We narrowed to 216 results for: 48138
-
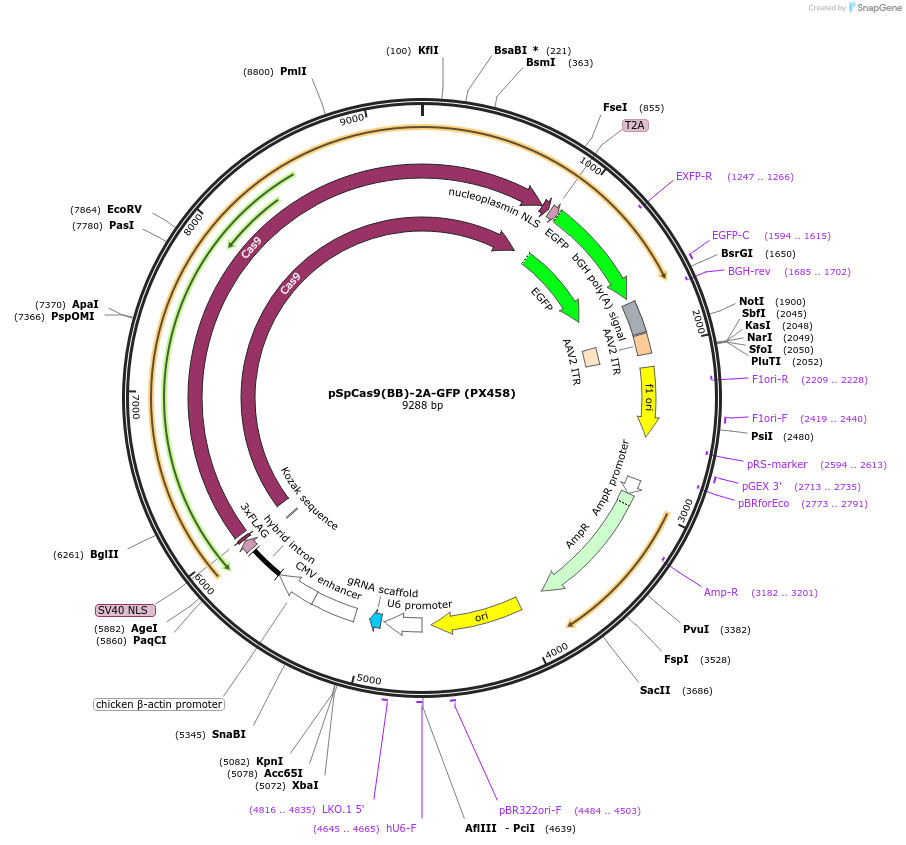
Plasmid#48138PurposeCas9 from S. pyogenes with 2A-EGFP, and cloning backbone for sgRNADepositorHas ServiceCloning Grade DNAInserthSpCas9
UseCRISPRTags3XFLAG and GFPExpressionMammalianPromoterCbhAvailable SinceOct. 29, 2013AvailabilityAcademic Institutions and Nonprofits only
These results may also be relevant.
-
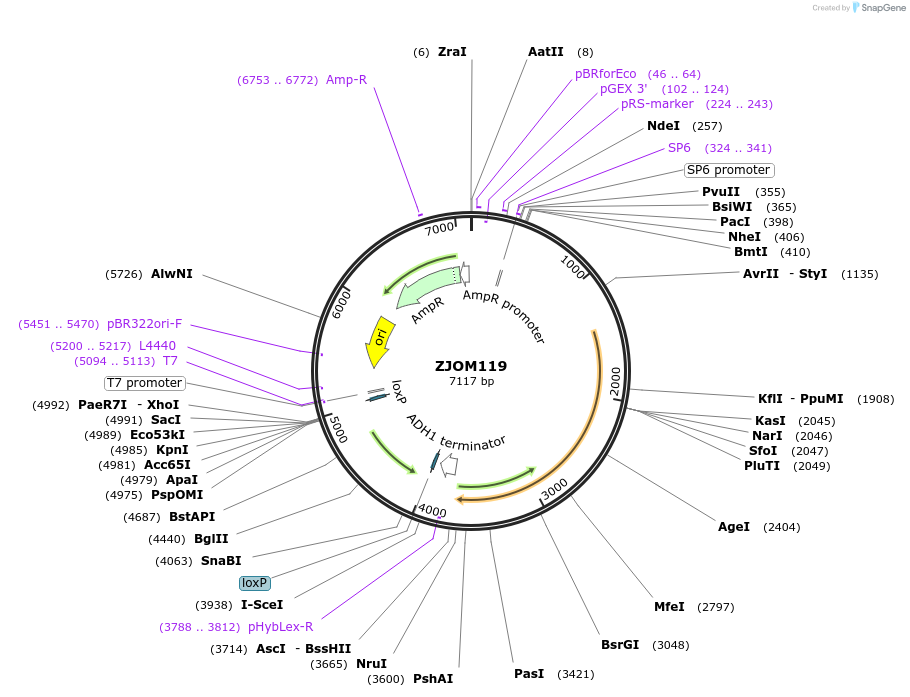
ZJOM119
