We narrowed to 81,735 results for: TRI
-
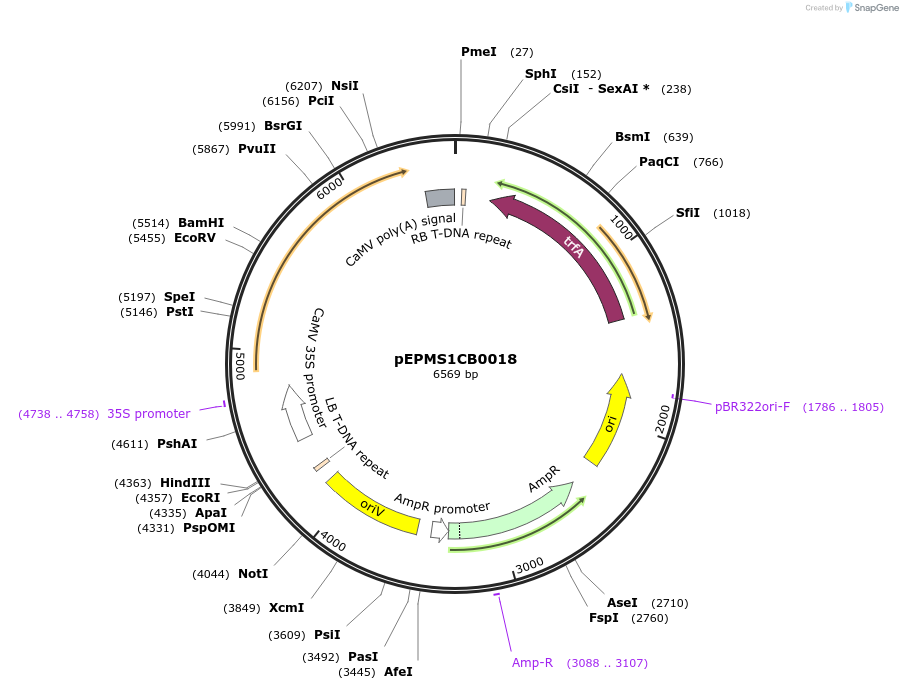
Plasmid#227543PurposeLevel 1 - Position 2 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CYP716A392_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
pEPMS1CB0019
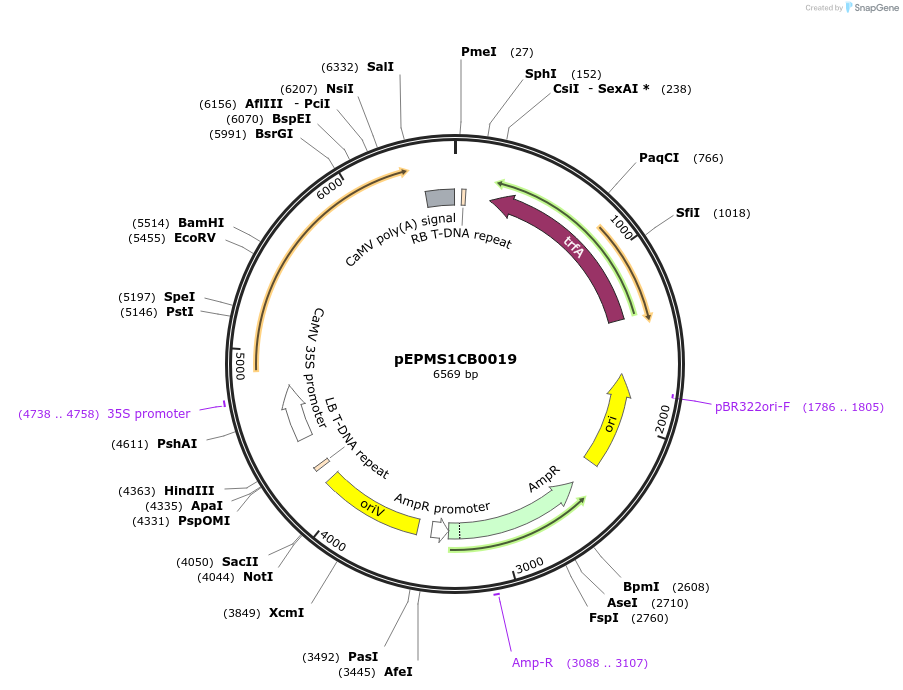
Plasmid#227544PurposeLevel 1 - Position 2 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CYP716A393_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
pEPMS1CB0020
Plasmid#227545PurposeLevel 1 - Position 2 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CYP716A429_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
pEPMS1CB0021
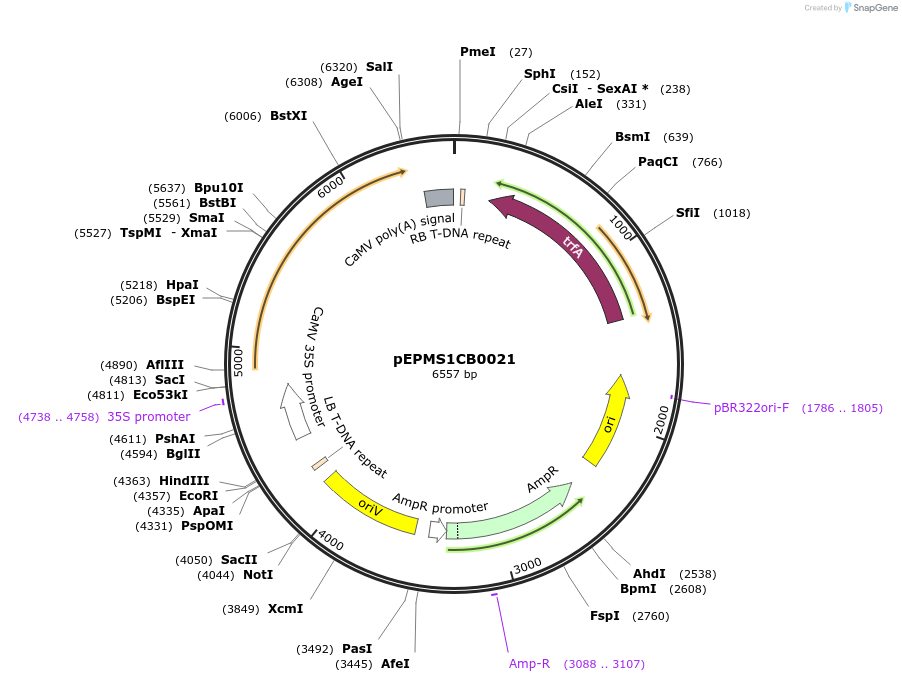
Plasmid#227546PurposeLevel 1 - Position 2 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CYP716A430_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
pEPMS1CB0022
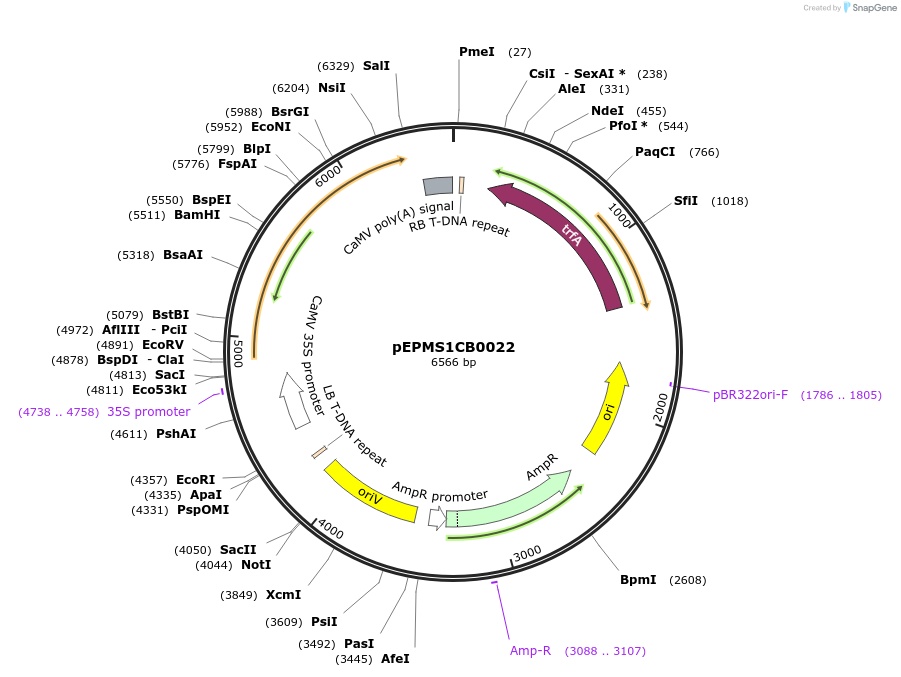
Plasmid#227547PurposeLevel 1 - Position 2 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CYP716A431_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
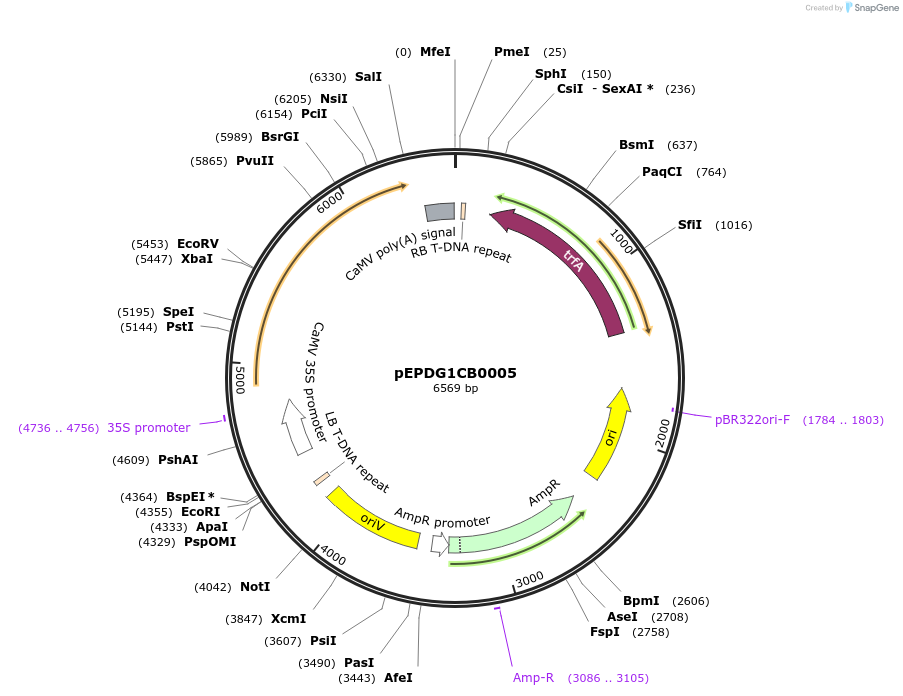
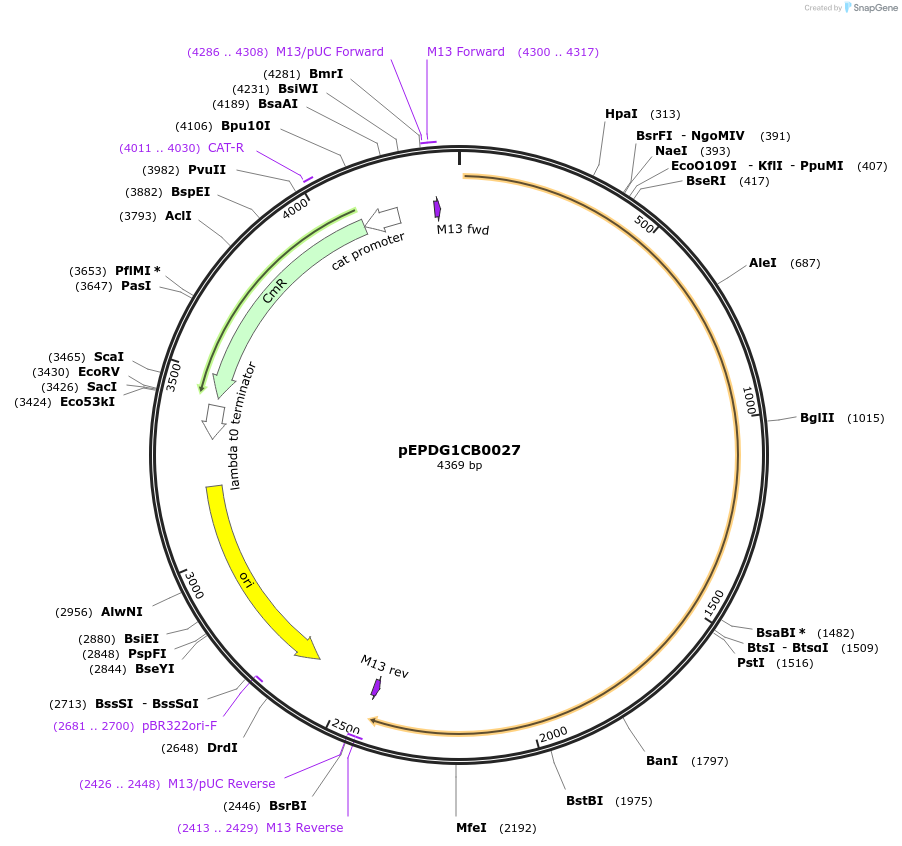
pEPDG1CB0005
Plasmid#227548PurposeLevel 1 - Position 1 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CYP716A392a_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
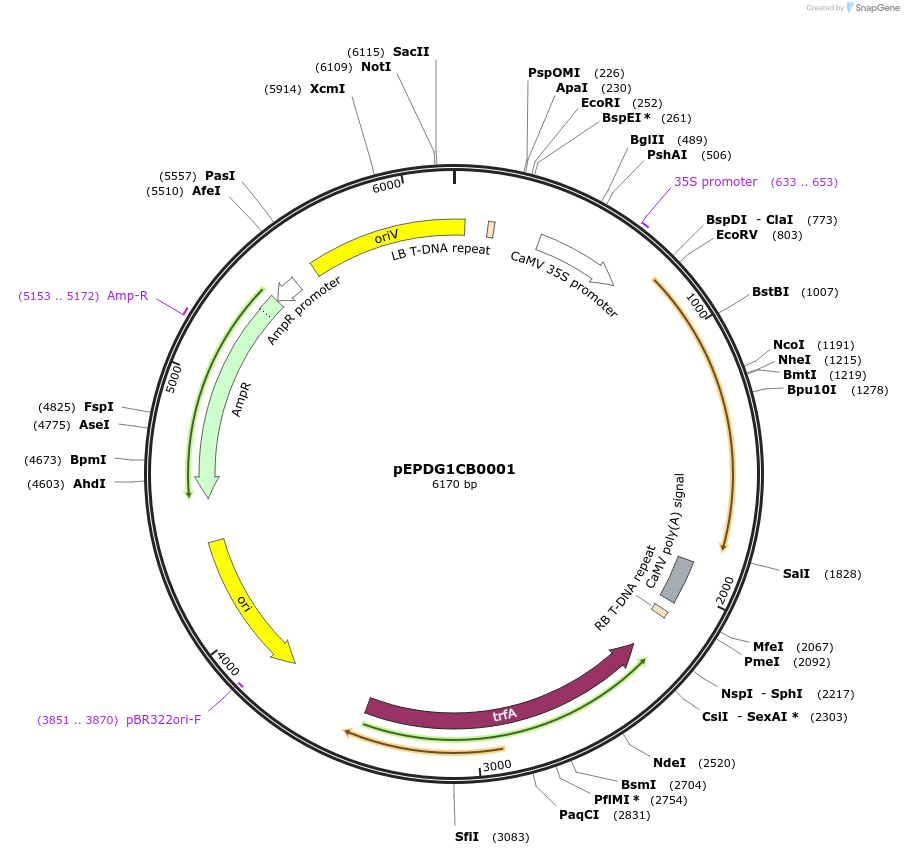
pEPDG1CB0001
Plasmid#227549PurposeLevel 1 - Position 1 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CoACT1_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
