We narrowed to 216 results for: 48138
-
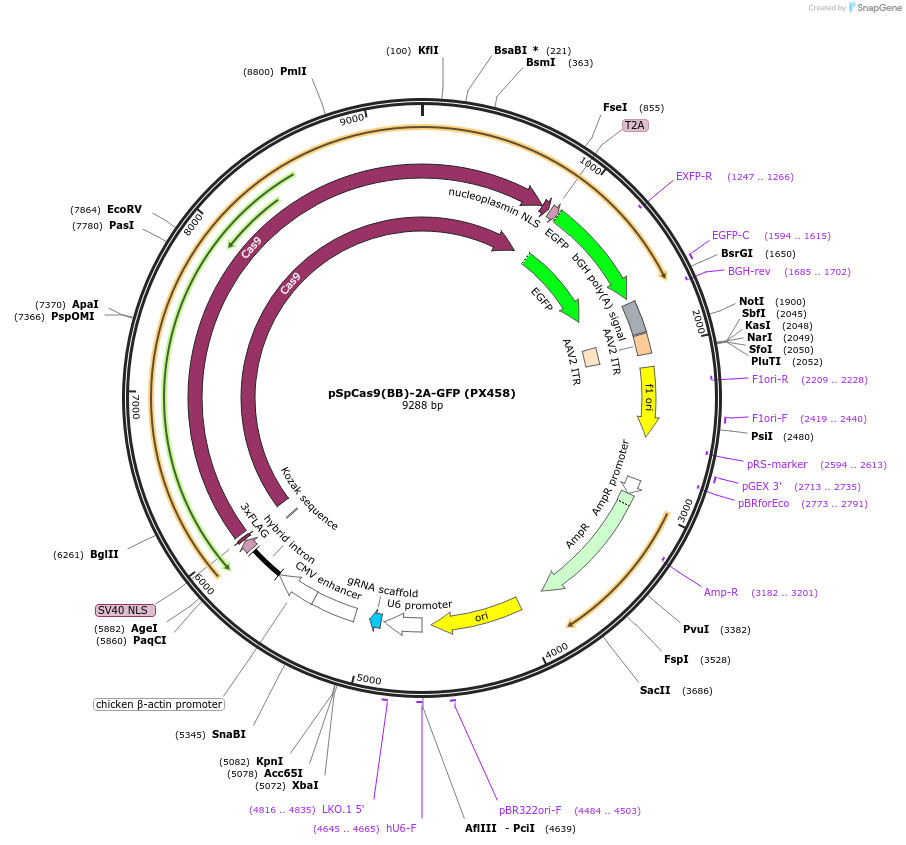
Plasmid#48138PurposeCas9 from S. pyogenes with 2A-EGFP, and cloning backbone for sgRNADepositorHas ServiceCloning Grade DNAInserthSpCas9
UseCRISPRTags3XFLAG and GFPExpressionMammalianPromoterCbhAvailable SinceOct. 29, 2013AvailabilityAcademic Institutions and Nonprofits only
These results may also be relevant.
-
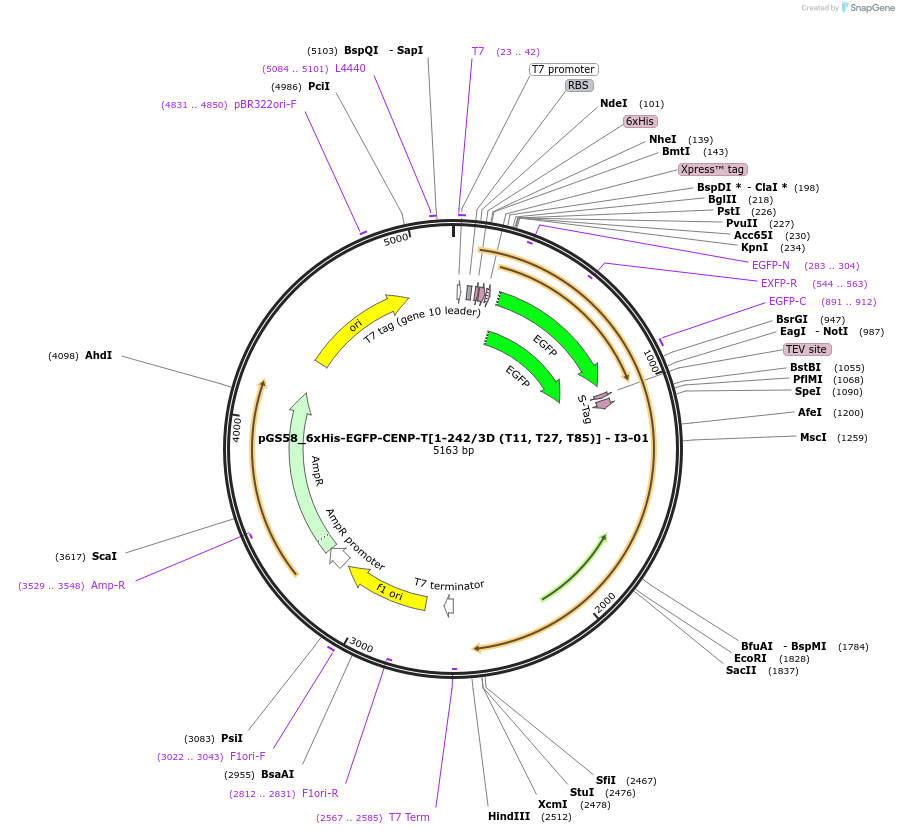
pGS58_6xHis-EGFP-CENP-T[1-242/3D (T11, T27, T85)] - I3-01
Plasmid#211890PurposeBacterial expression of CENP-T[1-242/3D]-I3-01DepositorInsertCENP-T[1-242/3D]-I3-01 (CENPT Human, Synthetic)
ExpressionBacterialMutationT11D, T27D, T85DAvailable SinceMay 7, 2024AvailabilityAcademic Institutions and Nonprofits only -