Plasmid#133661PurposeIntegration of NAB2-mTagBFP2 as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast nucleus.DepositorAvailable SinceDec. 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
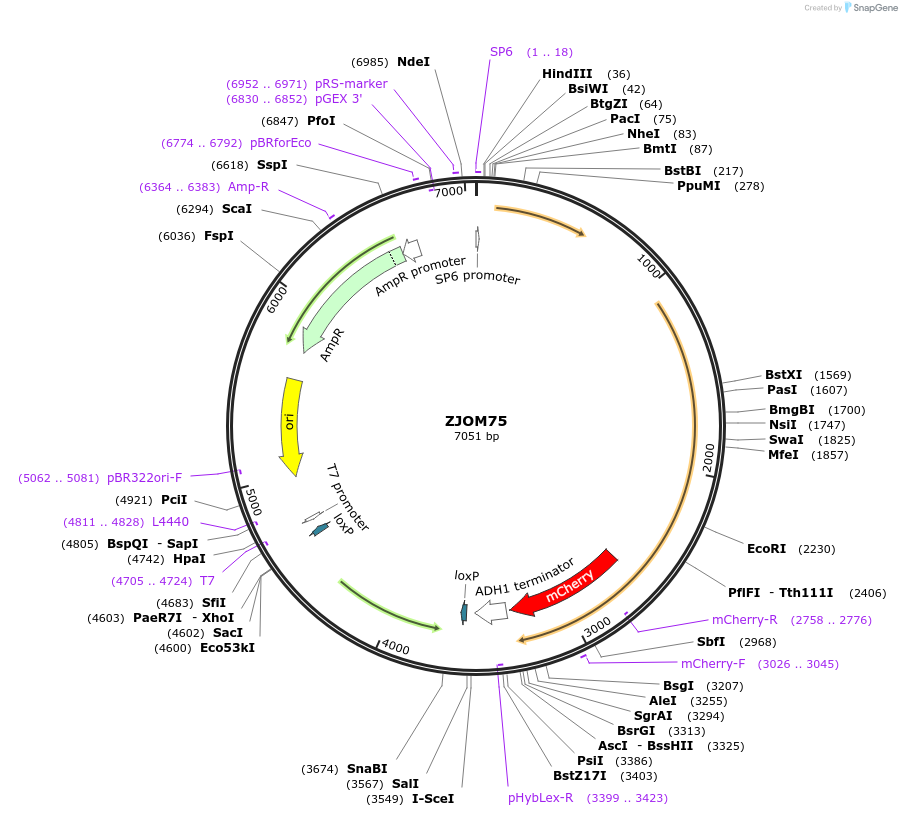
ZJOM75
Plasmid#133649PurposeIntegration of ANP1-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast early golgi.DepositorAvailable SinceJan. 2, 2020AvailabilityAcademic Institutions and Nonprofits only -
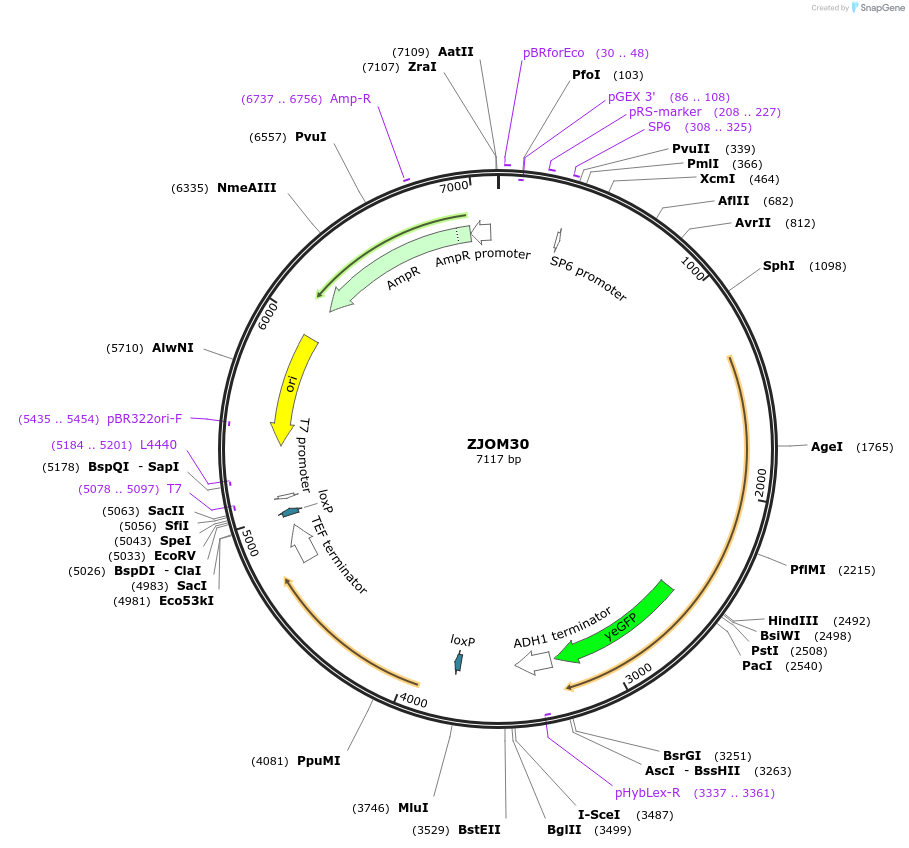
ZJOM30
