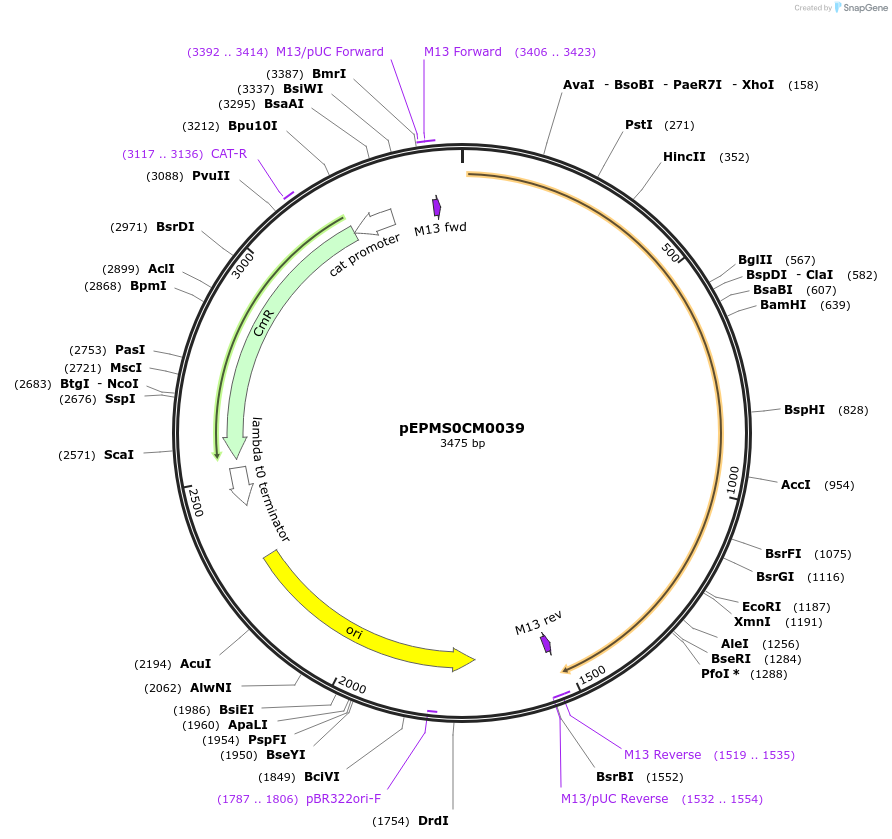
pEPMS0CM0039
Plasmid#227521PurposeL0 part - CDS (without STOP codon)DepositorInsertCYP716A393 (Calendula officinalis cytochrome P450 2)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
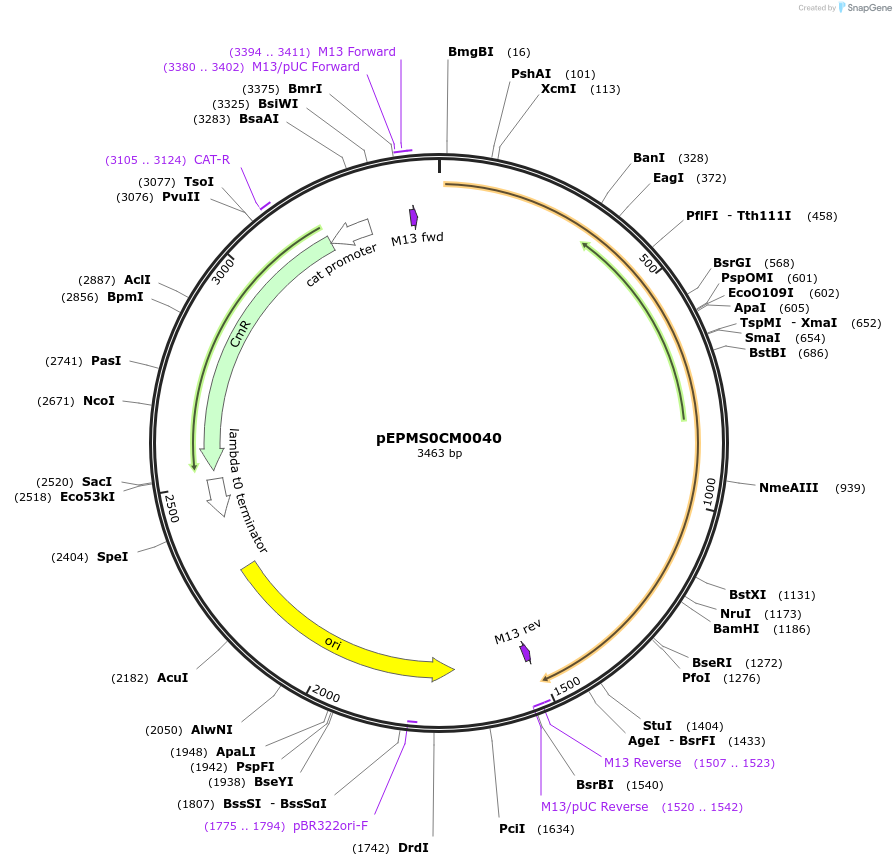
pEPMS0CM0040
Plasmid#227522PurposeL0 part - CDS (without STOP codon)DepositorInsertCYP716A429 (Calendula officinalis cytochrome P450 3)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
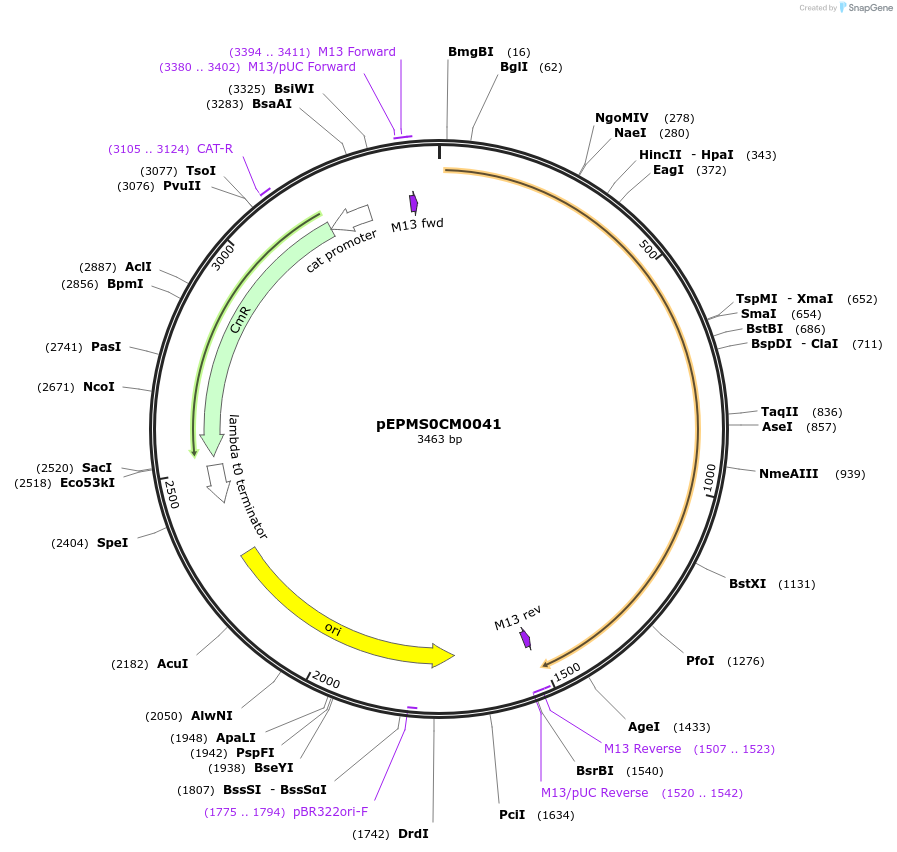
pEPMS0CM0041
Plasmid#227523PurposeL0 part - CDS (without STOP codon)DepositorInsertCYP716A430 (Calendula officinalis cytochrome P450 4)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
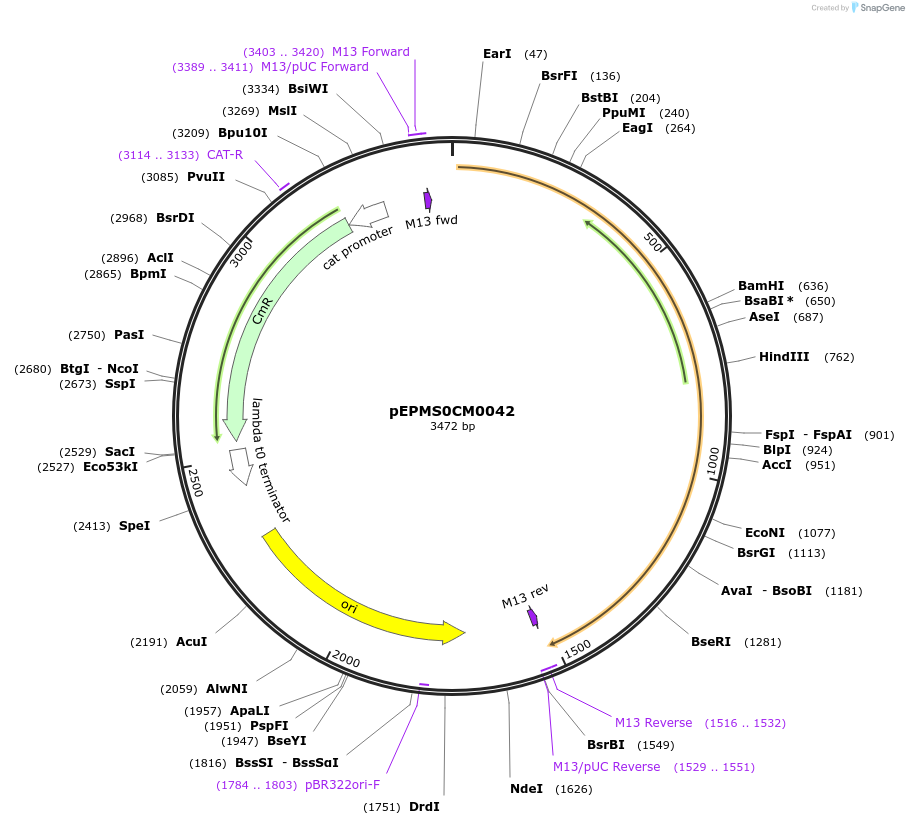