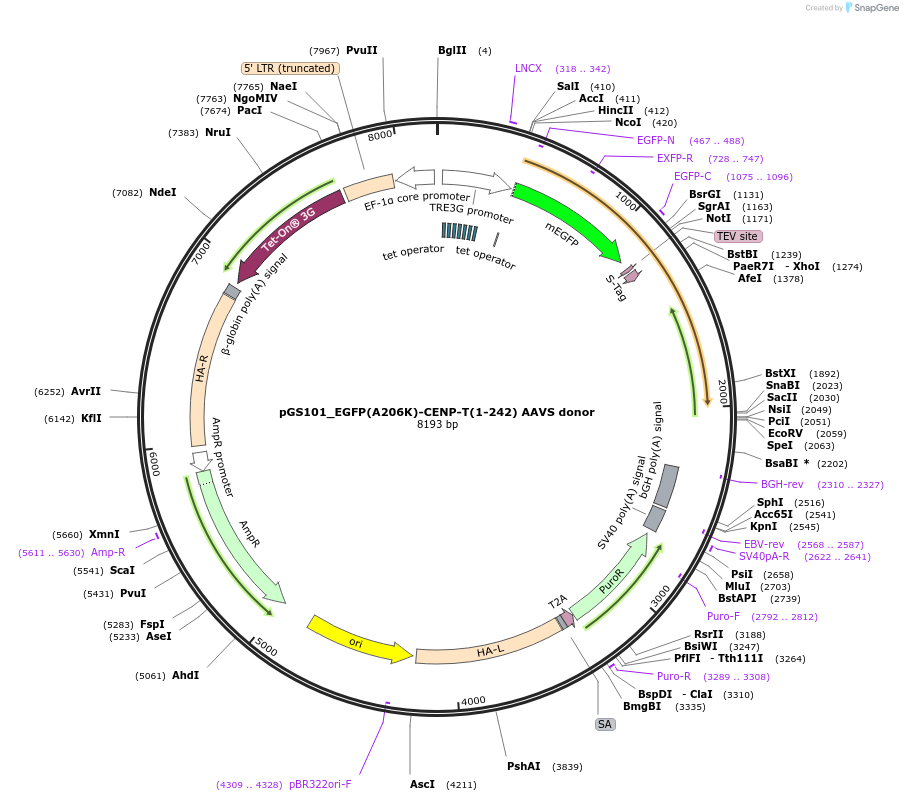
pGS101_EGFP(A206K)-CENP-T(1-242) AAVS donor
Plasmid#211867PurposeCENP-T(1-242) transgene integration at AAVS1 safe harbor locusDepositorAvailable SinceMarch 20, 2024AvailabilityAcademic Institutions and Nonprofits only -
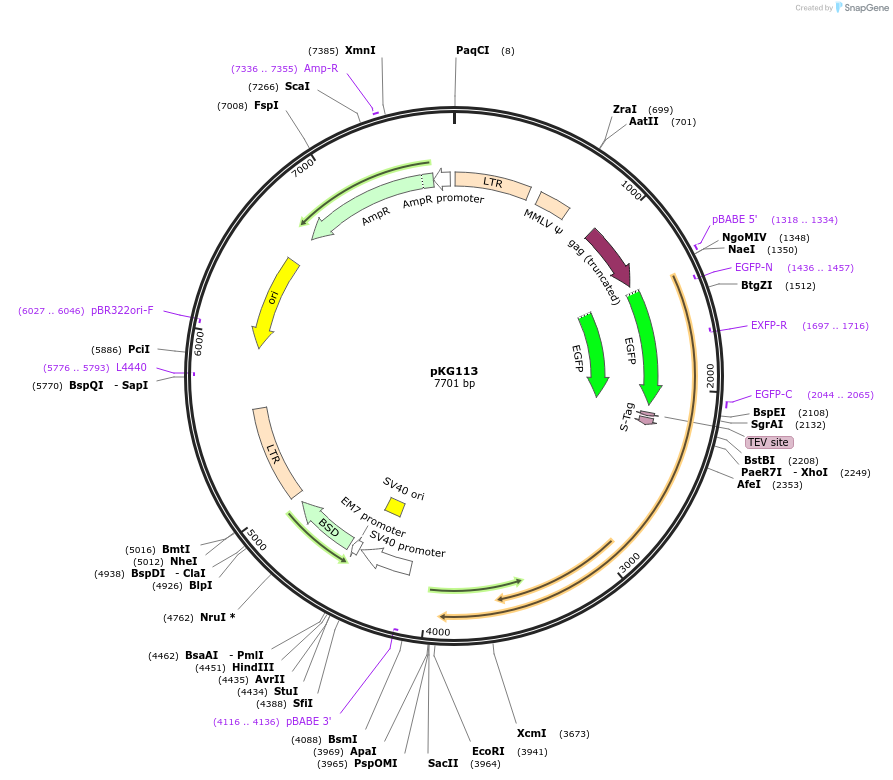
pKG113
Plasmid#63073PurposeRetroviral expression of EGFP-TEV-S-tag CENP-T for protein localization and affinity purification (LAP)DepositorInsertCENP-T (CENPT Human)
UseRetroviralTagsEGFP and TEV-SExpressionMammalianMutationE. coli codon optimized. G217R mutation compared …Available SinceJan. 13, 2016AvailabilityAcademic Institutions and Nonprofits only -
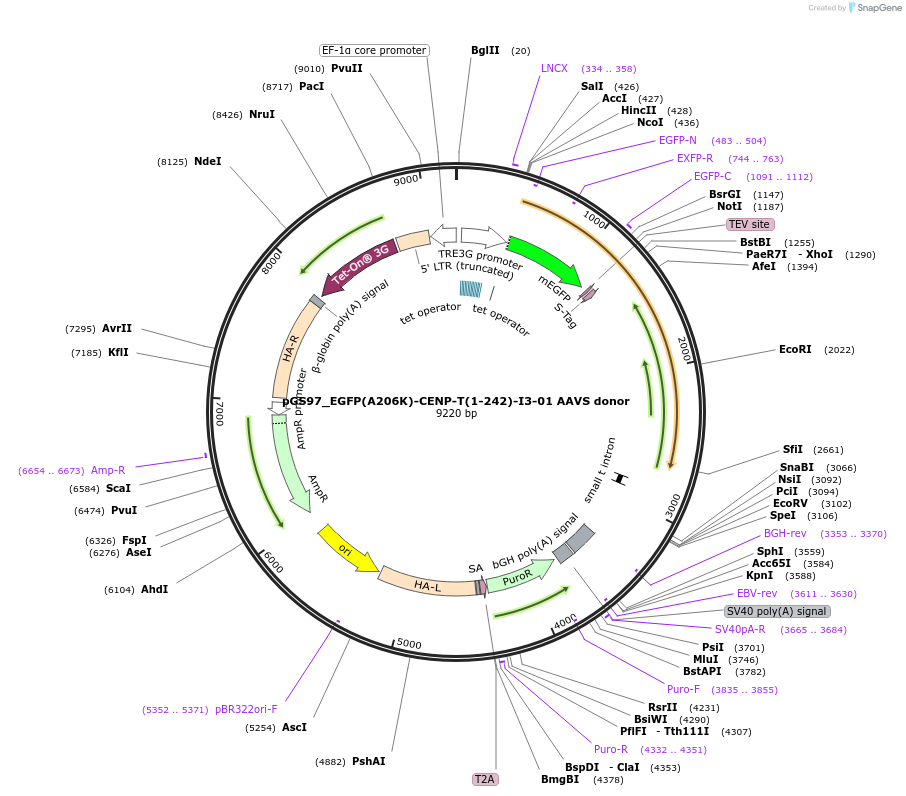