Plasmid#133673PurposeIntegration of ERG6-GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Present on ER and lipid droplets.DepositorAvailable SinceApril 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
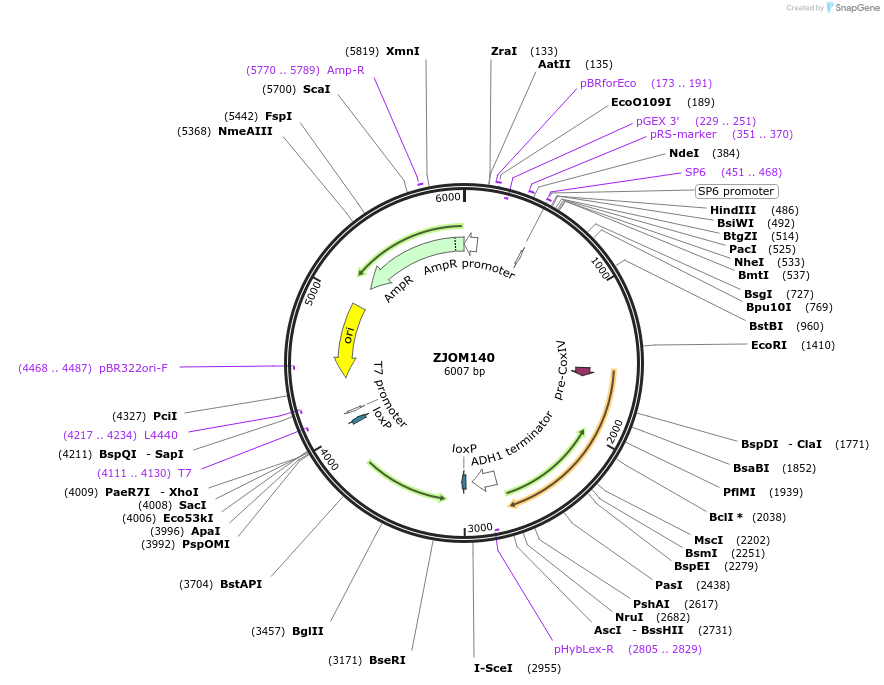
ZJOM140
Plasmid#133668PurposeIntegration of COX4-mTagBFP2 as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast mitochondria.DepositorAvailable SinceFeb. 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