pEPMS0CM0042
Plasmid#227524PurposeL0 part - CDS (without STOP codon)DepositorInsertCYP716A431 (Calendula officinalis cytochrome P450 5)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
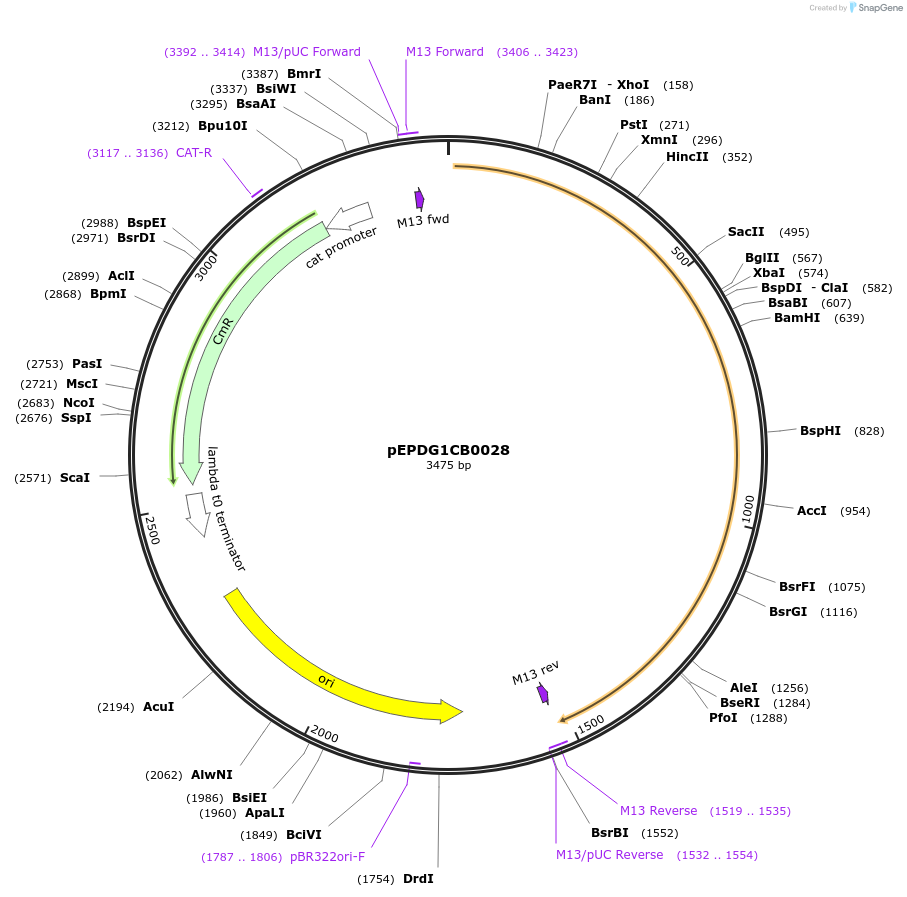
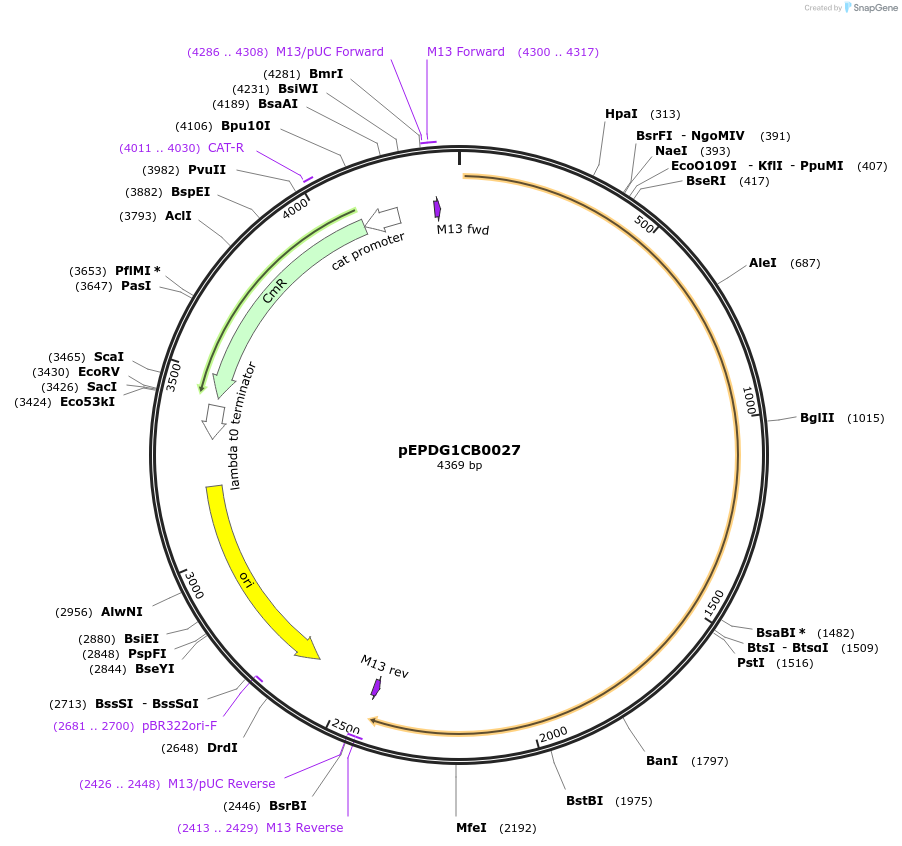
pEPDG1CB0028
Plasmid#227525PurposeL0 part - CDS (without STOP codon)DepositorInsertCYP716A392a (Calendula arvensis cytochrome P450 1)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
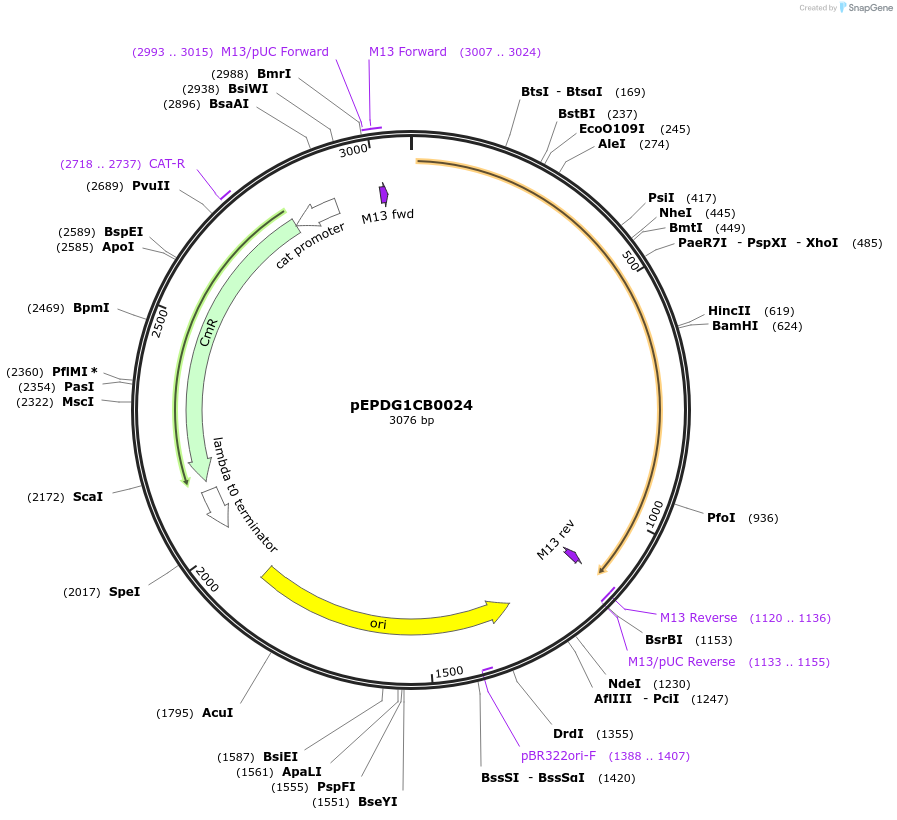
pEPDG1CB0024
Plasmid#227526PurposeL0 part - CDS (without STOP codon)DepositorInsertCoACT1 (Calendula officinalis acyltransferase 1)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
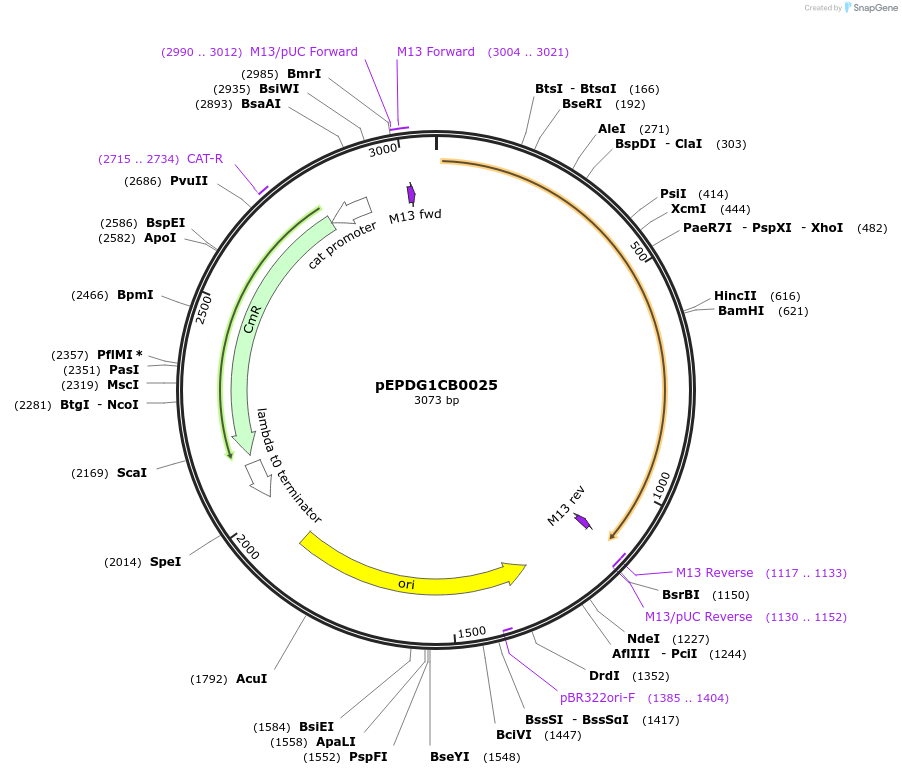