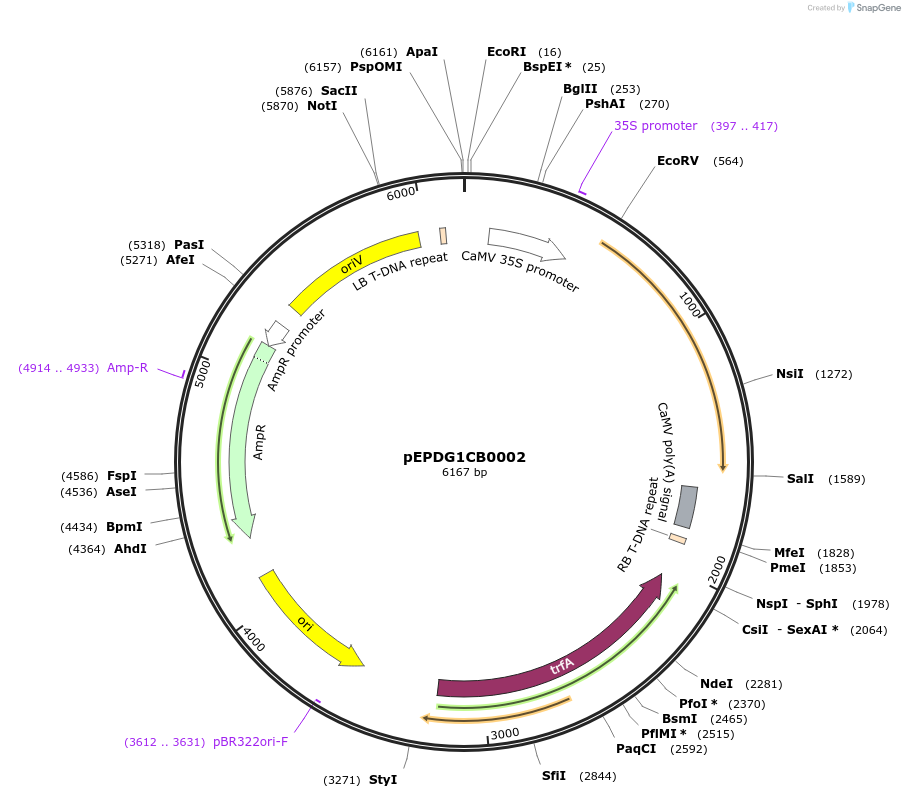
pEPDG1CB0002
Plasmid#227550PurposeLevel 1 - Position 1 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CoACT2_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
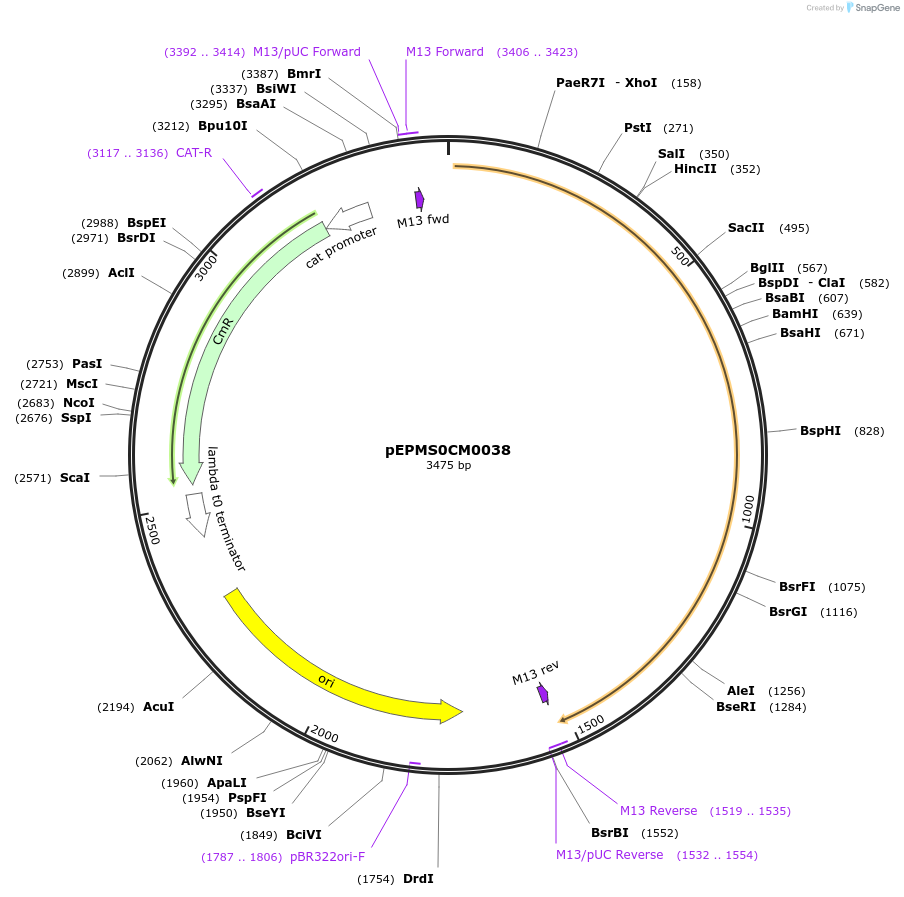
pEPMS0CM0038
Plasmid#227520PurposeL0 part - CDS (without STOP codon)DepositorInsertCYP716A392 (Calendula officinalis cytochrome P450 1)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
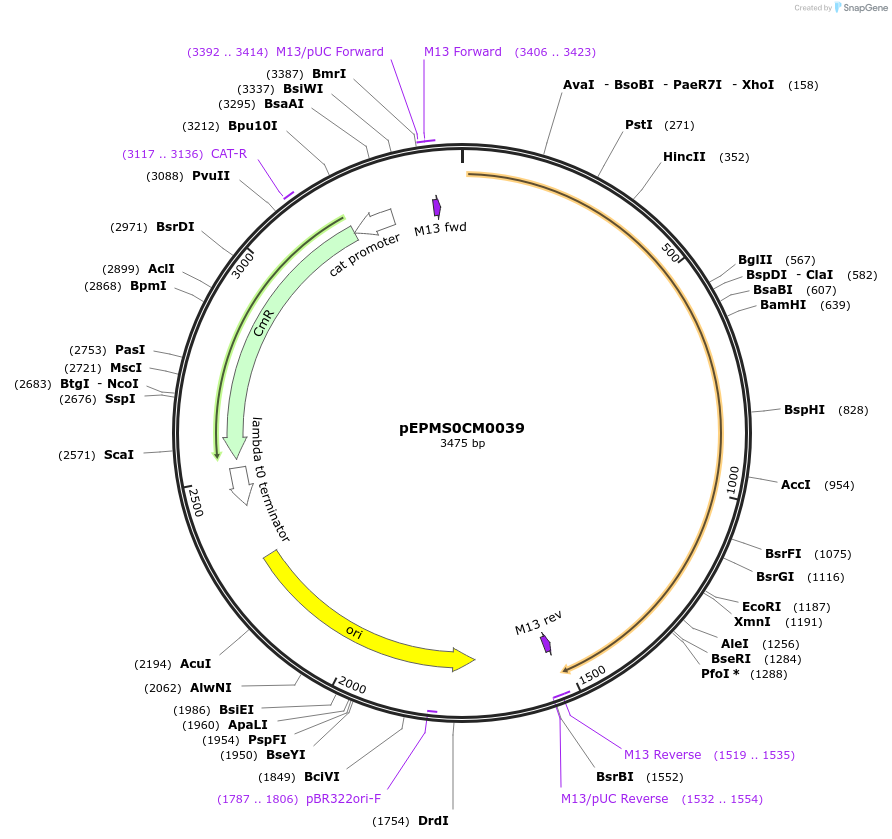
pEPMS0CM0039
Plasmid#227521PurposeL0 part - CDS (without STOP codon)DepositorInsertCYP716A393 (Calendula officinalis cytochrome P450 2)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
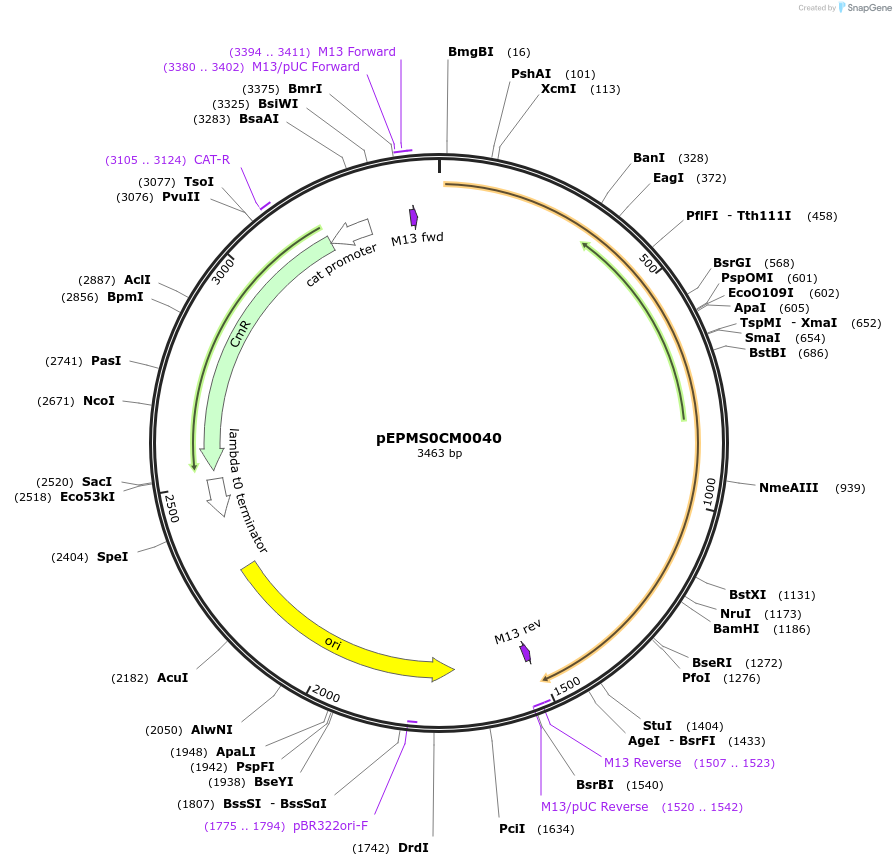
pEPMS0CM0040
Plasmid#227522PurposeL0 part - CDS (without STOP codon)DepositorInsertCYP716A429 (Calendula officinalis cytochrome P450 3)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
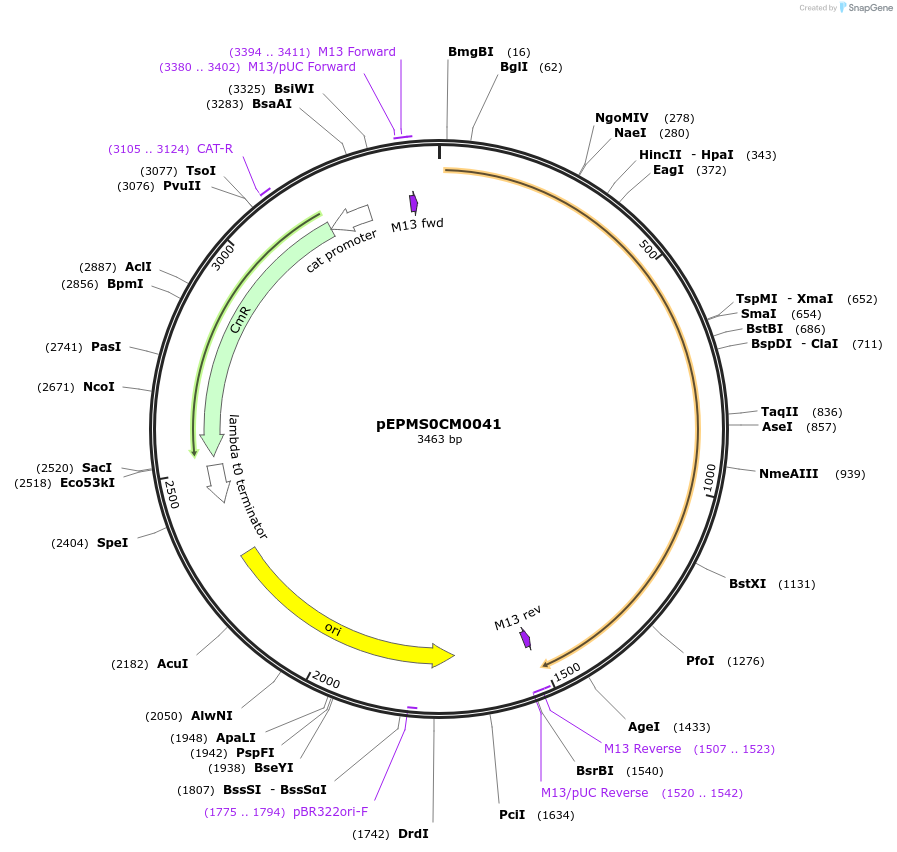
pEPMS0CM0041
Plasmid#227523PurposeL0 part - CDS (without STOP codon)DepositorInsertCYP716A430 (Calendula officinalis cytochrome P450 4)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
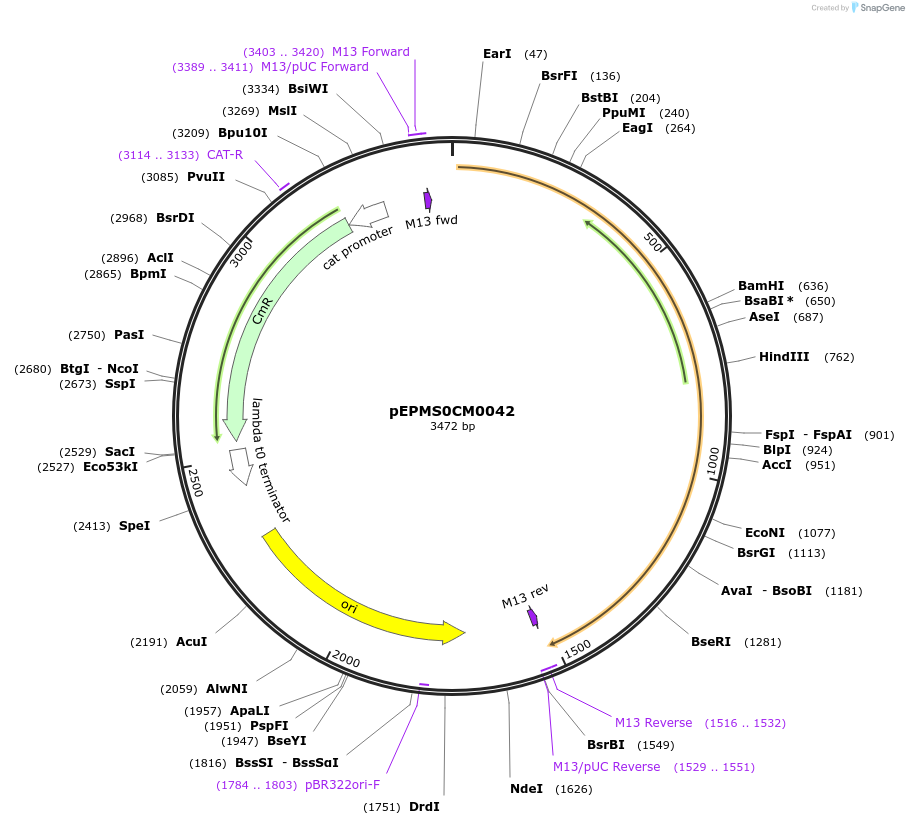
pEPMS0CM0042
Plasmid#227524PurposeL0 part - CDS (without STOP codon)DepositorInsertCYP716A431 (Calendula officinalis cytochrome P450 5)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
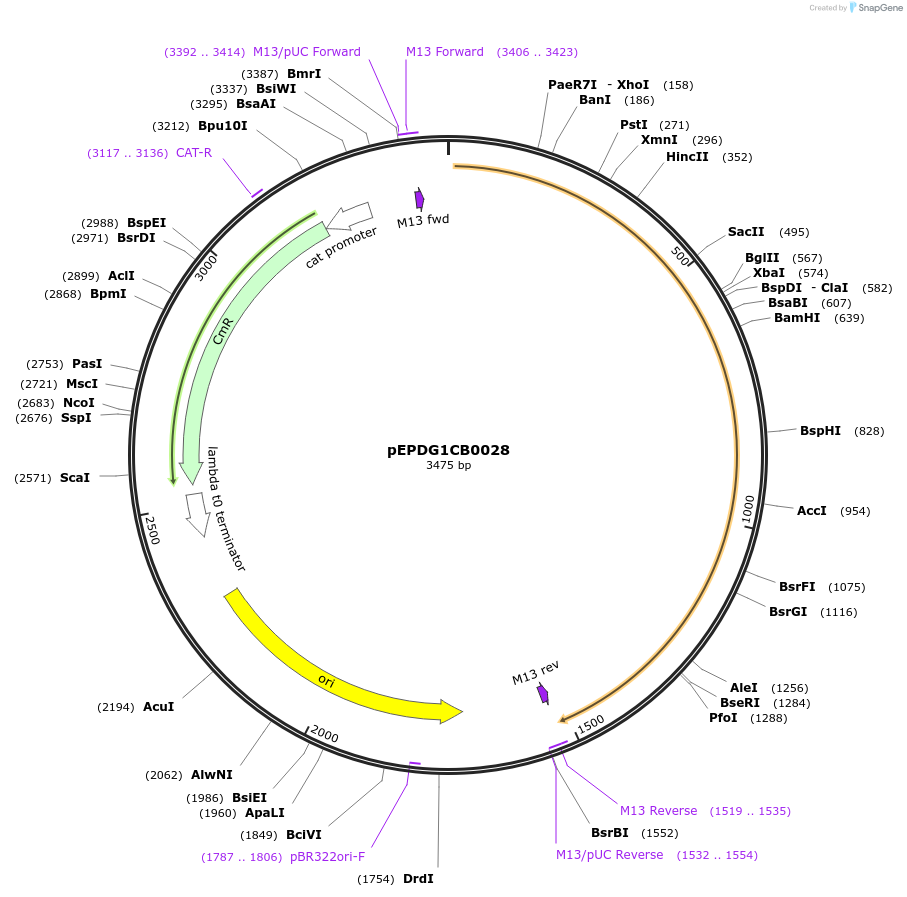
pEPDG1CB0028
Plasmid#227525PurposeL0 part - CDS (without STOP codon)DepositorInsertCYP716A392a (Calendula arvensis cytochrome P450 1)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