pGS97_EGFP(A206K)-CENP-T(1-242)-I3-01 AAVS donor
Plasmid#211866PurposeCENP-T(1-242)-I3-01 transgene integration at AAVS1 safe harbor locusDepositorAvailable SinceJune 17, 2024AvailabilityAcademic Institutions and Nonprofits only -
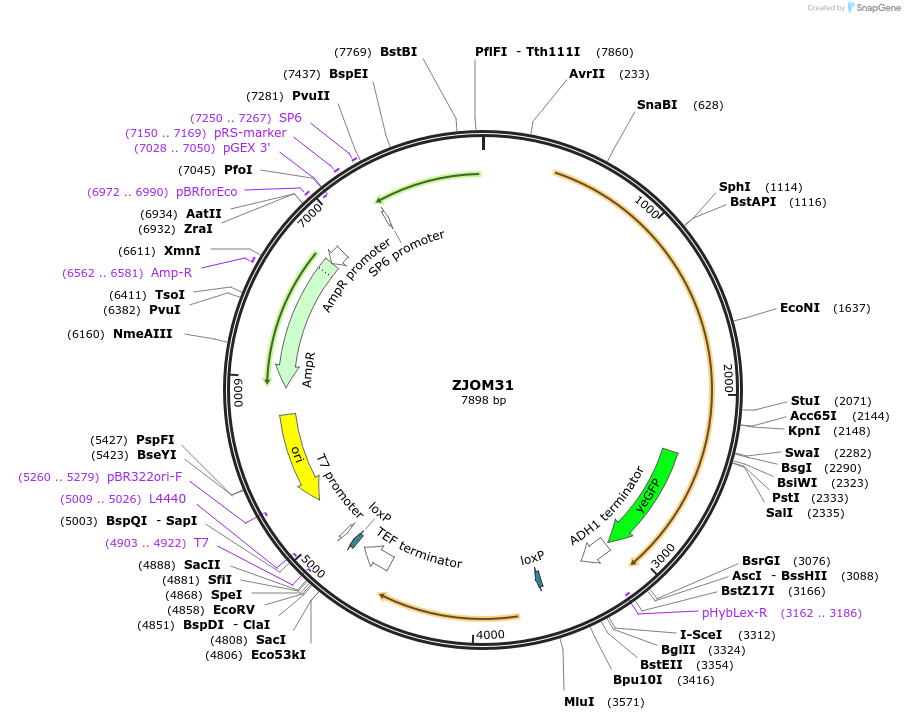
ZJOM31
Plasmid#133645PurposeIntegration of TGL3-GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Marker for yeast lipid droplet.DepositorAvailable SinceDec. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
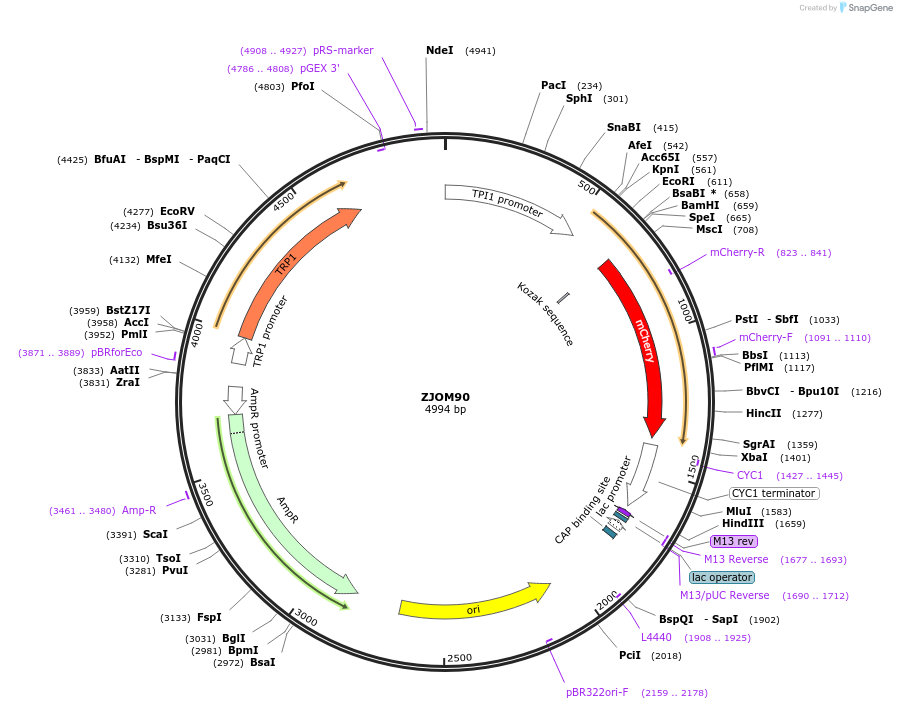
ZJOM90
Plasmid#133647PurposeIntegration of SS-mCherry-HDEL in genome, use auxotrophic marker TRP1(Saccharomyces cerevisiae). Marker for yeast endoplasmic reticulum (ER).DepositorInsertpTPI1-SS-mCherry-HDEL