pEPDG1CB0025
Plasmid#227527PurposeL0 part - CDS (without STOP codon)DepositorInsertCoACT2 (Calendula officinalis acyltransferase 2)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
pEPDG1CB0026
Plasmid#227528PurposeL0 part - CDS (without STOP codon)DepositorInsertCoACT3 (Calendula officinalis acyltransferase 3)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
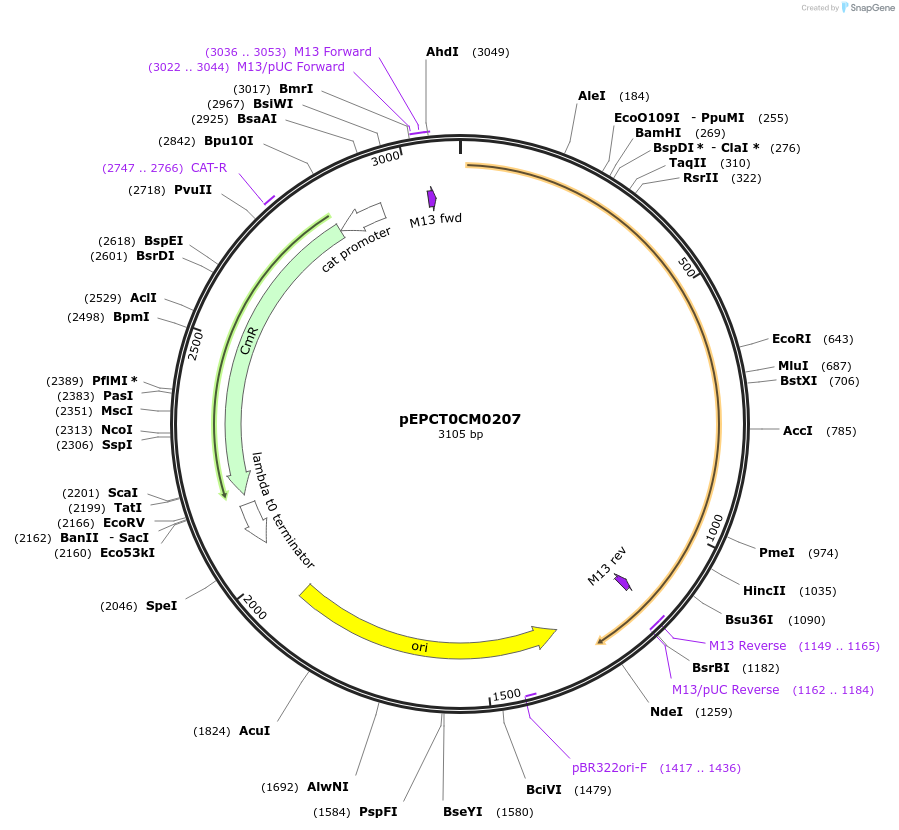
pEPCT0CM0207
Plasmid#227529PurposeL0 part - CDS (without STOP codon)DepositorInsertCoACT4 (Calendula officinalis acyltransferase 4)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
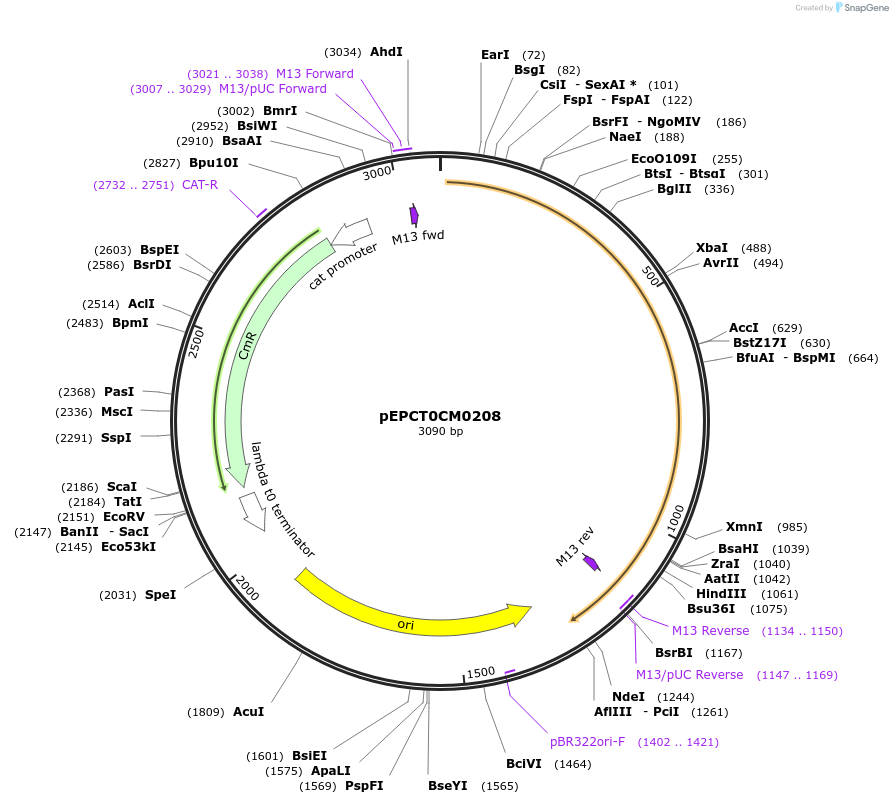
pEPCT0CM0208