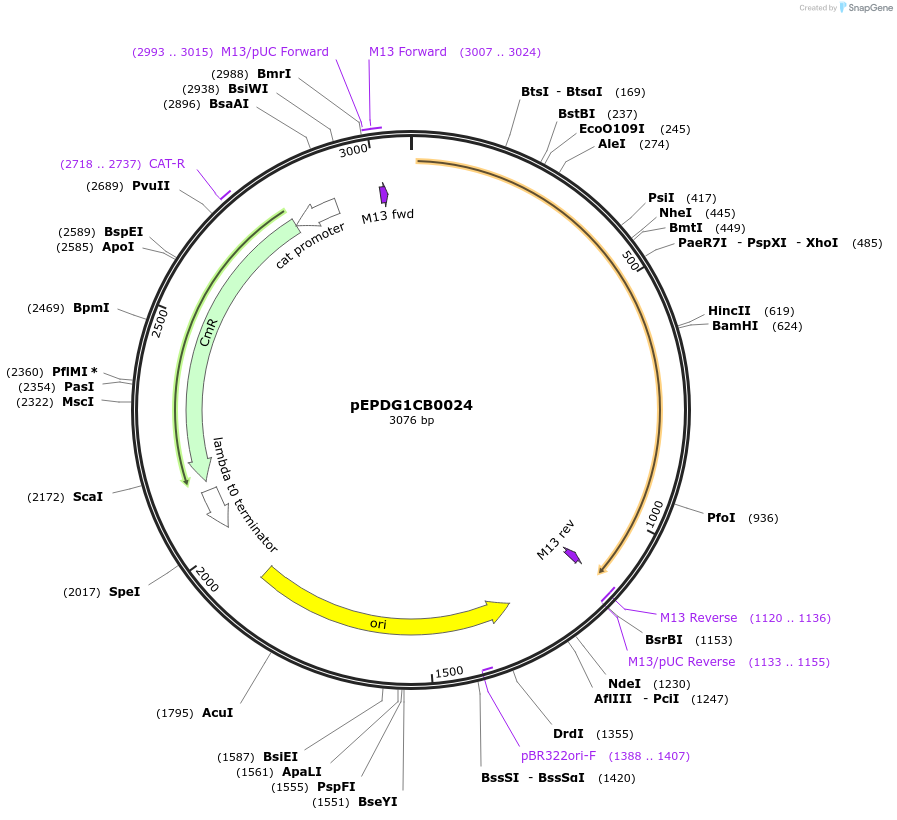
pEPDG1CB0024
Plasmid#227526PurposeL0 part - CDS (without STOP codon)DepositorInsertCoACT1 (Calendula officinalis acyltransferase 1)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
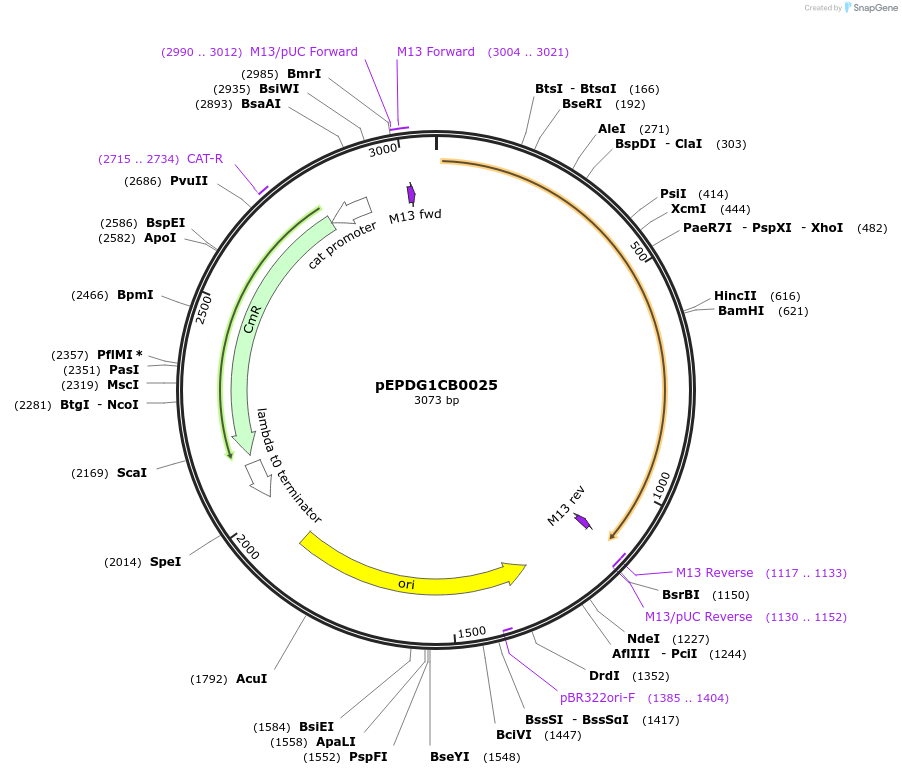
pEPDG1CB0025
Plasmid#227527PurposeL0 part - CDS (without STOP codon)DepositorInsertCoACT2 (Calendula officinalis acyltransferase 2)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
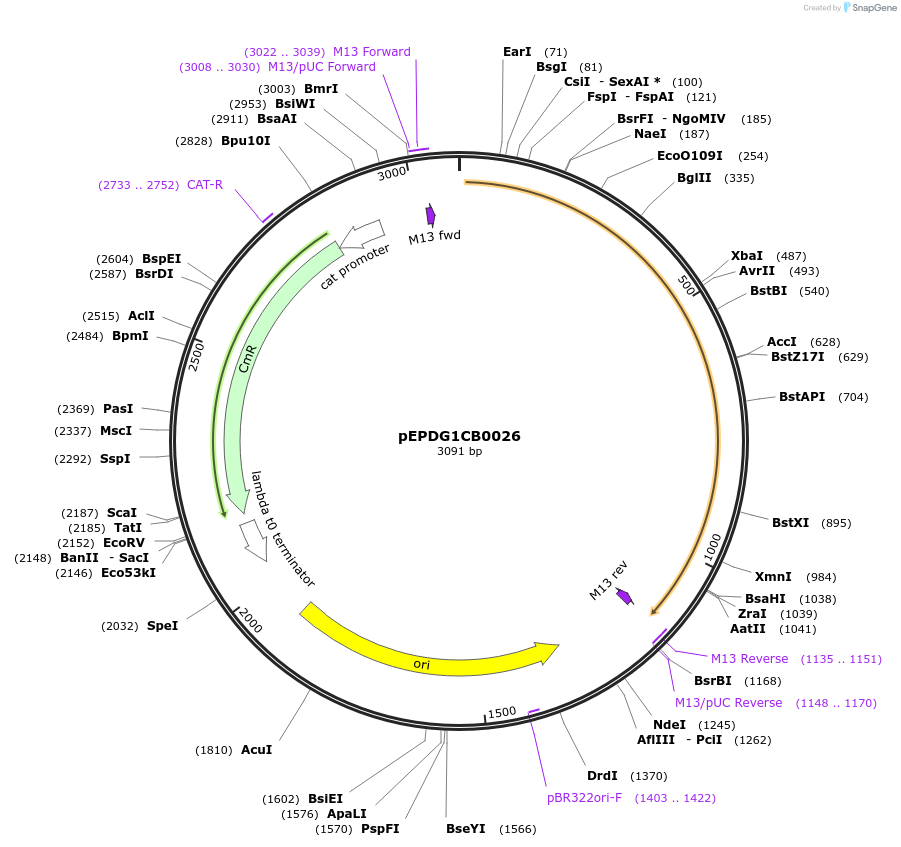
pEPDG1CB0026
Plasmid#227528PurposeL0 part - CDS (without STOP codon)DepositorInsertCoACT3 (Calendula officinalis acyltransferase 3)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
pEPCT0CM0207
Plasmid#227529PurposeL0 part - CDS (without STOP codon)DepositorInsertCoACT4 (Calendula officinalis acyltransferase 4)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
pEPCT0CM0208
Plasmid#227530PurposeL0 part - CDS (without STOP codon)DepositorInsertCoACT5 (Calendula officinalis acyltransferase 5)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
pEPCT0CM0209
Plasmid#227531PurposeL0 part - CDS (without STOP codon)DepositorInsertCoACT6 (Calendula officinalis acyltransferase 6)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
pEPHS0CM0054
Plasmid#227515PurposeL0 part - CDS (without STOP codon)DepositorInsertTdTXSS (Tragopogon dubius taraxasterol synthase)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
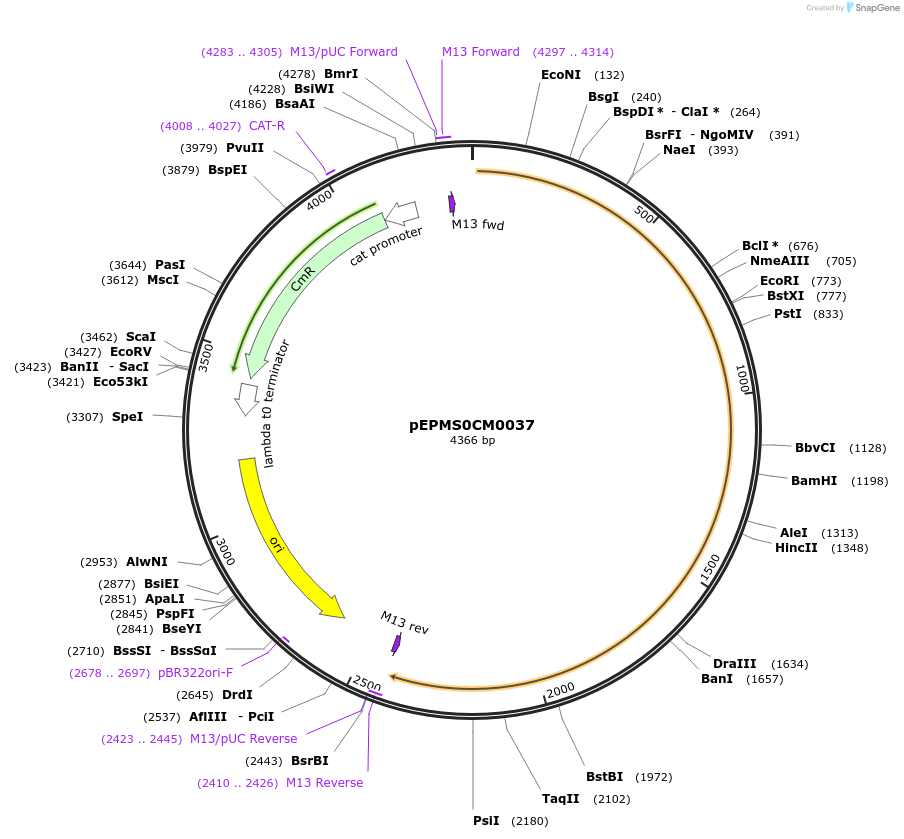
pEPMS0CM0037