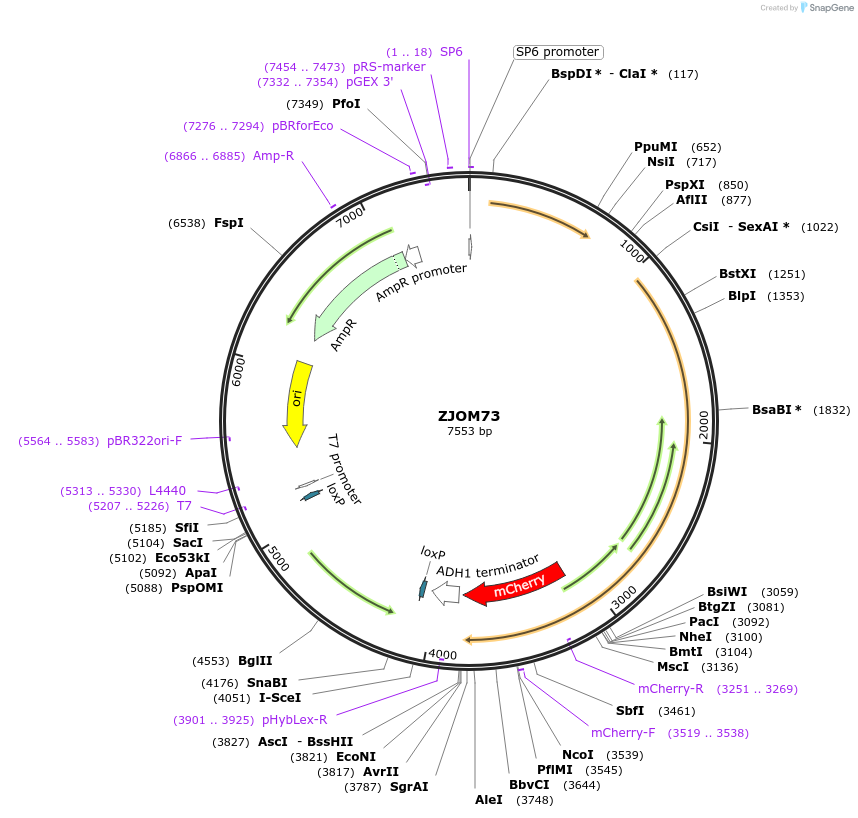
ZJOM73
Plasmid#133651PurposeIntegration of CHS5-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast late golgi/early endosome.DepositorAvailable SinceApril 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
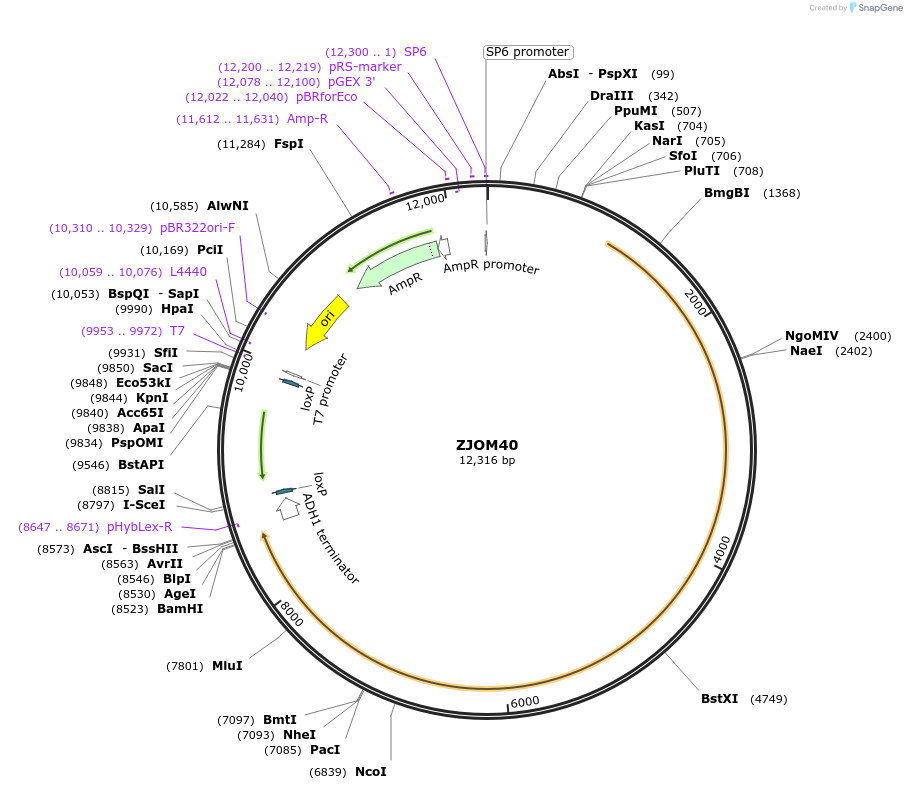
ZJOM40
Plasmid#133650PurposeIntegration of SEC7-DuDre as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast late golgi/early endosome.DepositorAvailable SinceApril 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