Plasmid#227530PurposeL0 part - CDS (without STOP codon)DepositorInsertCoACT5 (Calendula officinalis acyltransferase 5)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
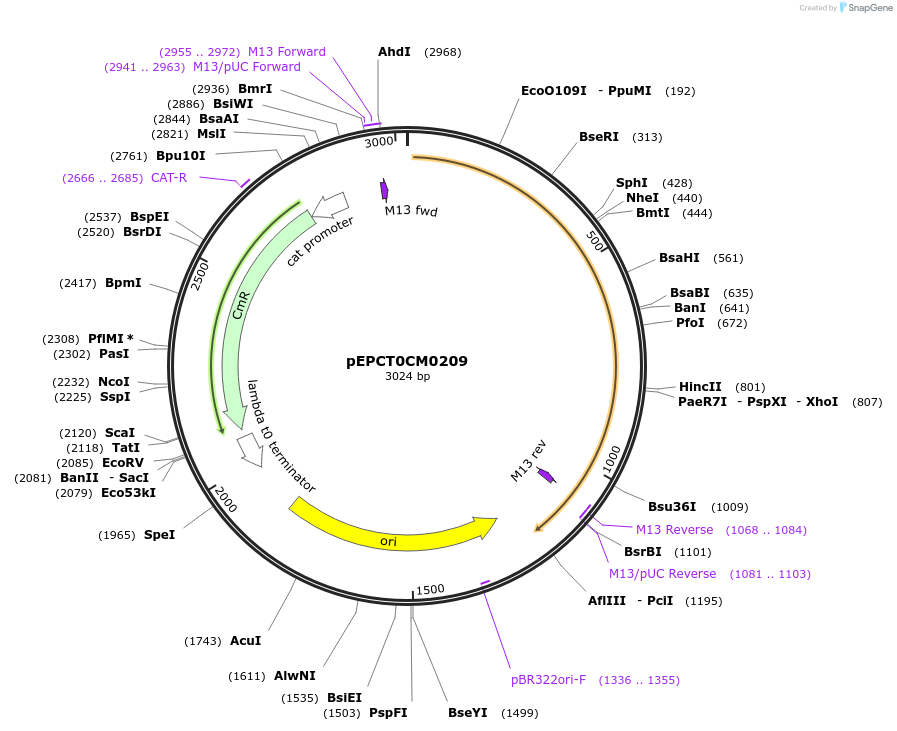
pEPCT0CM0209
Plasmid#227531PurposeL0 part - CDS (without STOP codon)DepositorInsertCoACT6 (Calendula officinalis acyltransferase 6)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
pEPMS0CM0024
Plasmid#227510PurposeL0 part - CDS (without STOP codon)DepositorInsertCoTXSS (Calendula officinalis taraxasterol synthase)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
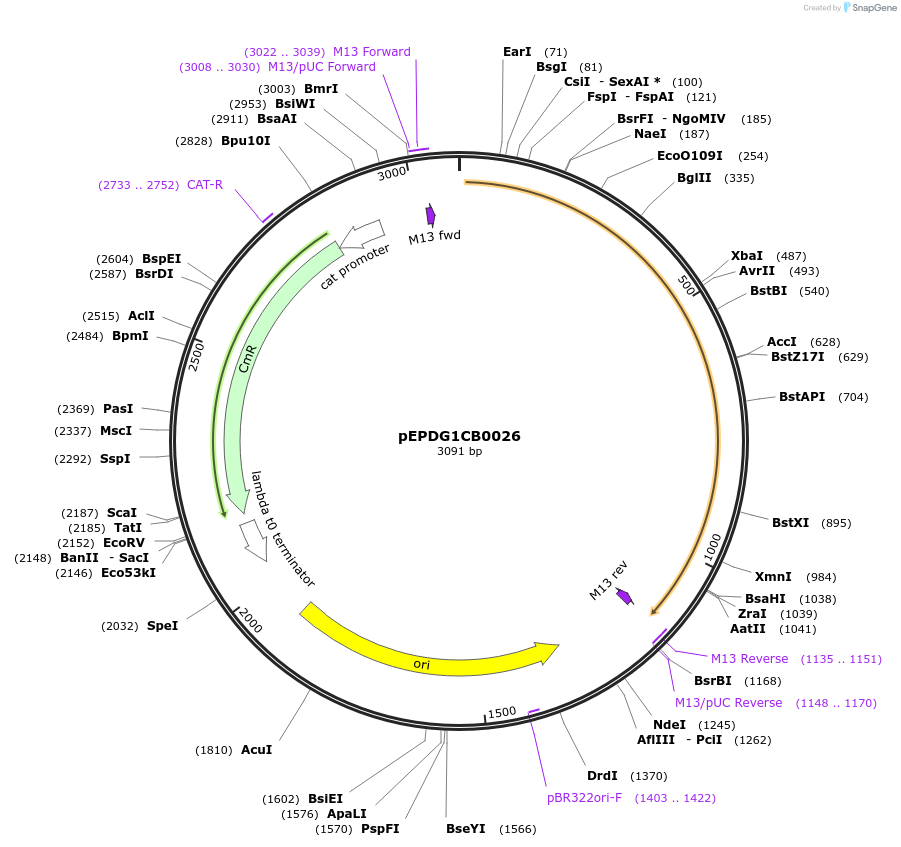
pEPDG1CB0027
Plasmid#227511PurposeL0 part - CDS (without STOP codon)DepositorInsertCaTXSS (Calendula arvensis taraxasterol synthase)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