TagsmCherryExpressionYeastAvailable SinceApril 22, 2020AvailabilityAcademic Institutions and Nonprofits only -
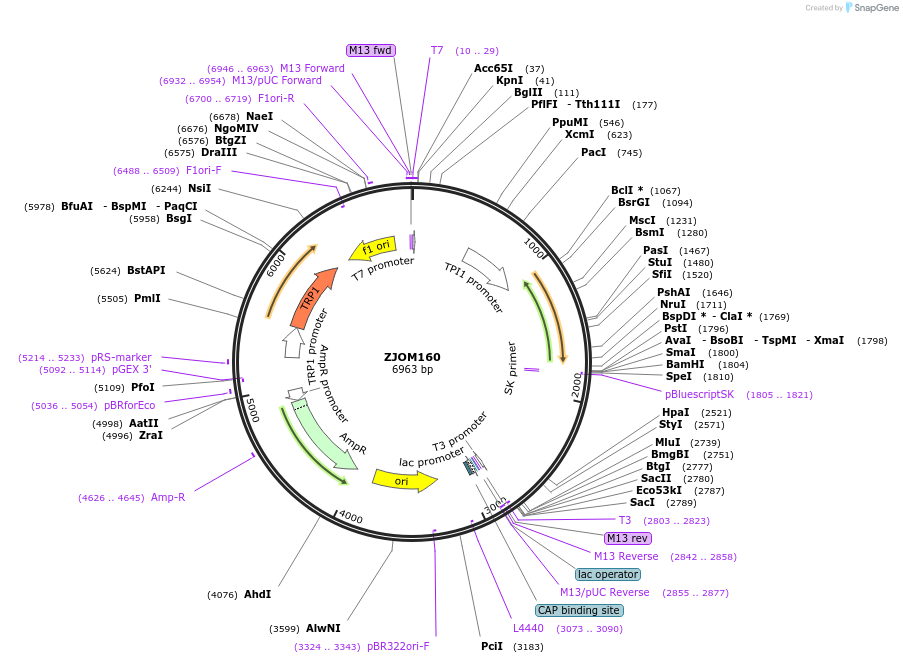
ZJOM160
Plasmid#133670PurposeIntegration of mTagBFP2-SKL as an additional copy in genome, use auxotrophic marker TRP1(Saccharomyces cerevisiae). Marker for yeast peroxisome.DepositorInsertpTPI1-mTagBFP2-SKL
TagsmTagBFP2ExpressionYeastAvailable SinceJan. 3, 2020AvailabilityAcademic Institutions and Nonprofits only -
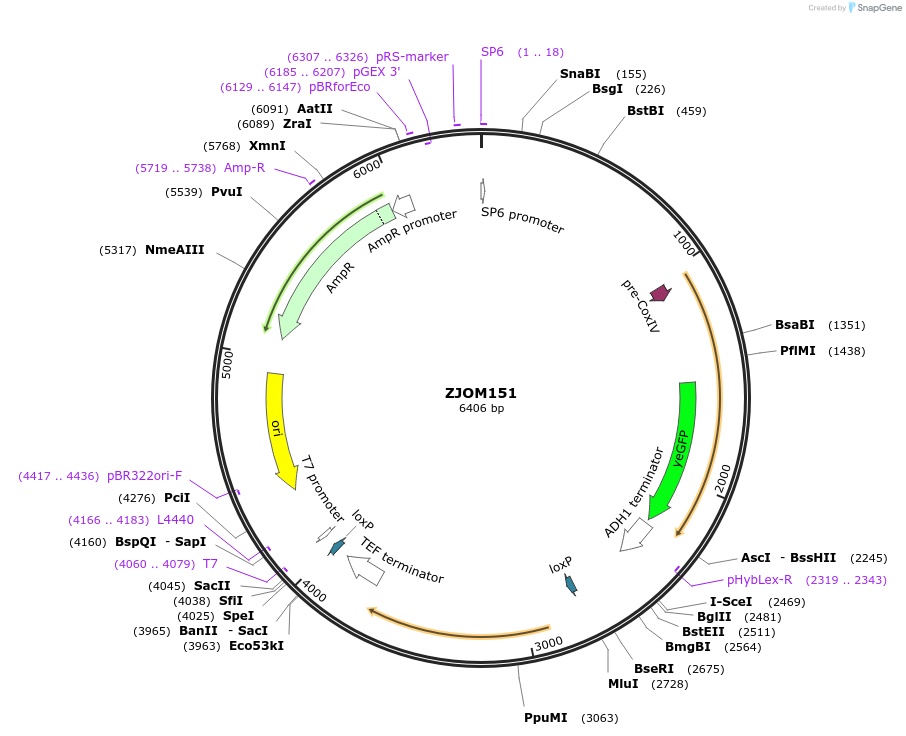
ZJOM151
Plasmid#133643PurposeIntegration of COX4-GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Marker for yeast mitochondria.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
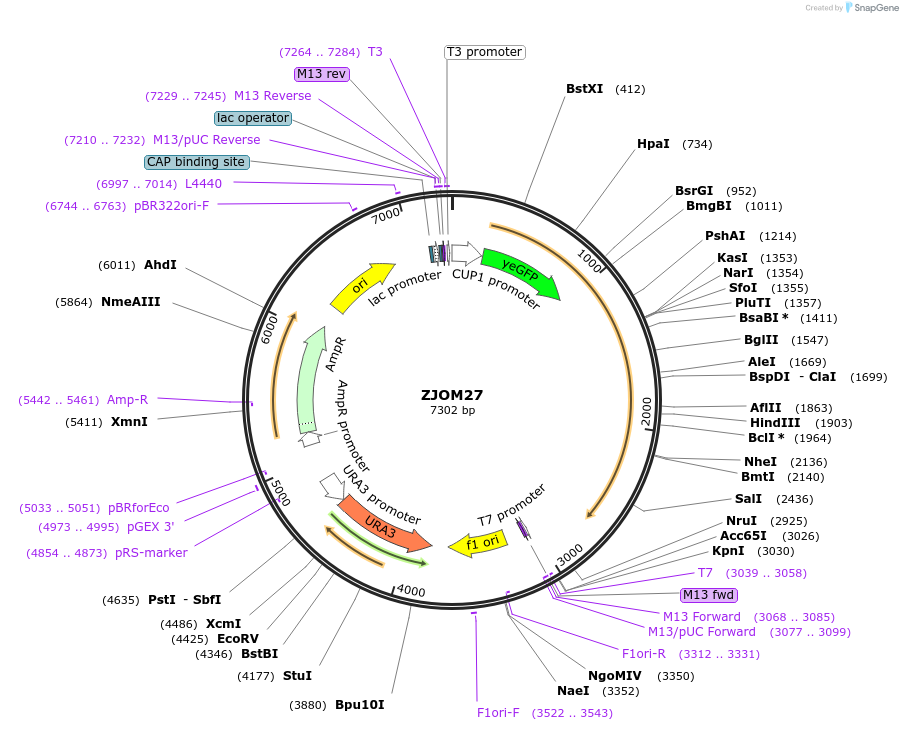
ZJOM27