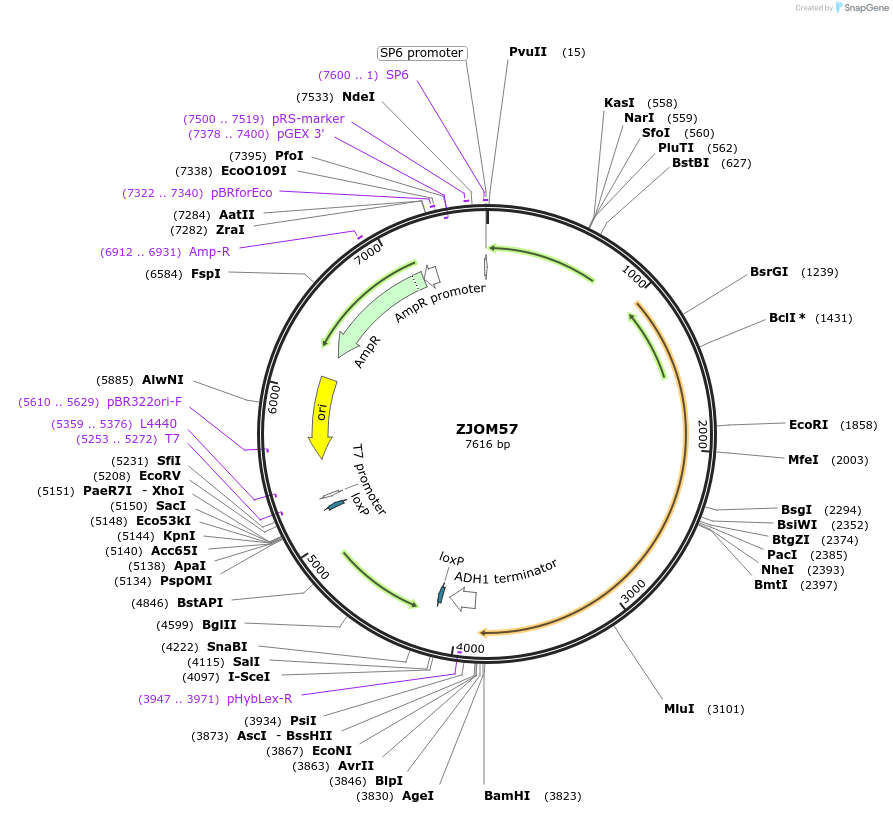
ZJOM57
Plasmid#133656PurposeIntegration of PEX3-DuDre as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast peroxisome.DepositorAvailable SinceNov. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
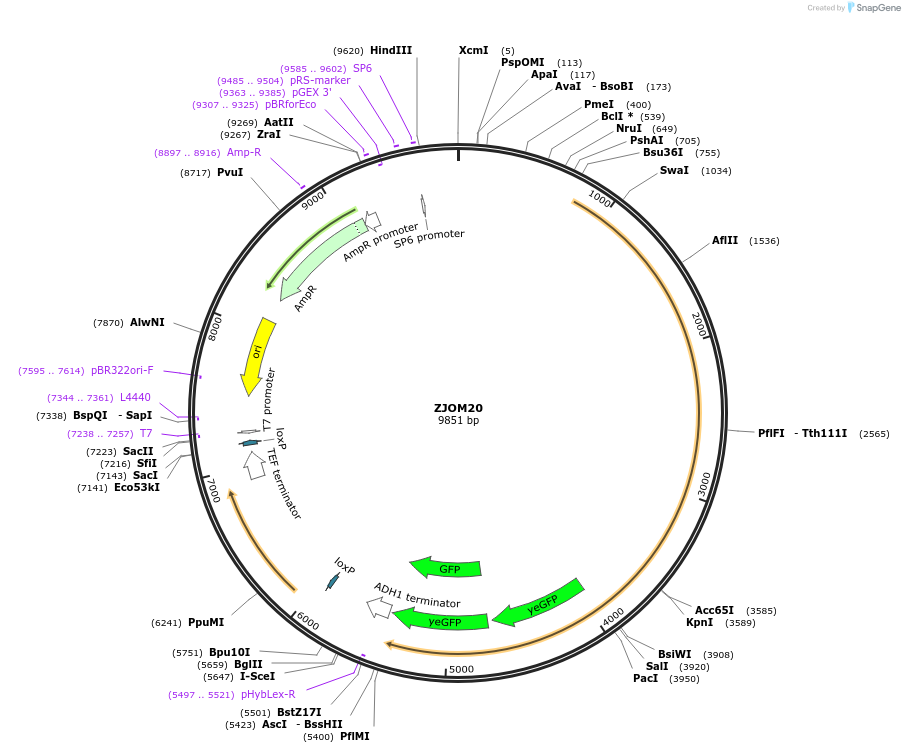
ZJOM20
Plasmid#133644PurposeIntegration of PEX1-2GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Marker for yeast peroxisome.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