Plasmid#227516PurposeL0 part - CDS (without STOP codon)DepositorInsertLsTXSS (Lactuca sativa taraxasterol synthase)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
pEPMS0CM0024
Plasmid#227510PurposeL0 part - CDS (without STOP codon)DepositorInsertCoTXSS (Calendula officinalis taraxasterol synthase)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
pEPDG1CB0027
Plasmid#227511PurposeL0 part - CDS (without STOP codon)DepositorInsertCaTXSS (Calendula arvensis taraxasterol synthase)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
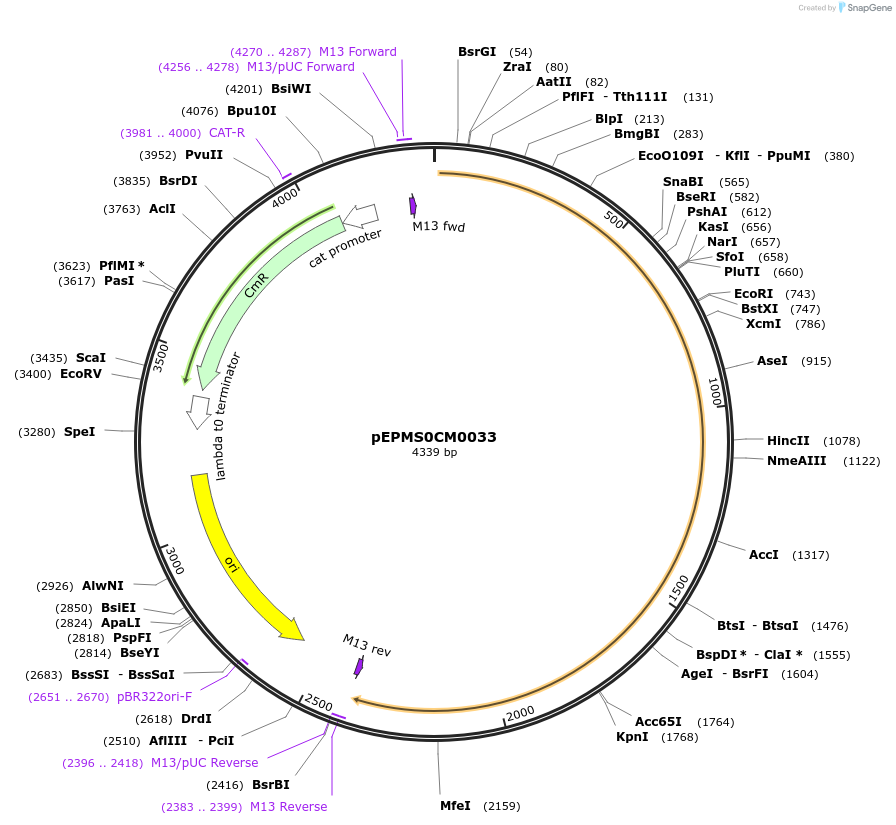
pEPMS0CM0033
Plasmid#227512PurposeL0 part - CDS (without STOP codon)DepositorInsertCcTXSS (Cynara cardunculus taraxasterol synthase)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
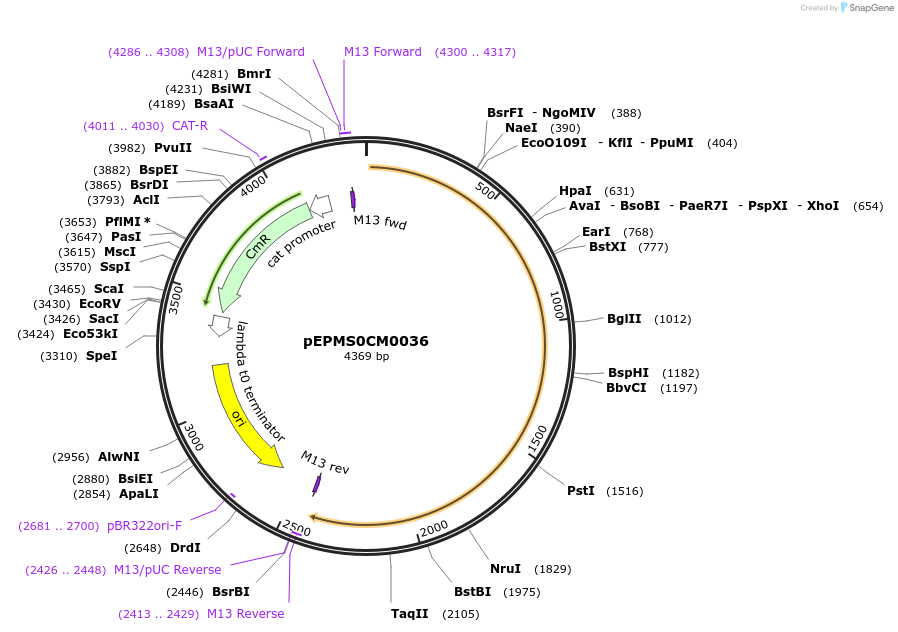
pEPMS0CM0036
Plasmid#227513PurposeL0 part - CDS (without STOP codon)DepositorInsertHaTXSS (Helianthus annuus taraxasterol synthase)
ExpressionPlantMutationBsaI sites removed by silent mutationAvailable SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
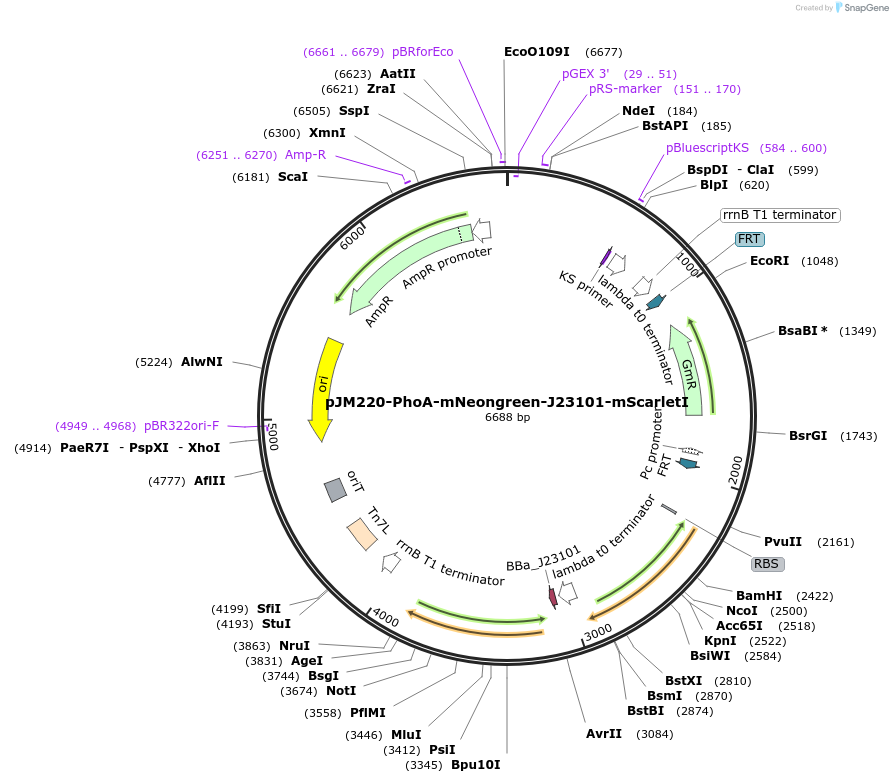
pJM220-PhoA-mNeongreen-J23101-mScarletI
Plasmid#240201PurposeMiniTn7 donor plasmid for constructing a ratiometric reporter for P limitation in Pseudomonas synxantha. A strong Anderson promoter (J23101) drives expression of mScarlet-IDepositorInsertsmNeongreen
mScarlet-I
PromoterJ23101 and PphoAAvailable SinceJuly 21, 2025AvailabilityAcademic Institutions and Nonprofits only -
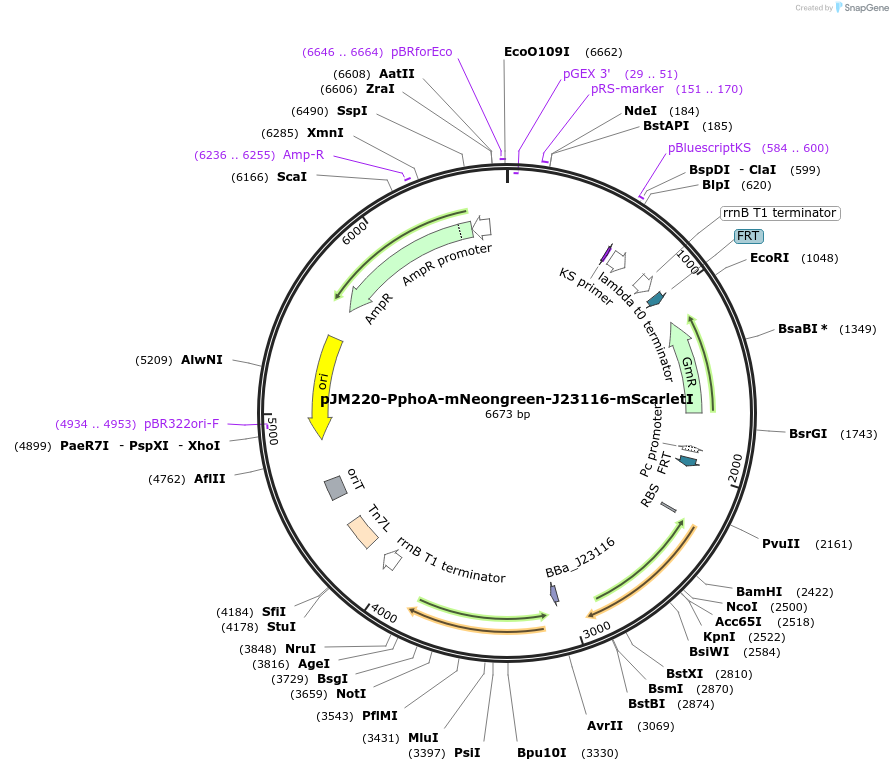