Plasmid#133641PurposeIntegration of GFP-PHO8 as an additional copy in genome, use auxotrophic marker URA3(Saccharomyces cerevisiae). Marker for yeast vacuole.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
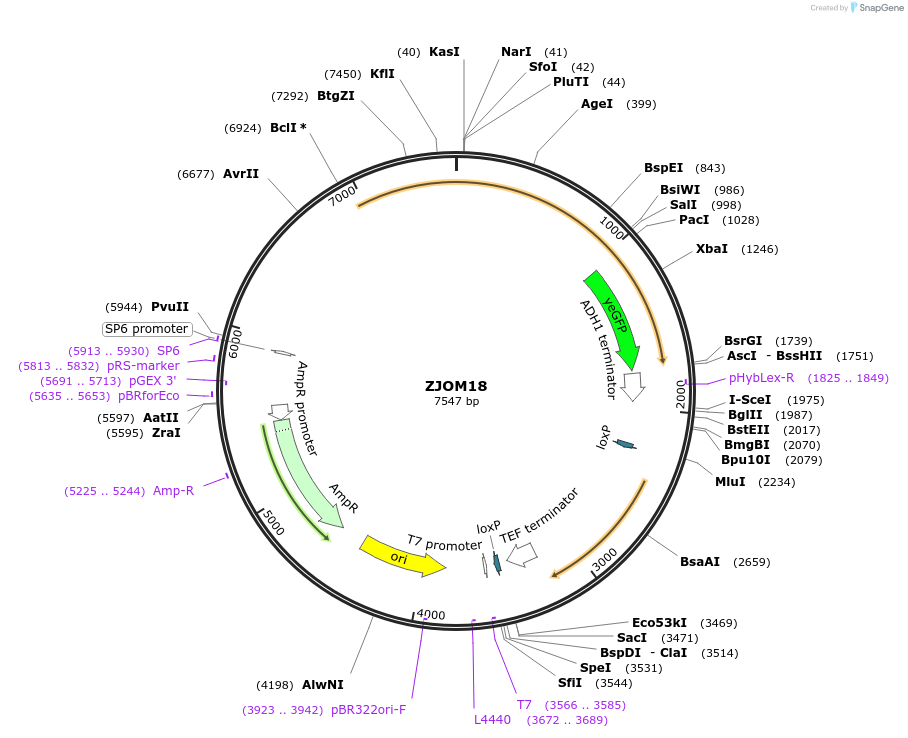
ZJOM18
Plasmid#133631PurposeIntegration of NAB2-GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Marker for yeast nucleus.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
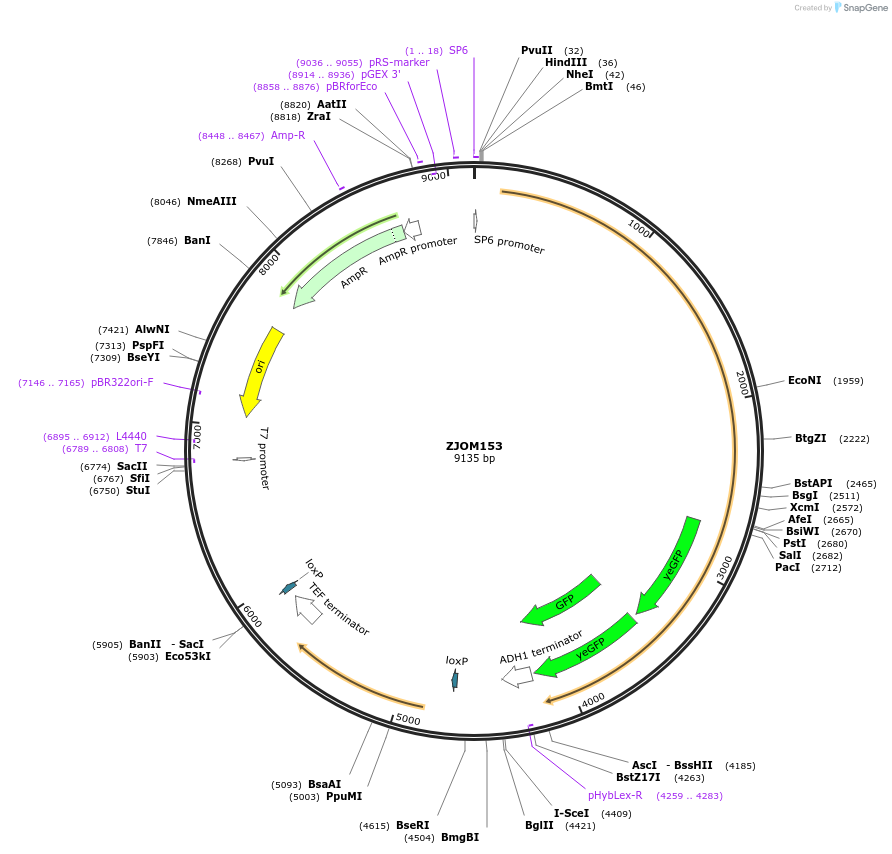
ZJOM153
Plasmid#133642PurposeIntegration of 2xGFP tag at VPH1 C terminus, use auxotrophic marker URA3(Candida albicans). Marker for yeast vacuole.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
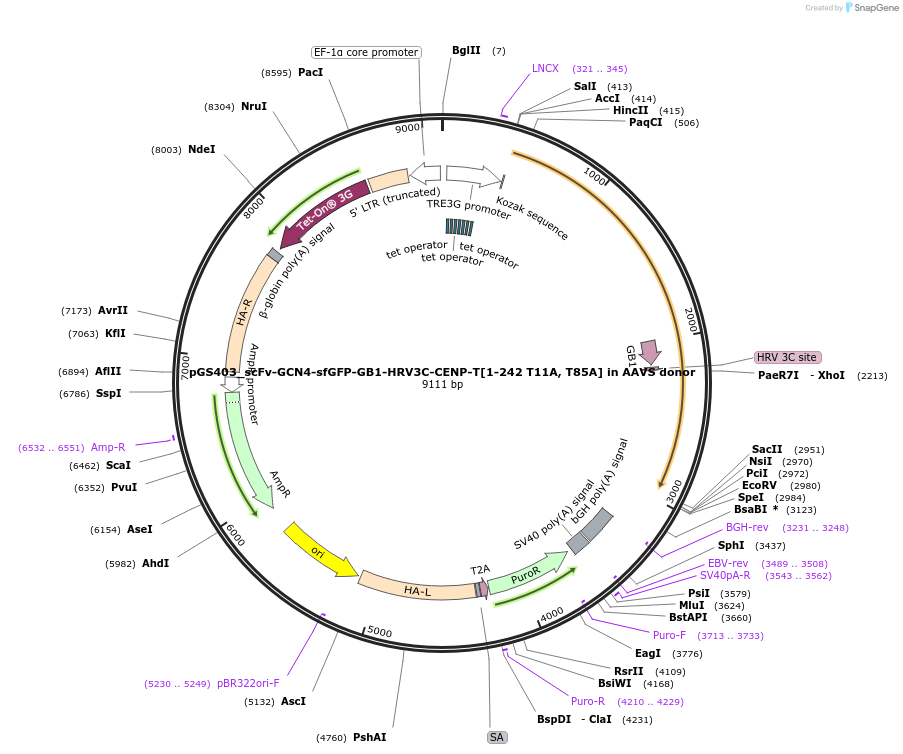