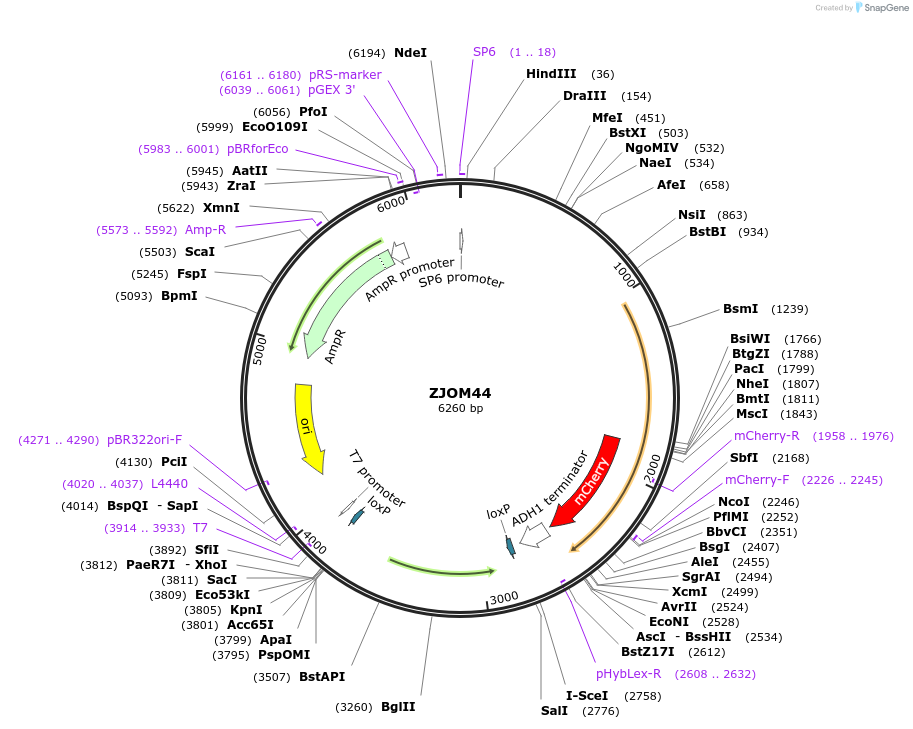
ZJOM44
Plasmid#133653PurposeIntegration of SNF7-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast late endosome.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
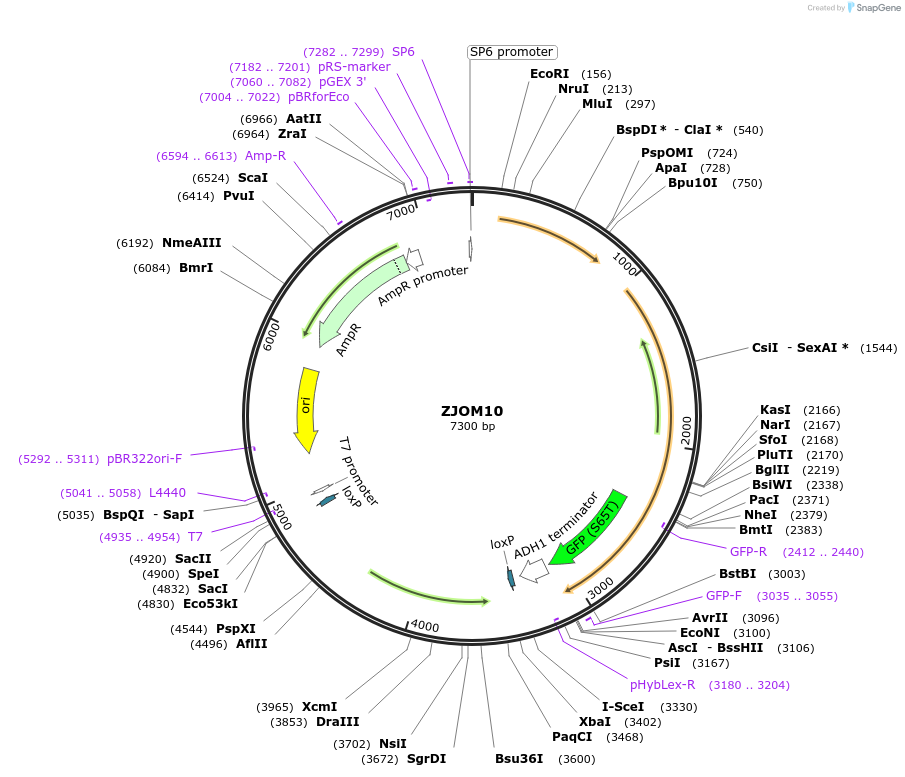
ZJOM10
Plasmid#133639PurposeIntegration of VPS4-GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Marker for yeast late endosome.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