pJM220-PphoA-mNeongreen-J23116-mScarletI
Plasmid#240200PurposeMiniTn7 donor plasmid for constructing a ratiometric reporter for P limitation in Pseudomonas synxantha. A weak Anderson promoter (J23116) drives expression of mScarlet-IDepositorInsertsmNeongreen
mScarlet-I
PromoterJ23116 and PphoAAvailable SinceJuly 21, 2025AvailabilityAcademic Institutions and Nonprofits only -
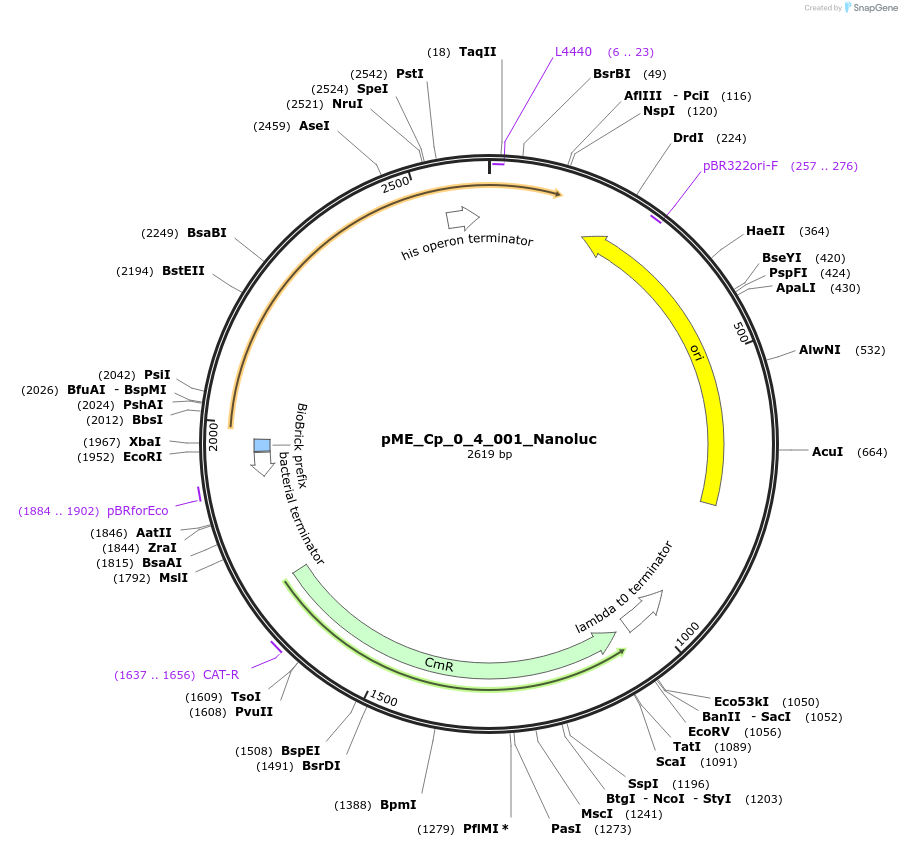
pME_Cp_0_4_001_Nanoluc
Plasmid#235893PurposeNanoluc luciferase reporter gene, codon optimized for Chlamydomonas reinhardtiiDepositorInsertNanoLuc
UseSynthetic BiologyAvailable SinceMay 5, 2025AvailabilityAcademic Institutions and Nonprofits only -
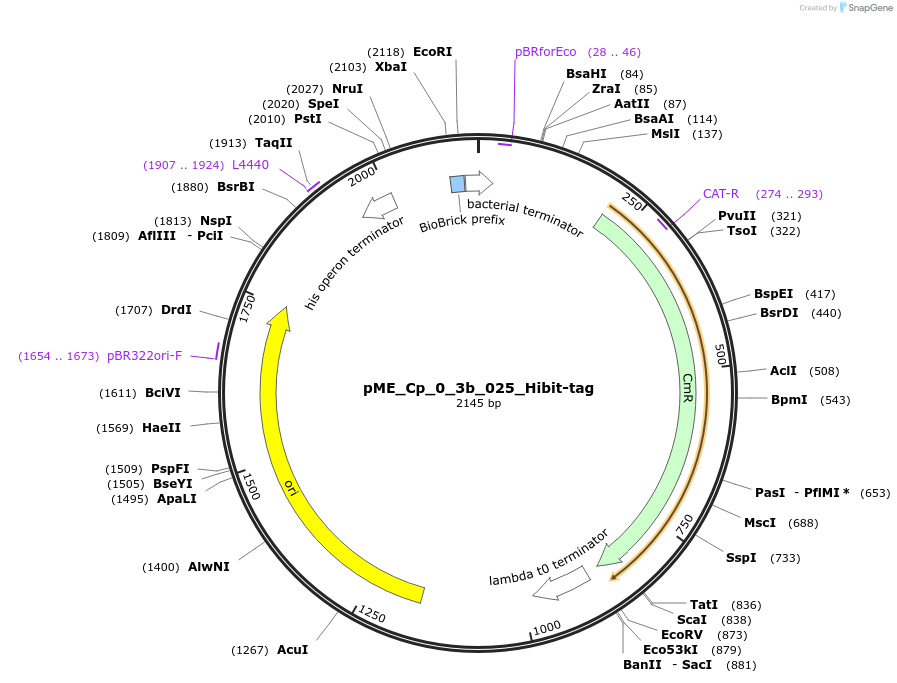
pME_Cp_0_3b_025_Hibit-tag
Plasmid#235878PurposeHiBiT N-tagDepositorInsertHiBiT N-tag
UseSynthetic BiologyAvailable SinceMay 5, 2025AvailabilityAcademic Institutions and Nonprofits only -
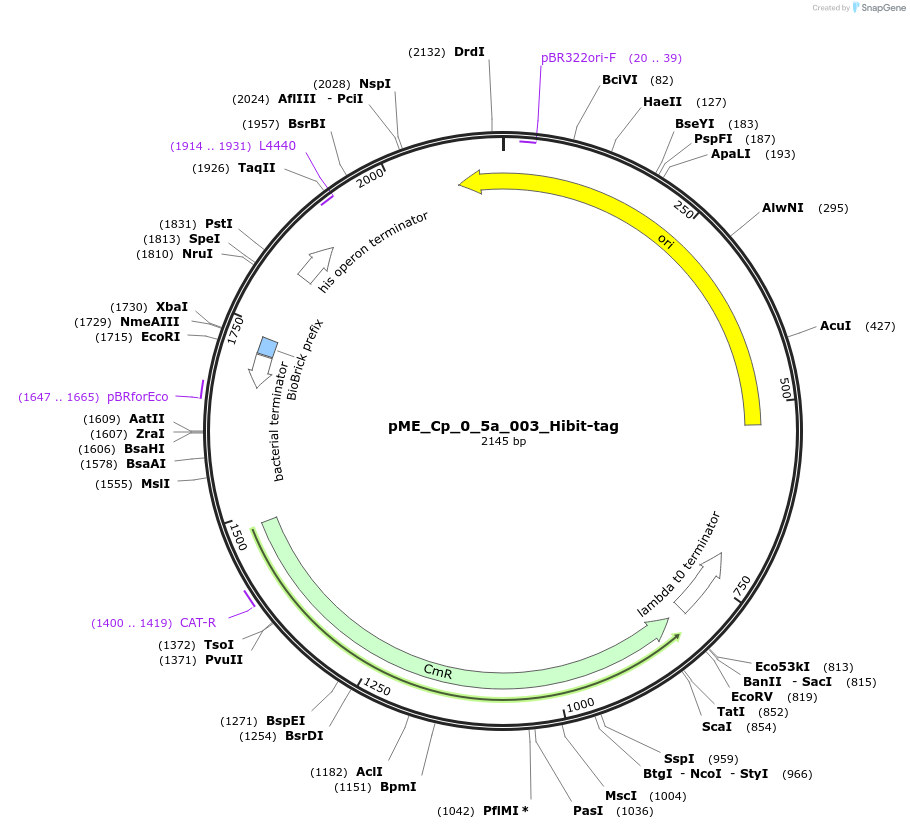
pME_Cp_0_5a_003_Hibit-tag
Plasmid#235885PurposeHiBiT C-tagDepositorInsertHiBiT C-tag
UseSynthetic BiologyAvailable SinceMay 5, 2025AvailabilityAcademic Institutions and Nonprofits only -
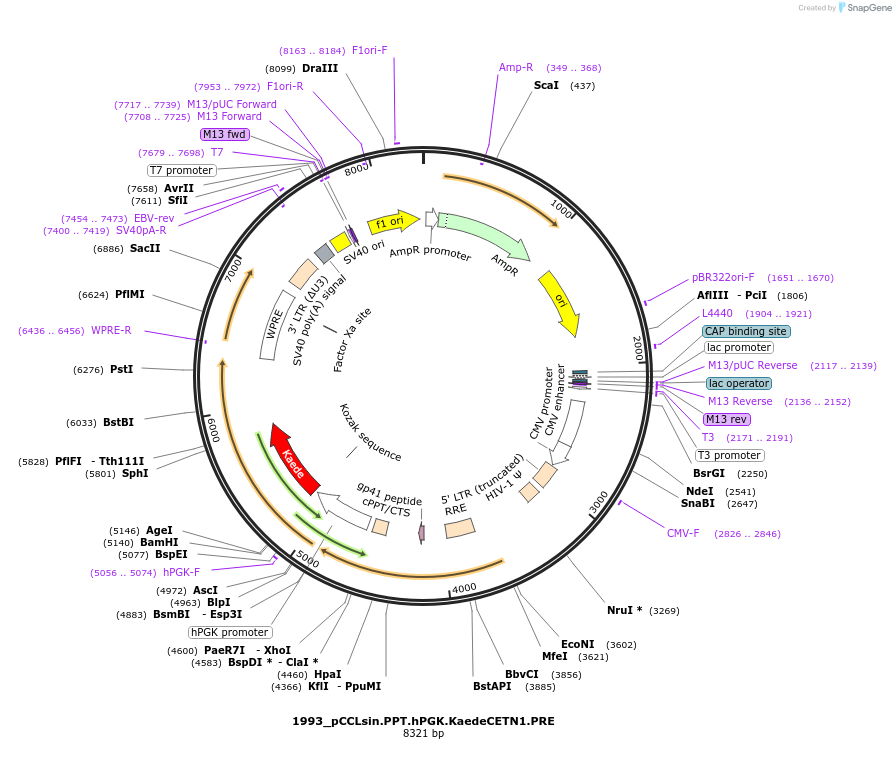
1993_pCCLsin.PPT.hPGK.KaedeCETN1.PRE
Plasmid#231804PurposeLabel centrosomes with lentivirus by encoding a fusion protein consisting of the photo-convertible protein Kaede fused to Centrin 1 (CETN1), an integral component of centrosomes.DepositorAvailable SinceFeb. 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
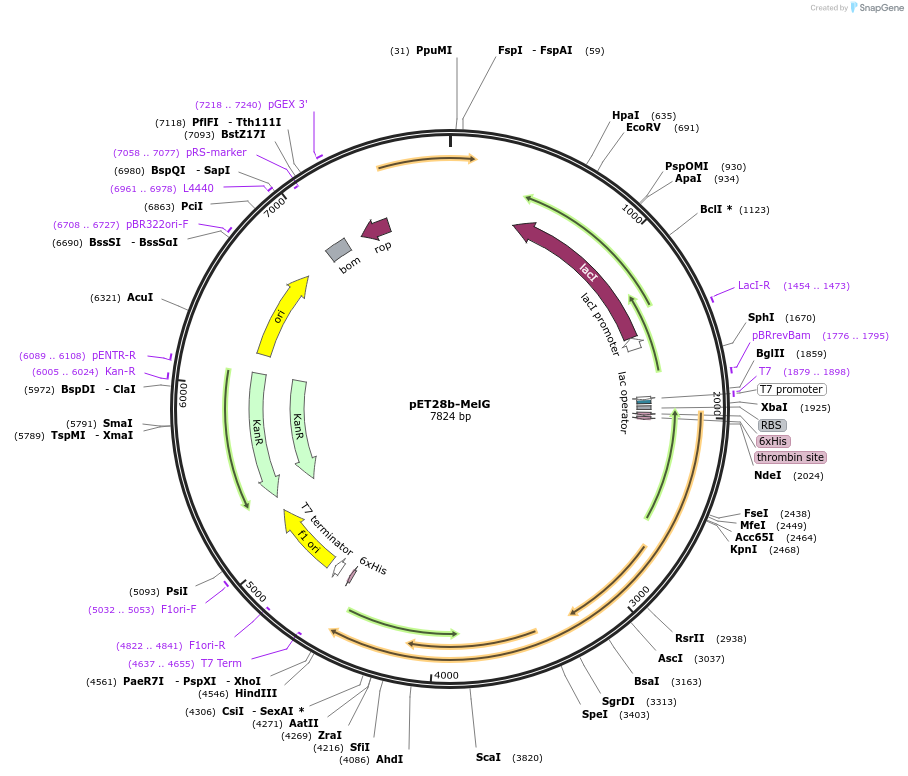
pET28b-MelG