pEPMS0CM0033
Plasmid#227512PurposeL0 part - CDS (without STOP codon)DepositorInsertCcTXSS (Cynara cardunculus taraxasterol synthase)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
pEPMS0CM0036
Plasmid#227513PurposeL0 part - CDS (without STOP codon)DepositorInsertHaTXSS (Helianthus annuus taraxasterol synthase)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
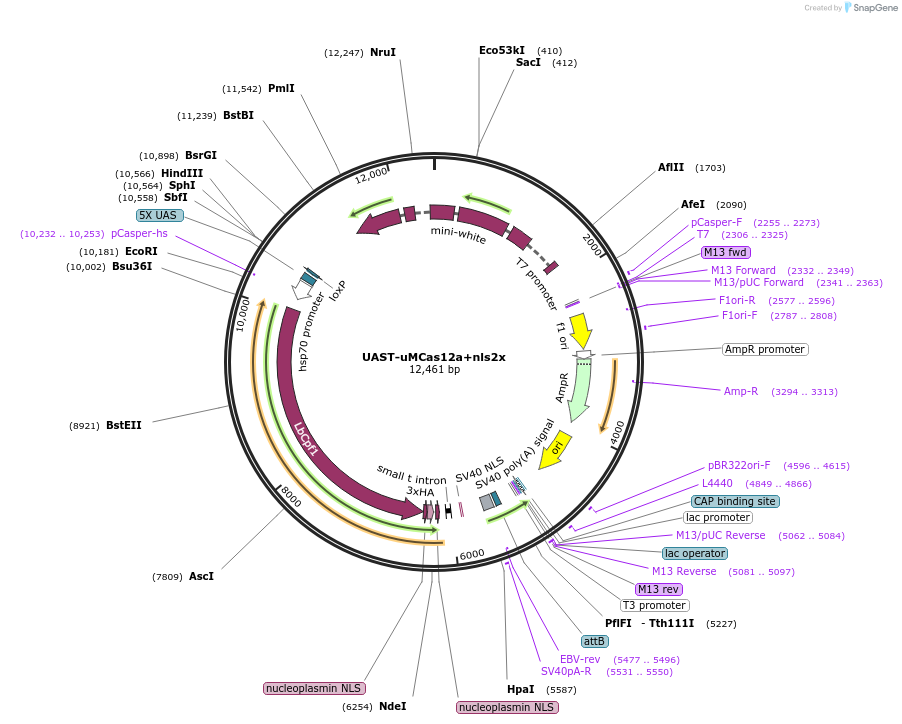
UAST-uMCas12a+nls2x
Plasmid#230909Purposeinducible expression of Cas12a+ with Gal4.DepositorInsertCas12a+
UseCRISPRAvailable SinceFeb. 11, 2025AvailabilityAcademic Institutions and Nonprofits only -
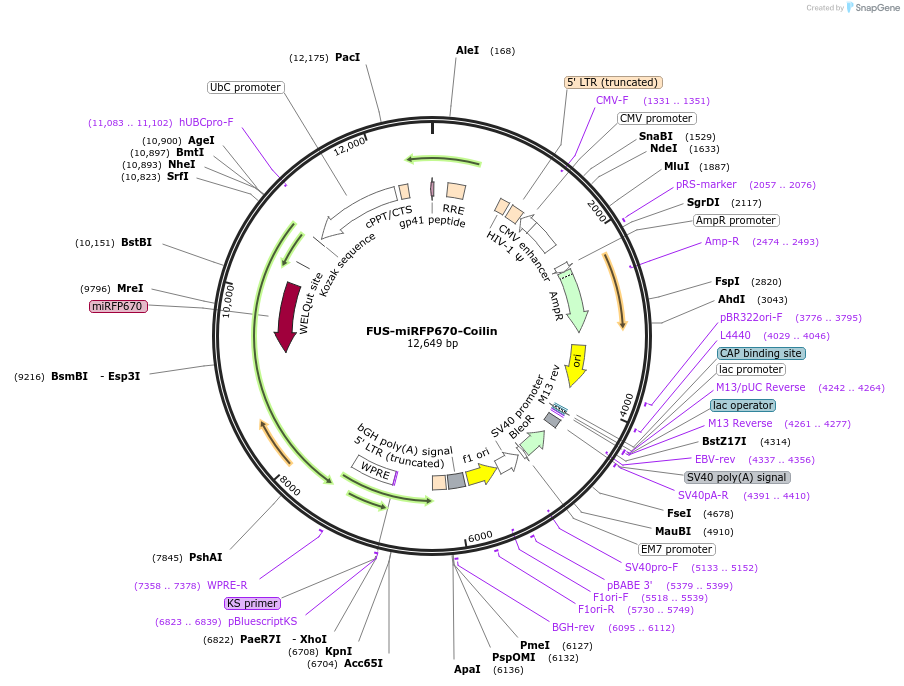
FUS-miRFP670-Coilin