pGS403_scFv-GCN4-sfGFP-GB1-HRV3C-CENP-T[1-242 T11A, T85A] in AAVS donor
Plasmid#211888PurposeCENP-T[1-242] transgene integration at AAVS1 safe harbor locusDepositorAvailable SinceMarch 20, 2024AvailabilityAcademic Institutions and Nonprofits only -
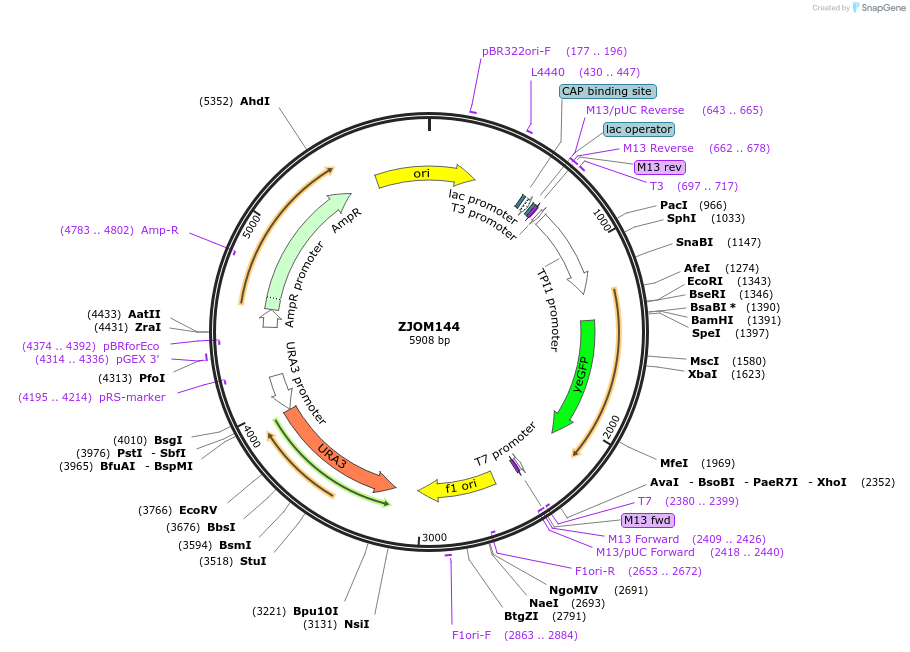
ZJOM144
Plasmid#133630PurposeIntegration of SS-GFP-HDEL in genome, use auxotrophic marker URA3(Saccharomyces cerevisiae) Marker for yeast endoplasmic reticulum (ER).DepositorInsertpTPI1-SS-GFP-HDEL
TagsEGFPExpressionYeastAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
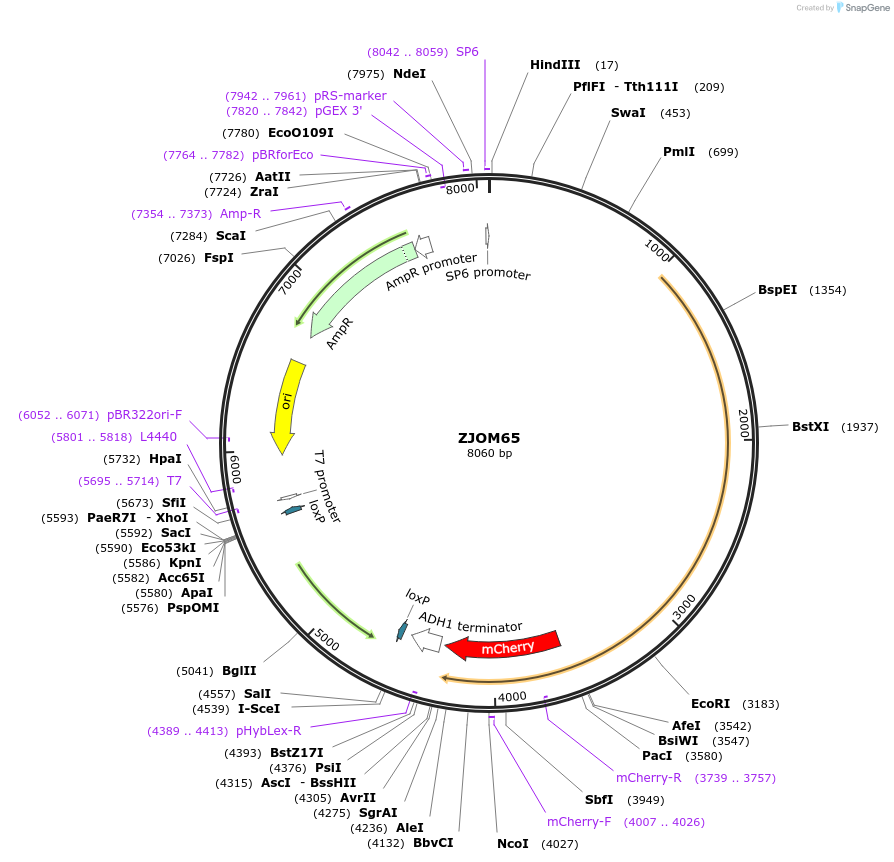
ZJOM65
Plasmid#133654PurposeIntegration of VPH1-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast vacuole.DepositorAvailable SinceJan. 3, 2020AvailabilityAcademic Institutions and Nonprofits only -