Plasmid#225729PurposeTo express MelG (Rv1937) in E. coliDepositorInsertmelG
Tags6xHisExpressionBacterialAvailable SinceJan. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
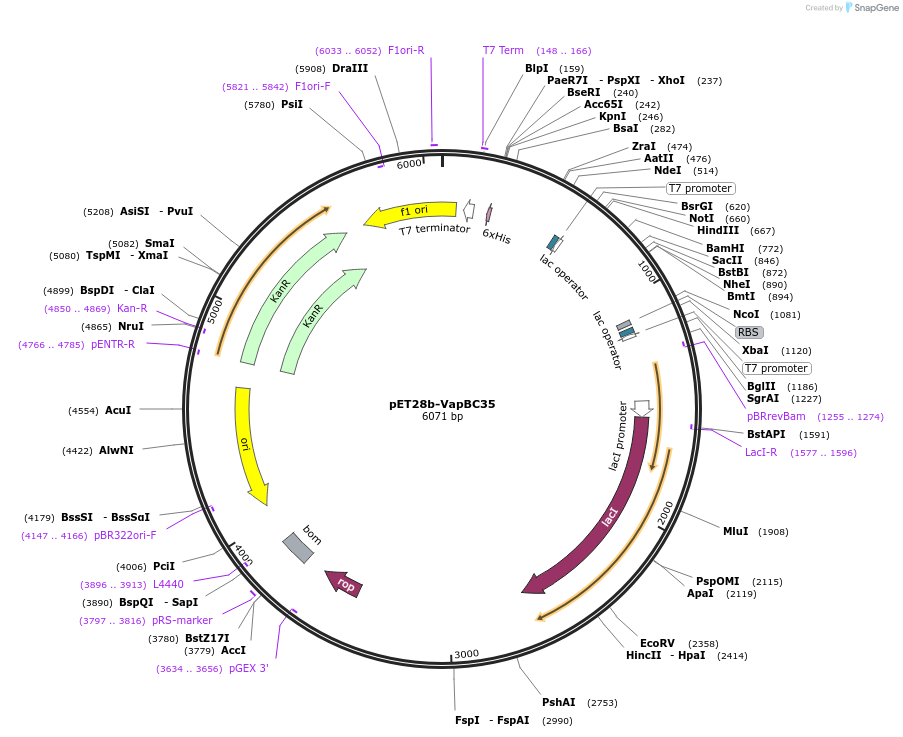
pET28b-VapBC35
Plasmid#225733PurposeTo express VapBC35(Rv1962) Toxin antitoxin in E. coliDepositorInsertvapBC35
Tags6xHisExpressionBacterialAvailable SinceJan. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
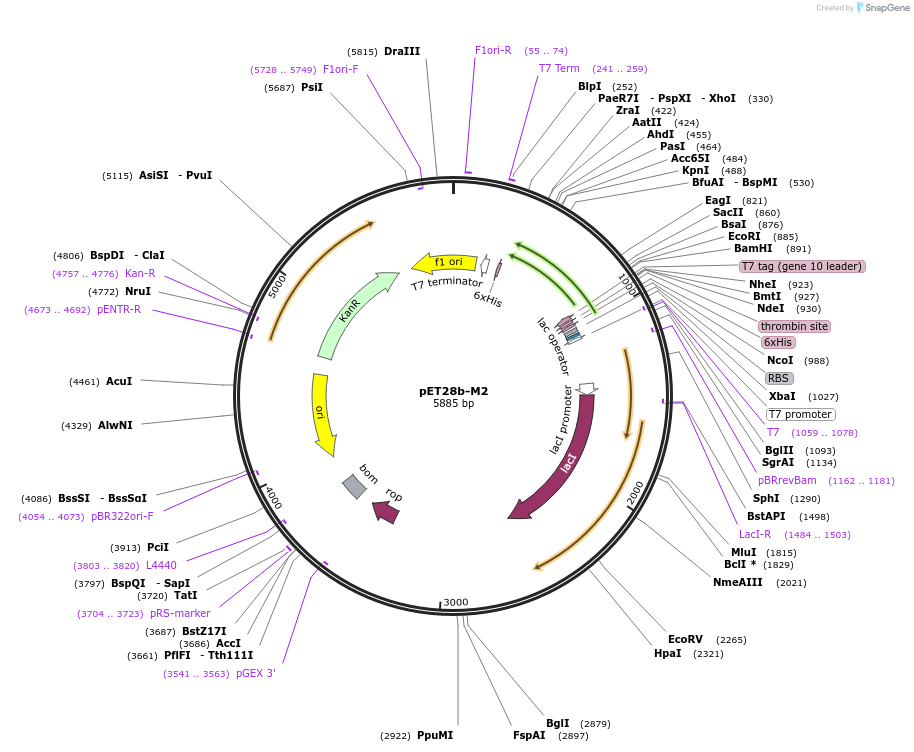
pET28b-M2
Plasmid#225735PurposeTo express M2 (219-399) in E. coliDepositorInsertmce3R
Tags6xHisExpressionBacterialMutationaa 219-399Available SinceJan. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
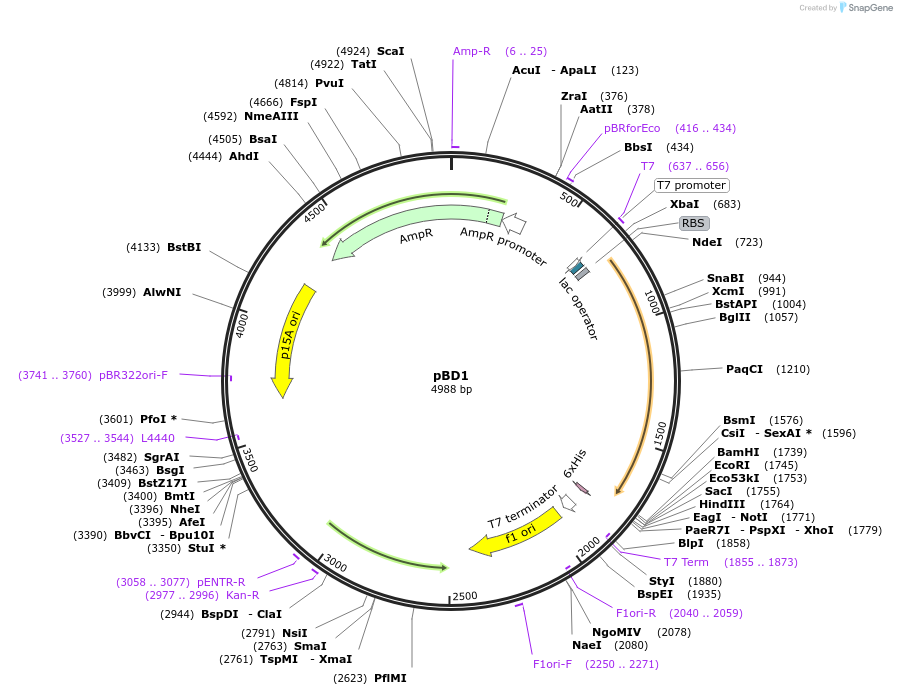
pBD1
Plasmid#229463PurposeExpresses RssBDepositorInsertRssB
ExpressionBacterialPromoterT7Available SinceJan. 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
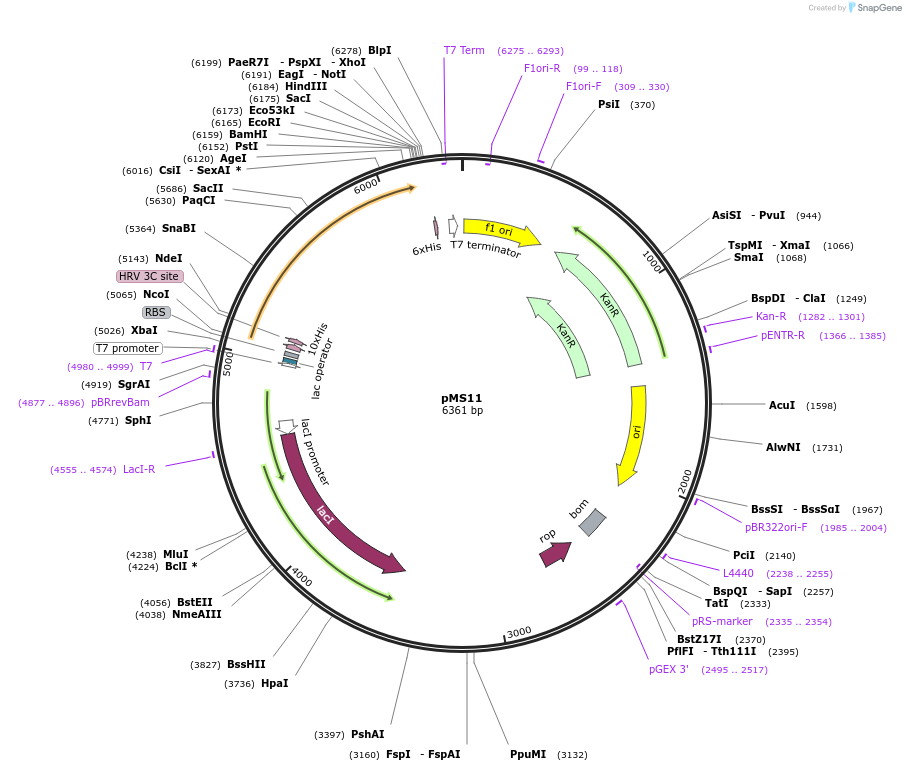
pMS11
Plasmid#229464PurposeExpresses RssB with His TagDepositorInsertRssB
Tags10xHisExpressionBacterialPromoterT7Available SinceJan. 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
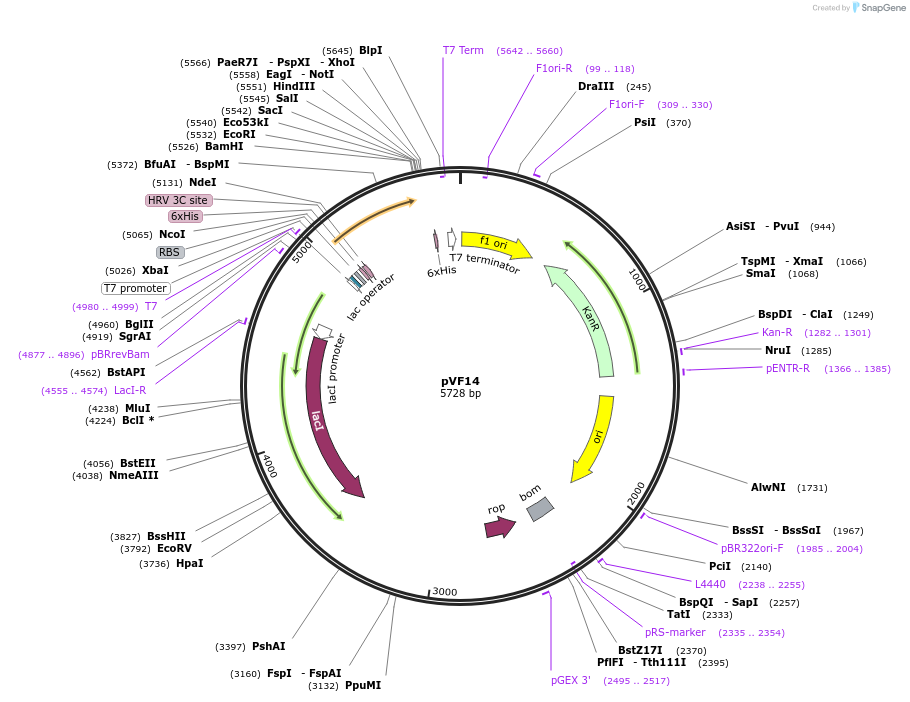
pVF14
Plasmid#229455PurposeExpresses IraD with His-tagDepositorInsertIraD
Tags6xHisExpressionBacterialPromoterT7Available SinceJan. 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
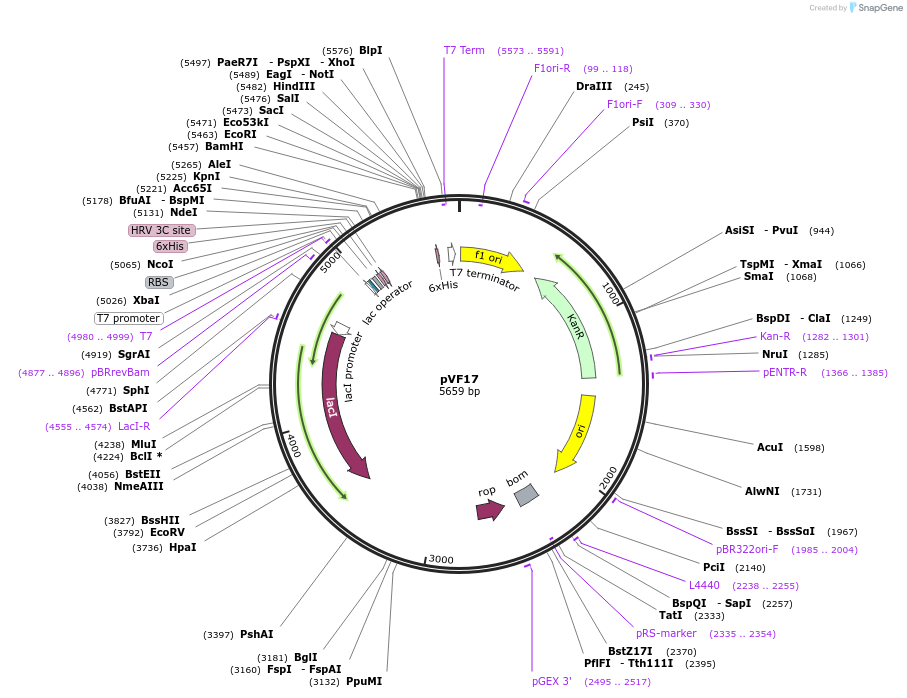
pVF17
Plasmid#229454PurposeExpresses IraM with His-tagDepositorInsertIraM
Tags6xHisExpressionBacterialPromoterT7Available SinceJan. 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