Plasmid#227386PurposeAdhesion module, creates 'seed' at Cajal bodyDepositorInsertCajal body FUS IDR
UseLentiviralAvailable SinceOct. 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
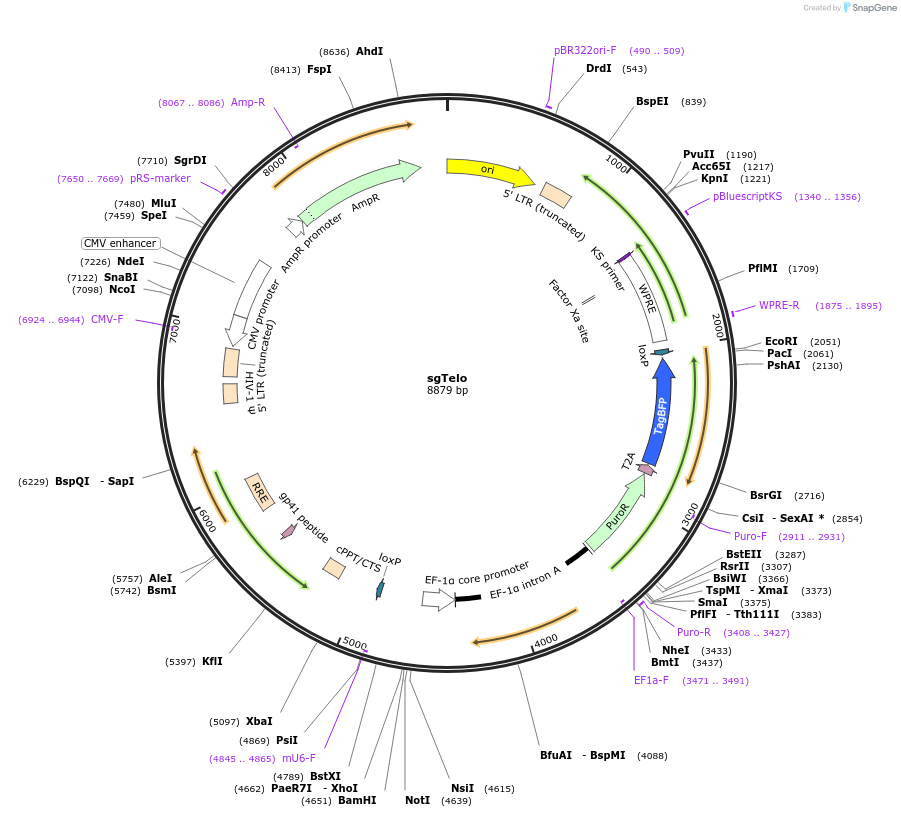
sgTelo
Plasmid#227392PurposeAdhesion module, guide RNA recognizes telomeresDepositorInsertguide RNA targeting telomeres
UseLentiviralAvailable SinceOct. 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
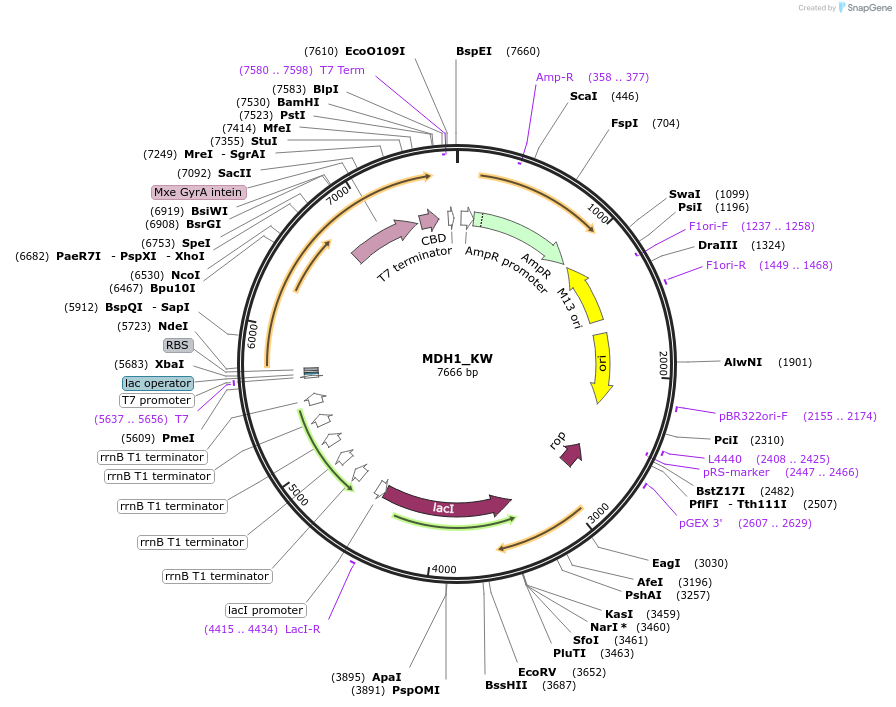
MDH1_KW
Plasmid#224145PurposeIPTG-inducible expression of C. elegans protein MDH-1 with a chitin tagDepositorAvailable SinceSept. 13, 2024AvailabilityAcademic Institutions and Nonprofits only -
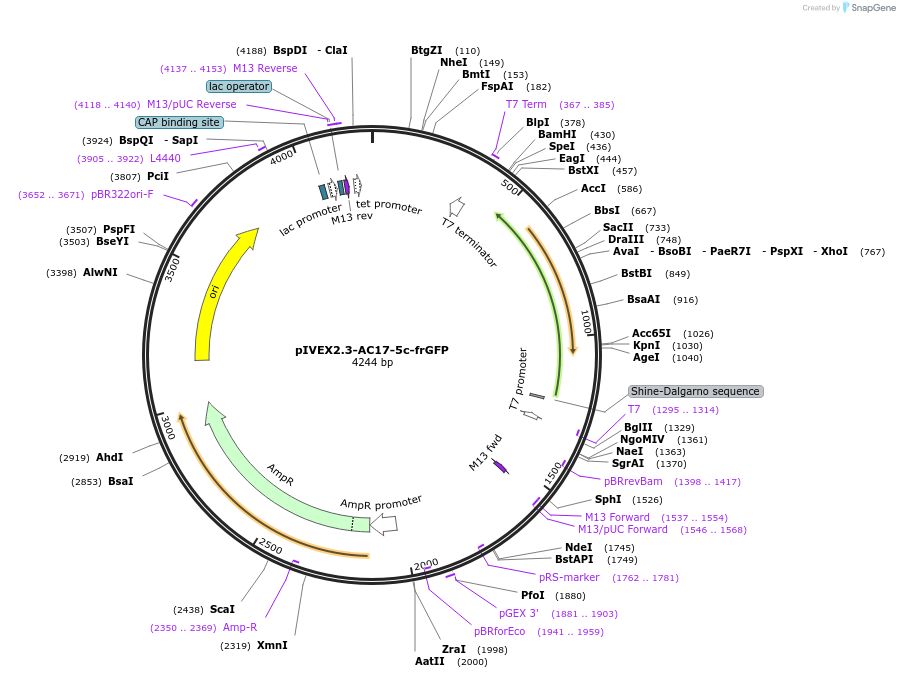