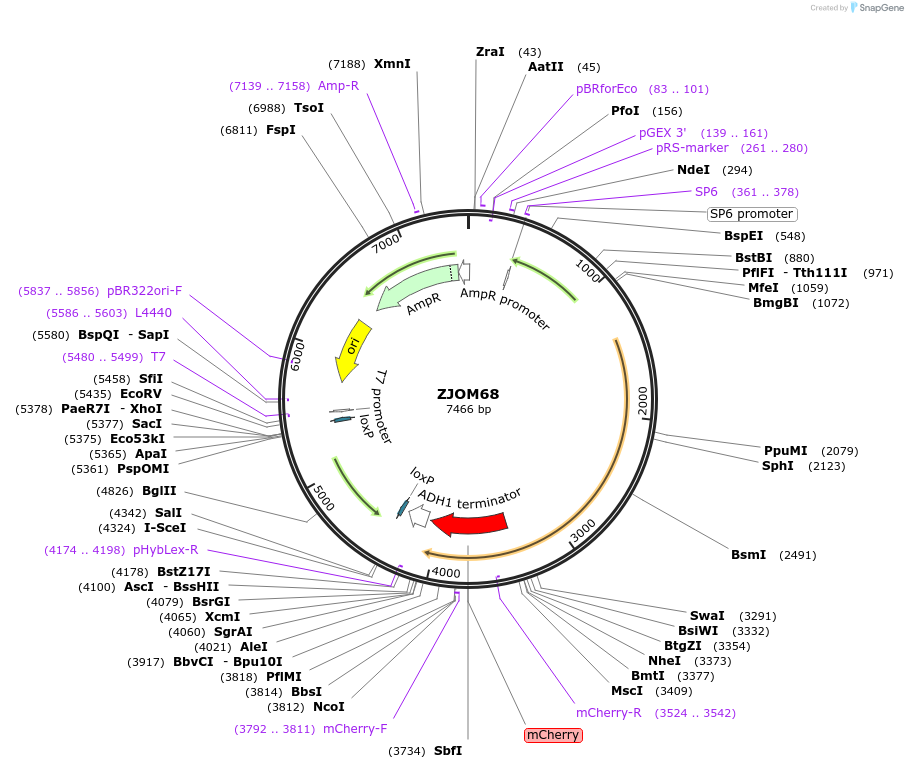
ZJOM68
Plasmid#133657PurposeIntegration of TGL3-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast lipid droplet.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
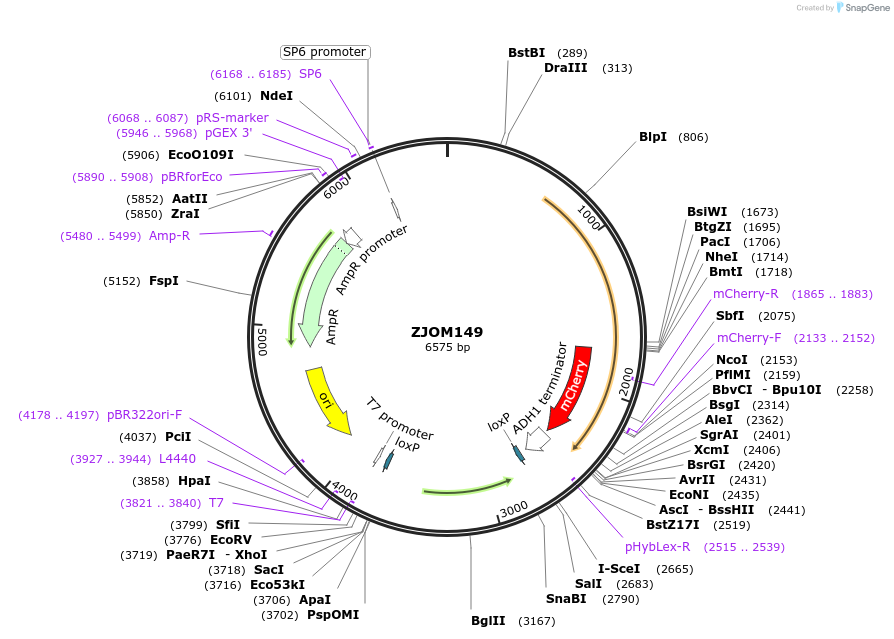
ZJOM149
Plasmid#133646PurposeIntegration of ELO3-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast endoplasmic reticulum (ER).DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
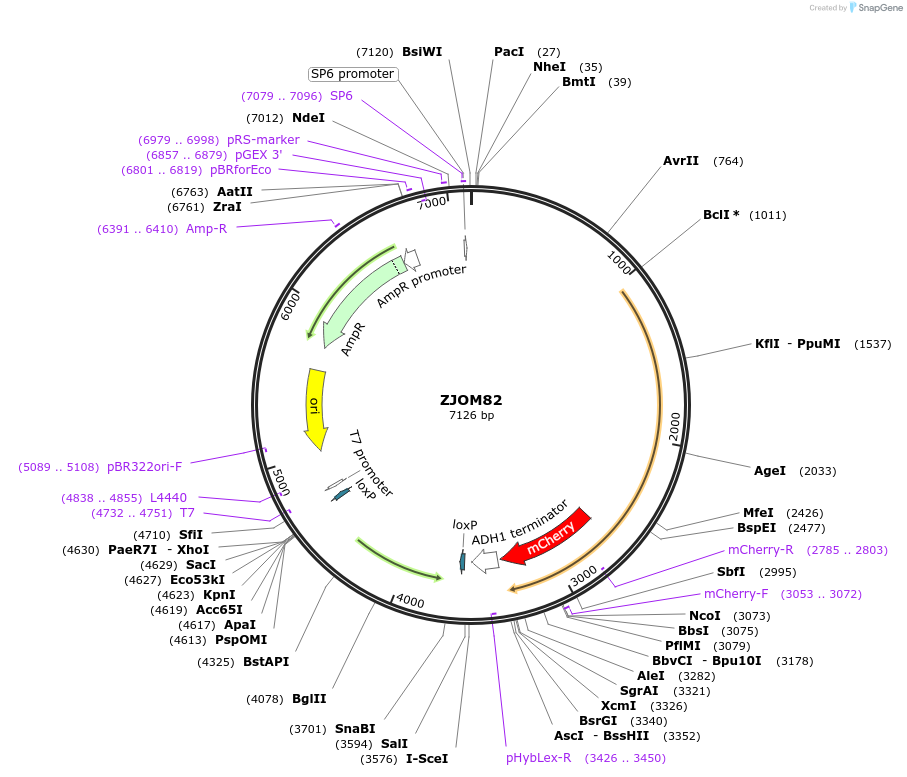
ZJOM82
Plasmid#133648PurposeIntegration of NAB2-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast nucleus.DepositorAvailable SinceJan. 22, 2020AvailabilityAcademic Institutions and Nonprofits only -