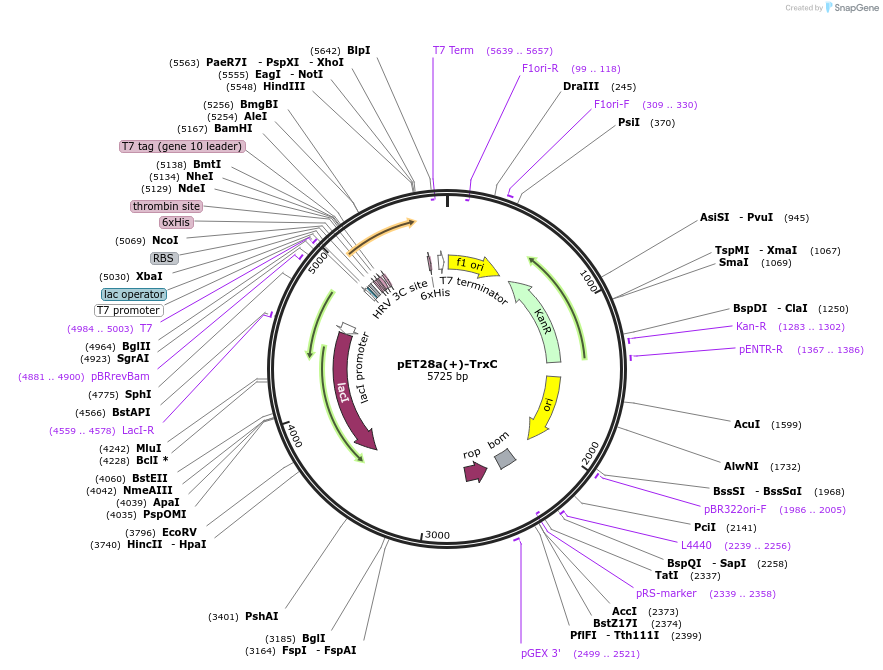
pET28a(+)-TrxC
Plasmid#225742PurposeTo express TrxC(Rv3914) in E. coliDepositorInsertTrxC
Tags6xHisExpressionBacterialMutationTrxCAvailable SinceOct. 28, 2024AvailabilityAcademic Institutions and Nonprofits only -
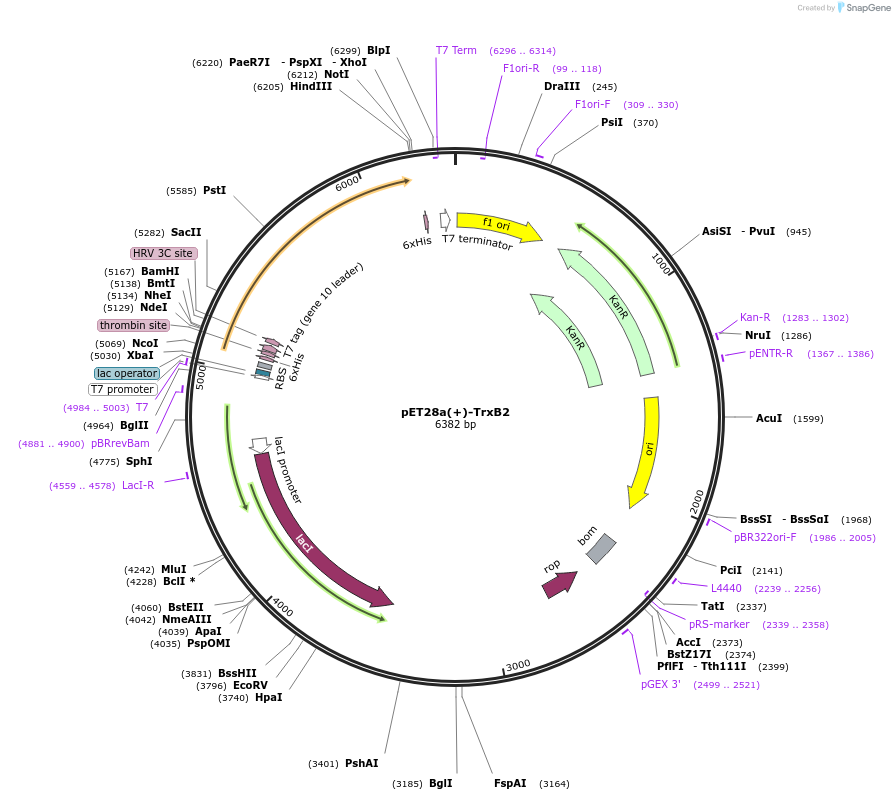
pET28a(+)-TrxB2
Plasmid#225741PurposeTo express TrxB2(Rv3913) in E. coliDepositorInsertTrxB2
Tags6xHisExpressionBacterialMutationTrxB2Available SinceOct. 28, 2024AvailabilityAcademic Institutions and Nonprofits only -
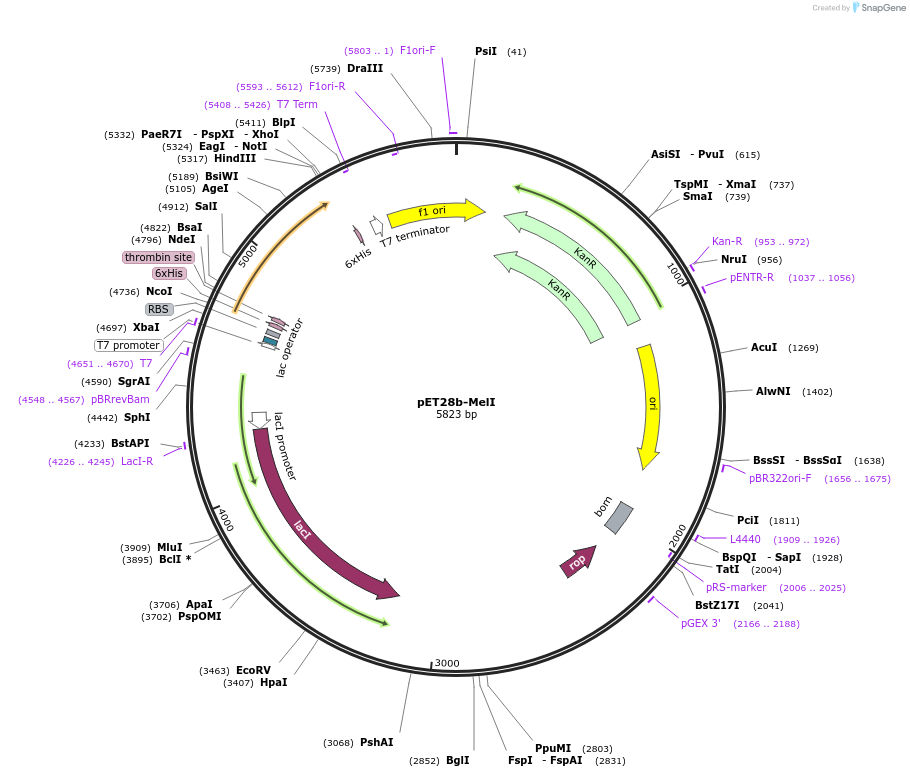
pET28b-MelI
Plasmid#225730PurposeTo express MelI (Rv1939) in E. coliDepositorInsertmelI
Tags6xHisExpressionBacterialAvailable SinceOct. 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
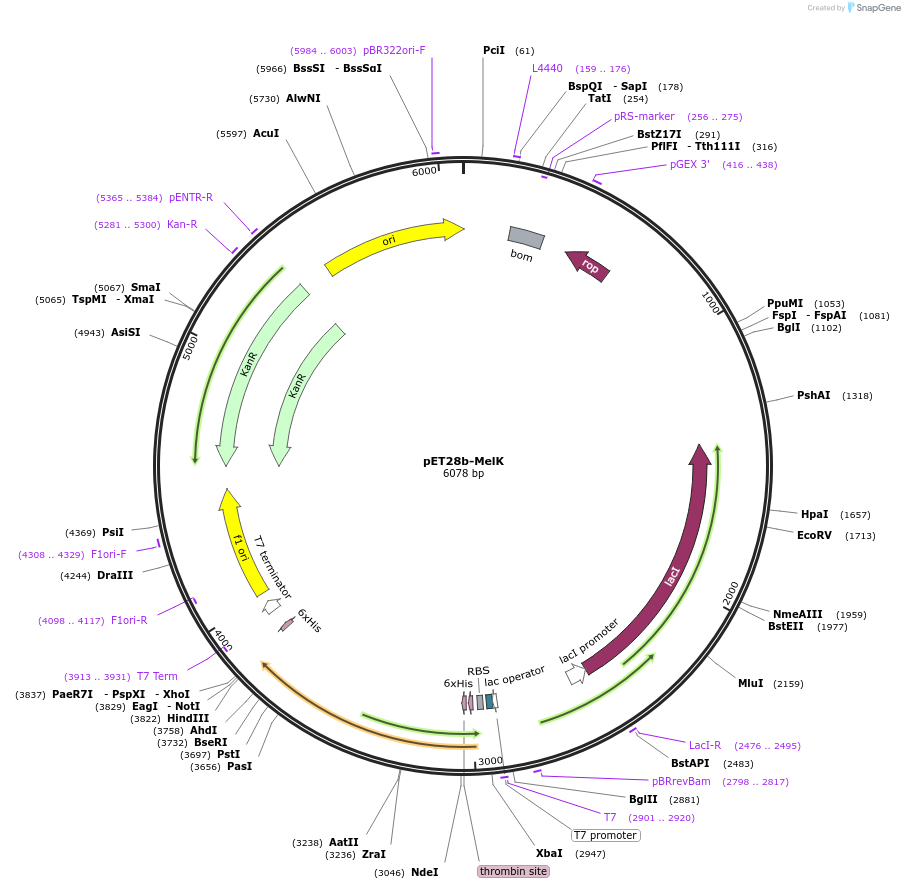
pET28b-MelK
Plasmid#225732PurposeTo express MelK (Rv1941) in E. coliDepositorInsertmelK
Tags6xHisExpressionBacterialAvailable SinceOct. 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
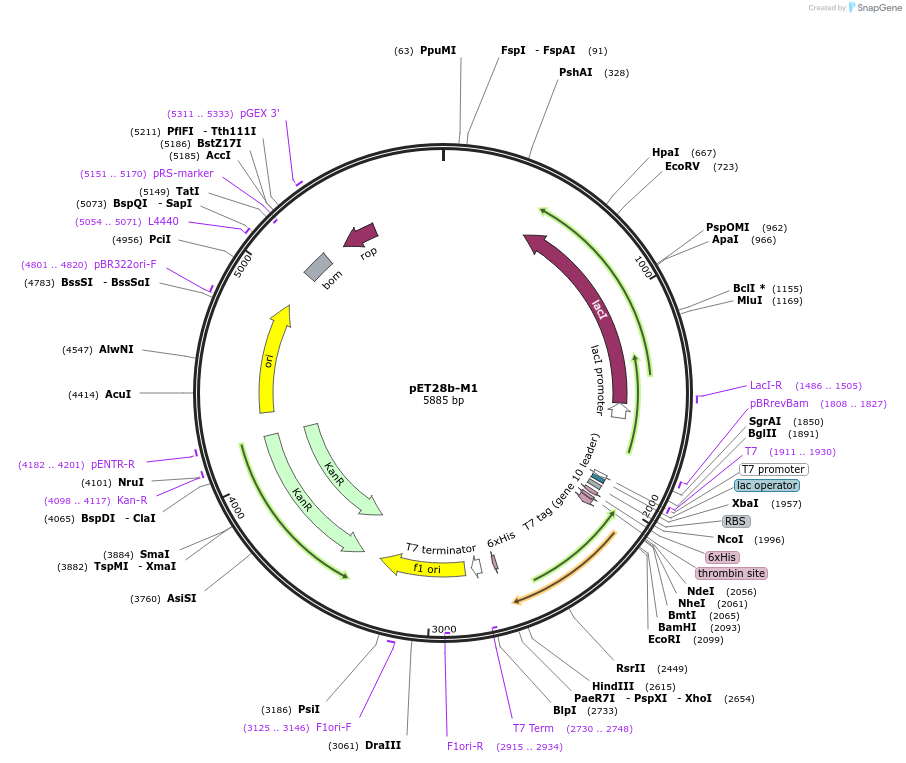
pET28b-M1
Plasmid#225734PurposeTo express M1 (13-193) in E. coliDepositorInsertmce3R
Tags6xHisExpressionBacterialMutationaa 13-193Available SinceOct. 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
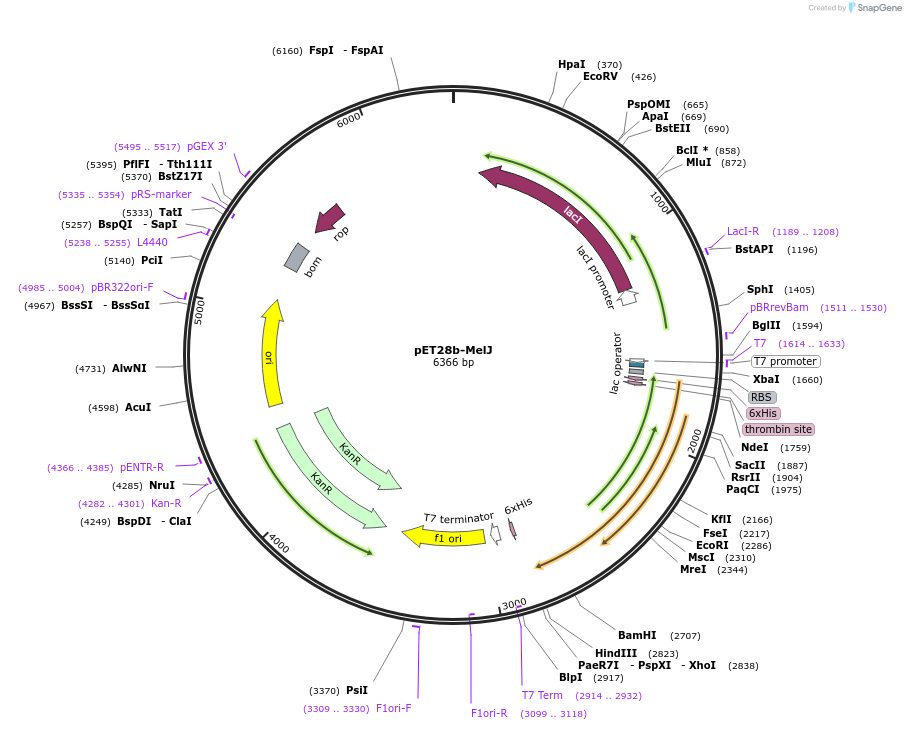
pET28b-MelJ
Plasmid#225731PurposeTo express MelJ (Rv1940) in E. coliDepositorInsertmelJ
Tags6xHisExpressionBacterialAvailable SinceOct. 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
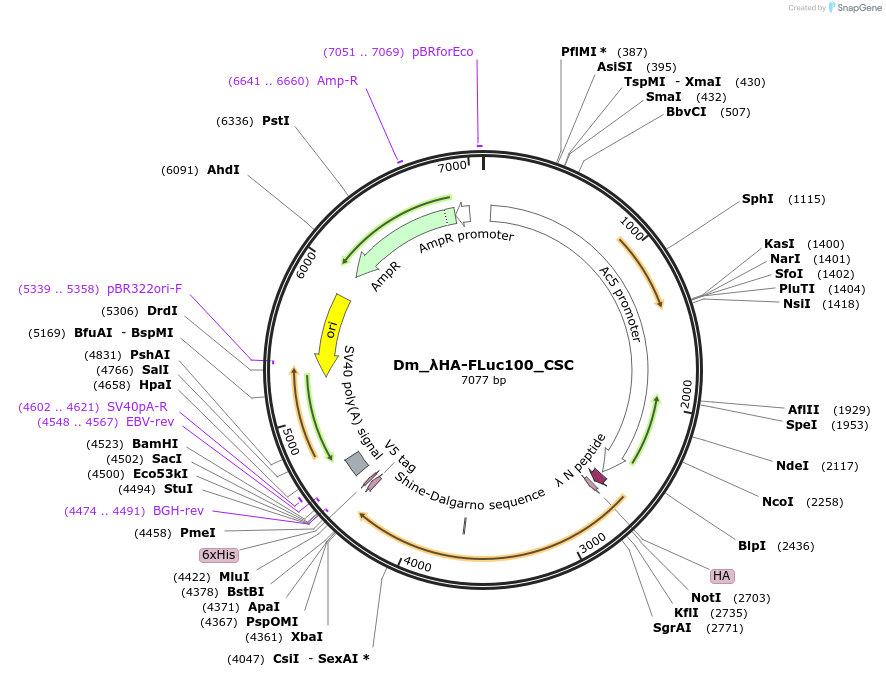