pIVEX2.3-AC17-5c-frGFP
Plasmid#217525PurposeAC17-5c riboswitch-regulated GFP reporter. The gene is inserted into downstream of T7 promoter.DepositorInsertAC17-5c riboswitch-regulated folding reporter GFP
UseIn vitro transcription-translationPromoterT7Available SinceMay 16, 2024AvailabilityAcademic Institutions and Nonprofits only -
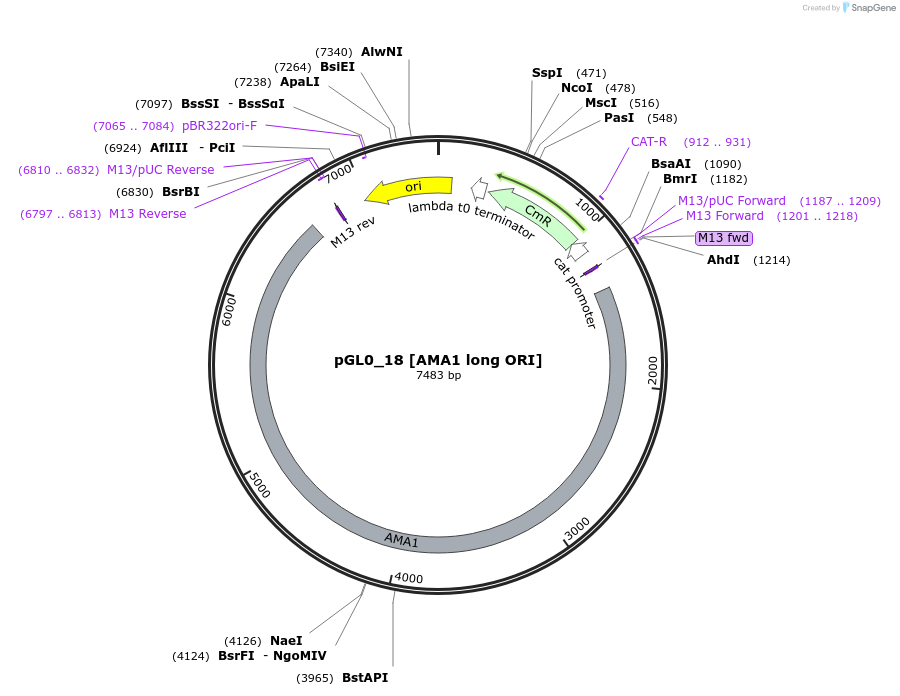
pGL0_18 [AMA1 long ORI]
Plasmid#198931PurposeLevel 0 partDepositorInsertAMA1 long ORI
UseSynthetic BiologyAvailable SinceSept. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
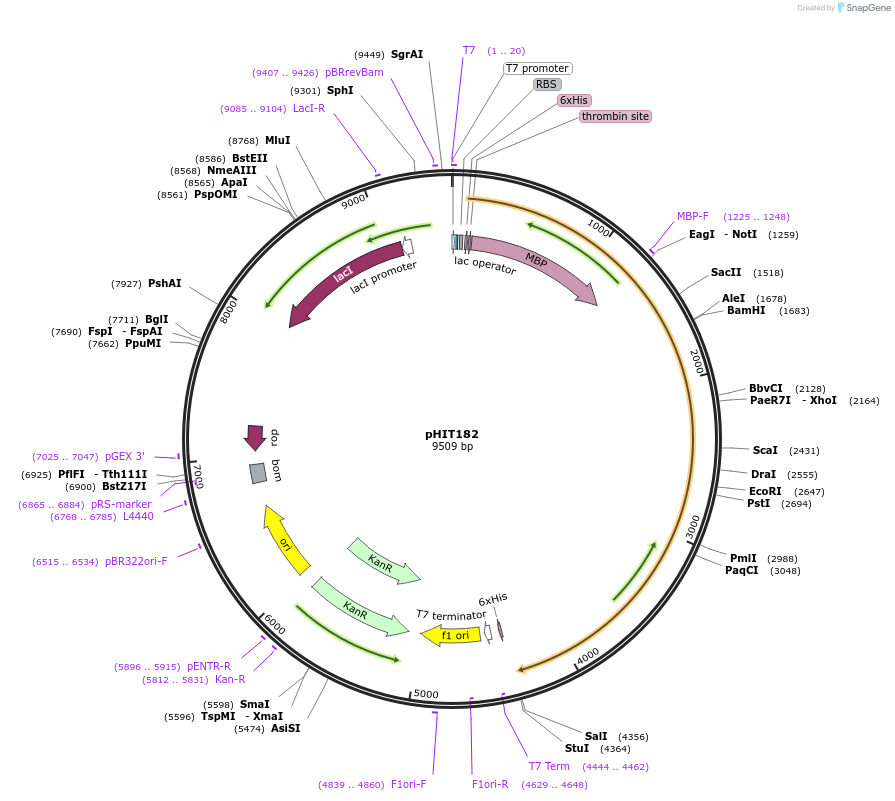
pHIT182
Plasmid#204511PurposeBacterial expression of catalytically defective CSR-1a with 6xHis and maltose binding protein tags for in vitro assaysDepositorInsertcsr-1a with D769A mutation (C. elegans) (csr-1 Nematode)