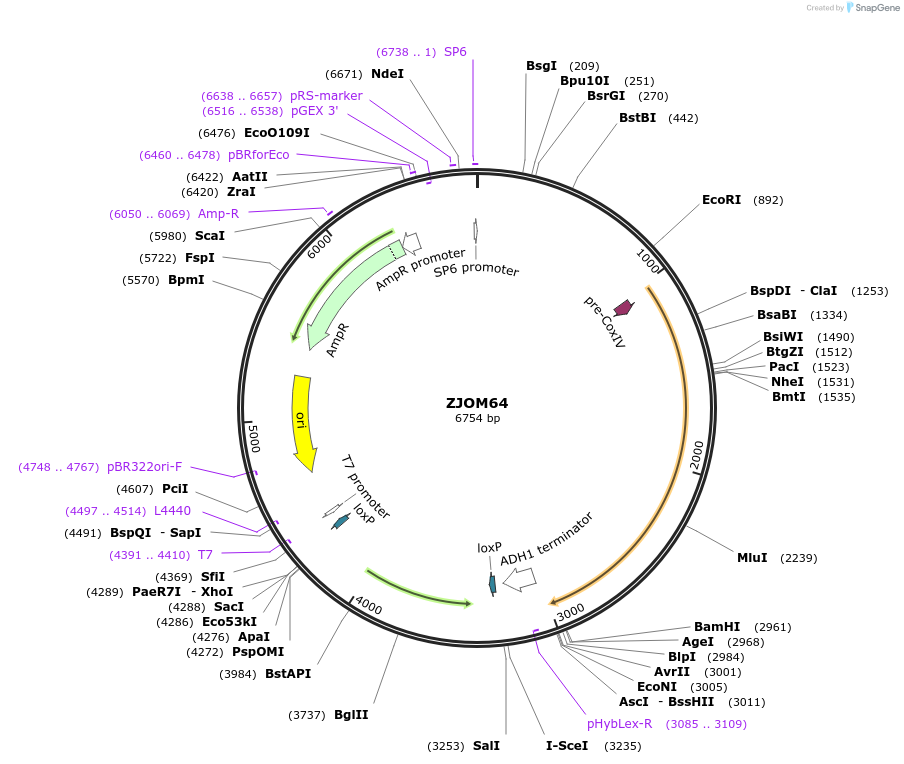
ZJOM64
Plasmid#133655PurposeIntegration of COX4-DuDre as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast mitochondria.DepositorAvailable SinceApril 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
ZJOM135
Plasmid#133659PurposeIntegration of SEC63-2mTagBFP2 as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast endoplasmic reticulum (ER).DepositorAvailable SinceDec. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
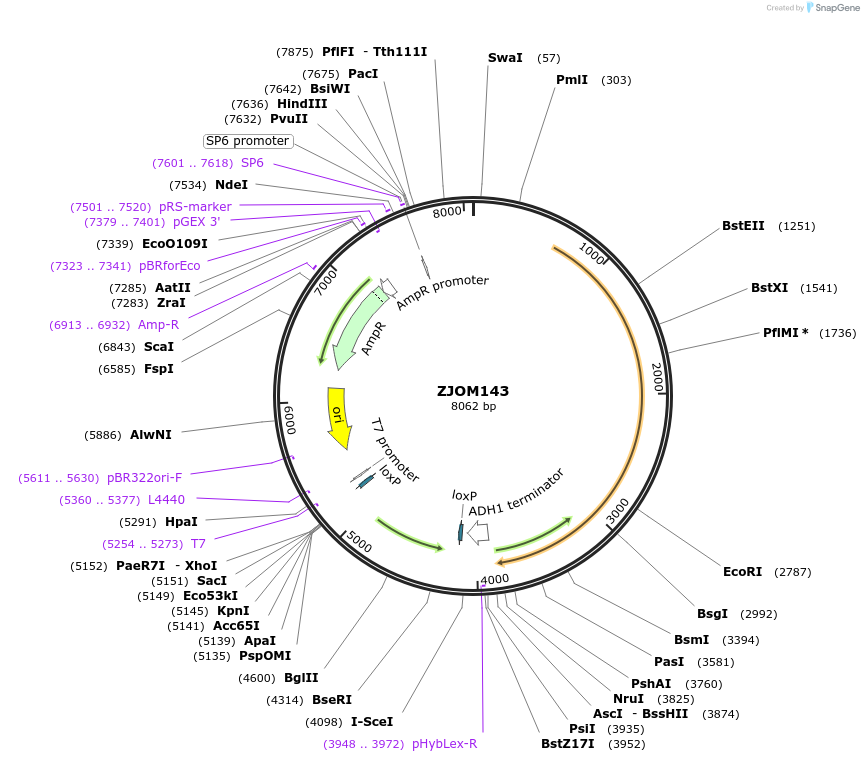
ZJOM143
Plasmid#133667PurposeIntegration of VPH1-mTagBFP2 as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast vacuole.DepositorAvailable SinceDec. 5, 2019AvailabilityAcademic Institutions and Nonprofits only