Dm_λHA-FLuc100_CSC
Plasmid#210592PurposeExpresses lambdaHA-optimal Firefly (CSC metric; Drosophila)DepositorInsertLambdaHA-optimal Firefly luciferase (Dm optimization, CSC metric)
Tagslambda HAExpressionInsectPromoterAc5Available SinceOct. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
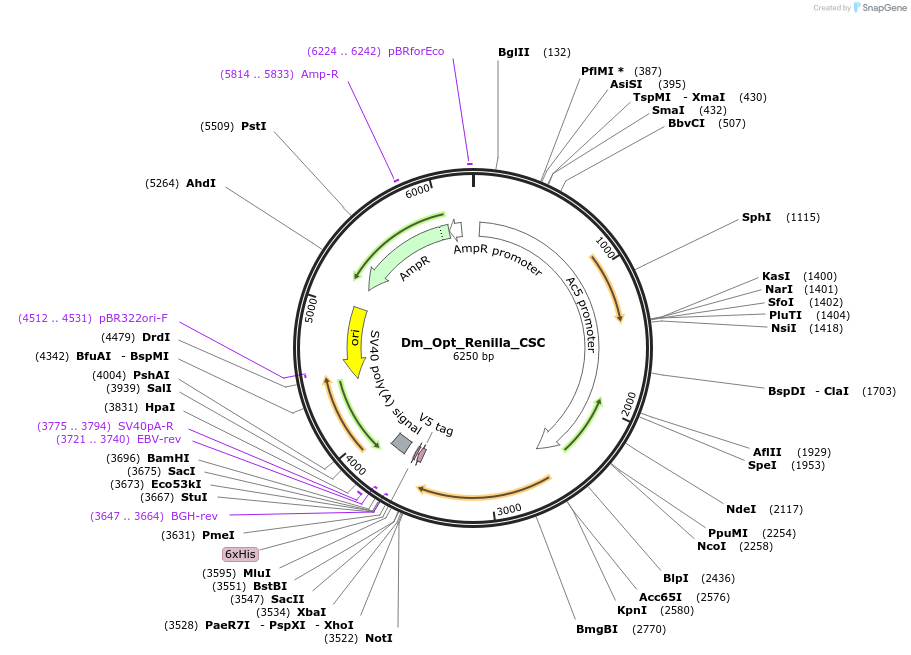
Dm_Opt_Renilla_CSC
Plasmid#210593PurposeExpresses Optimal Renilla luciferase (CSC metric; Drosophila)DepositorInsertOptimal Renilla luciferase (Optimized for Drosophila, CSC metric)
ExpressionInsectPromoterAc5Available SinceOct. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
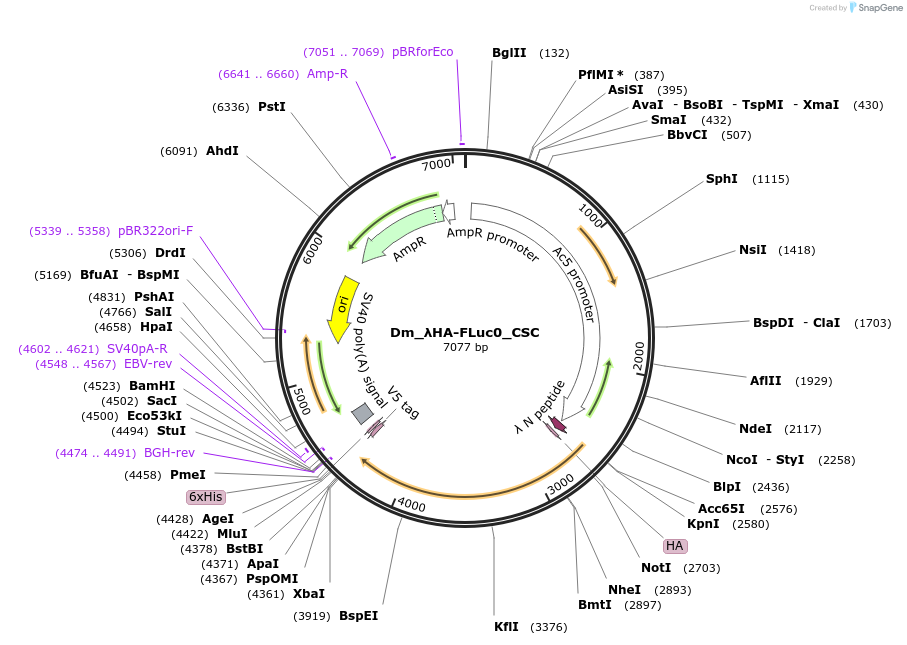
Dm_λHA-FLuc0_CSC
Plasmid#210589PurposeExpresses lambdaHA-nonoptimal Firefly (CSC metric; Drosophila)DepositorInsertLambdaHA-nonoptimal Firefly luciferase (Dm optimization, CSC metric)
UseLuciferaseTagslambda HAExpressionInsectPromoterAc5Available SinceOct. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
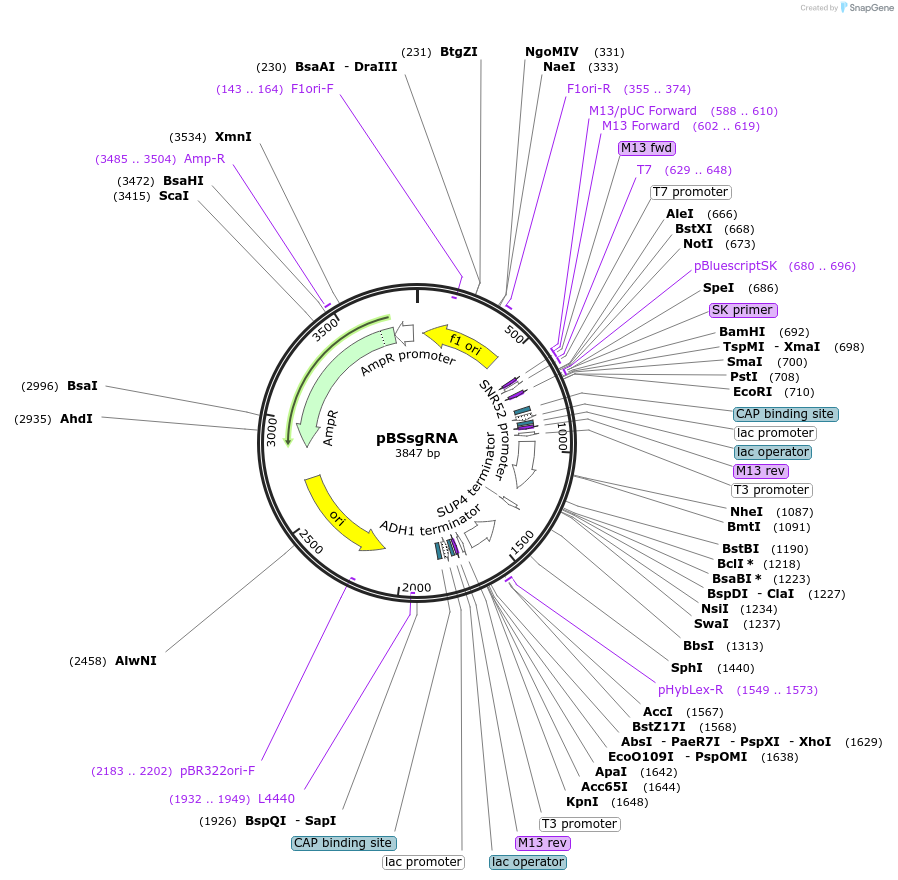
pBSsgRNA
Plasmid#224867PurposeYeast genome editing, Insertion of 20 bp oligo into sgRNA geneDepositorTypeEmpty backboneUseCRISPRExpressionYeastAvailable SinceOct. 17, 2024AvailabilityAcademic Institutions and Nonprofits only -
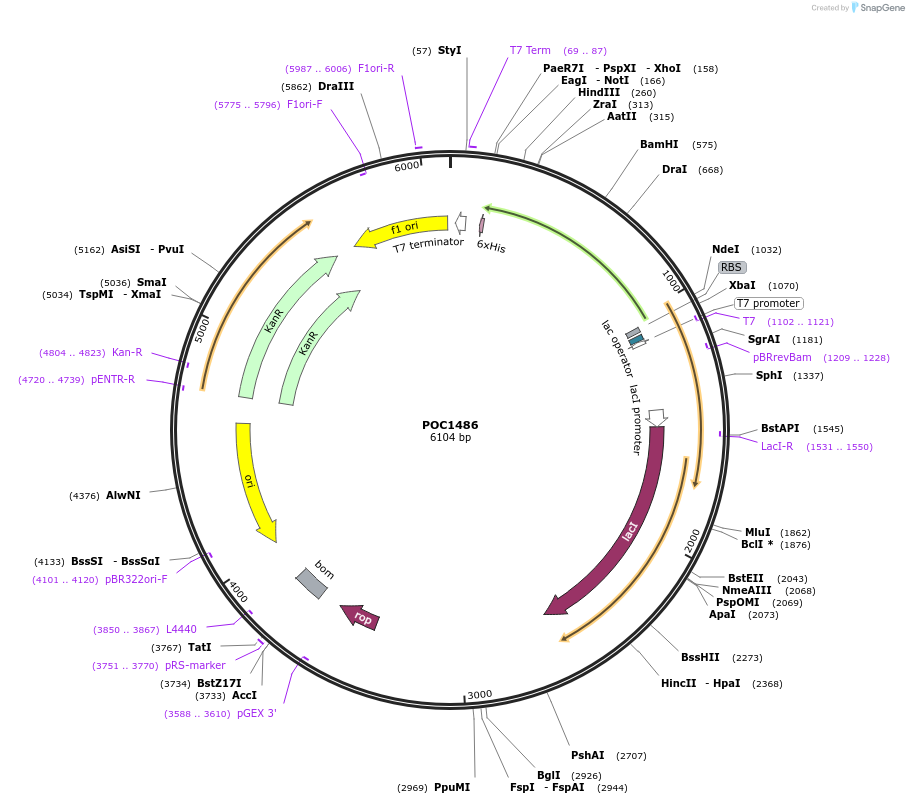
POC1486
Plasmid#213811PurposeBpiI-associated non-switchable methylaseDepositorInsertBpiI-associated non-switchable methylase M2.HpyAII
TagsHis6-tagExpressionBacterialPromoterT7Available SinceAug. 22, 2024AvailabilityAcademic Institutions and Nonprofits only