Tags6xHis, MBPExpressionBacterialMutationD769APromoterT7lacAvailable SinceAug. 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
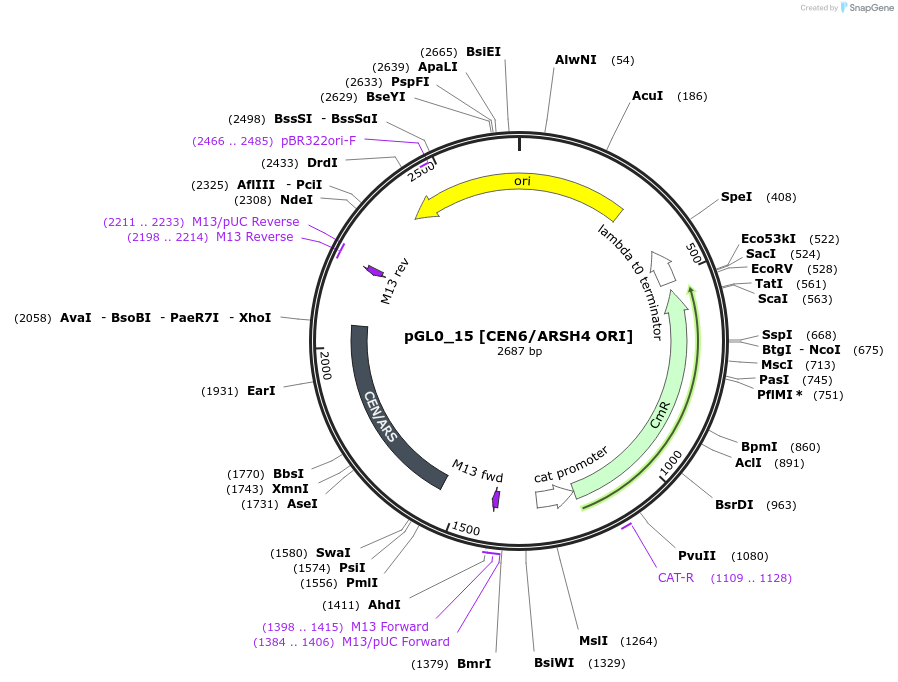
pGL0_15 [CEN6/ARSH4 ORI]
Plasmid#198928PurposeLevel 0 partDepositorInsertCEN6/ARSH4 ORI
UseSynthetic BiologyAvailable SinceJuly 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
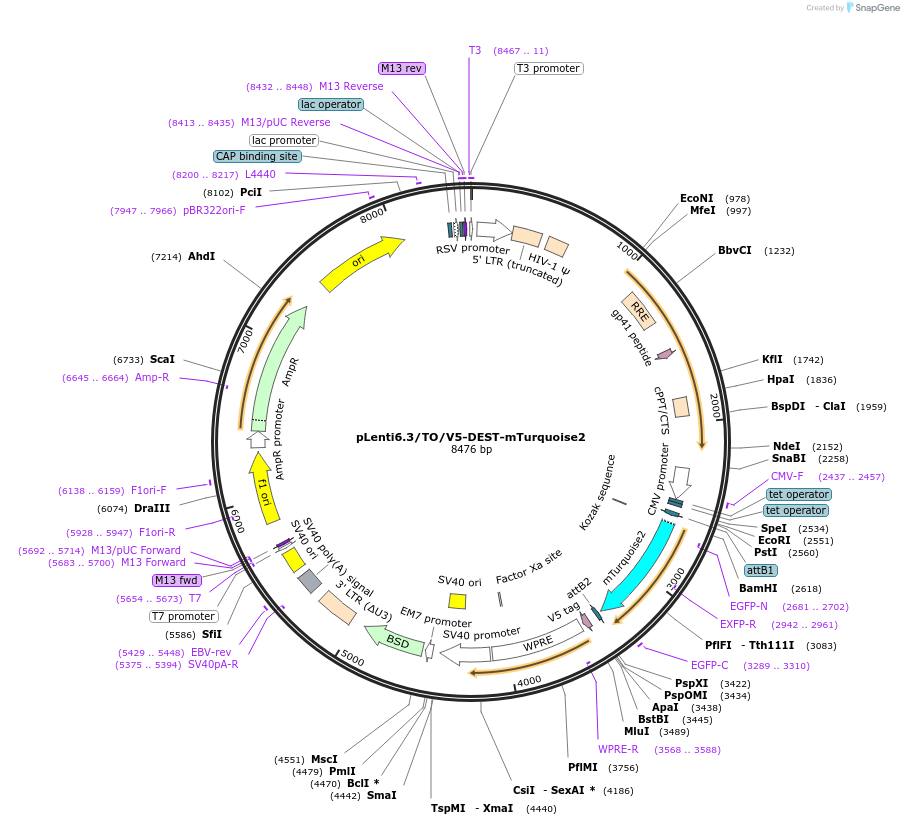
pLenti6.3/TO/V5-DEST-mTurquoise2
Plasmid#191010PurposeLentiviral expression vector for mTurquoise2DepositorInsertmTurquoise2
UseLentiviralPromoterCMVAvailable SinceJuly 12, 2023AvailabilityAcademic Institutions and Nonprofits only