ZJOM5
Plasmid#133635PurposeIntegration of CHS5-GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Marker for yeast late golgi/early endosome.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
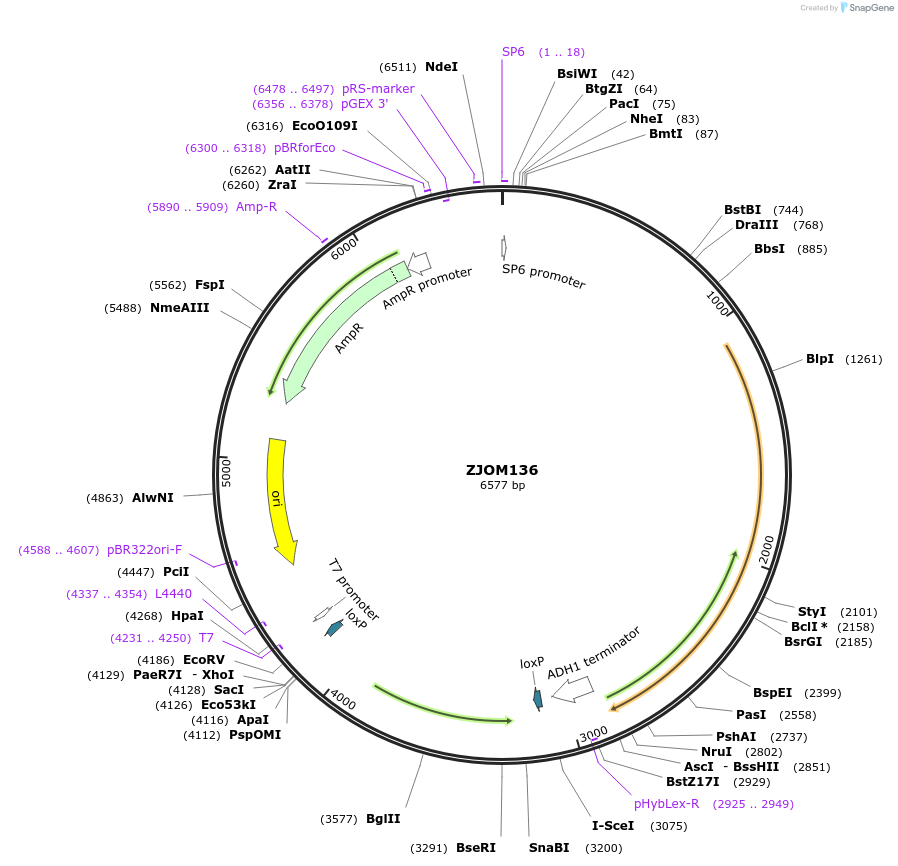
ZJOM136
Plasmid#133660PurposeIntegration of ELO3-mTagBFP2 as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast endoplasmic reticulum (ER).DepositorAvailable SinceDec. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
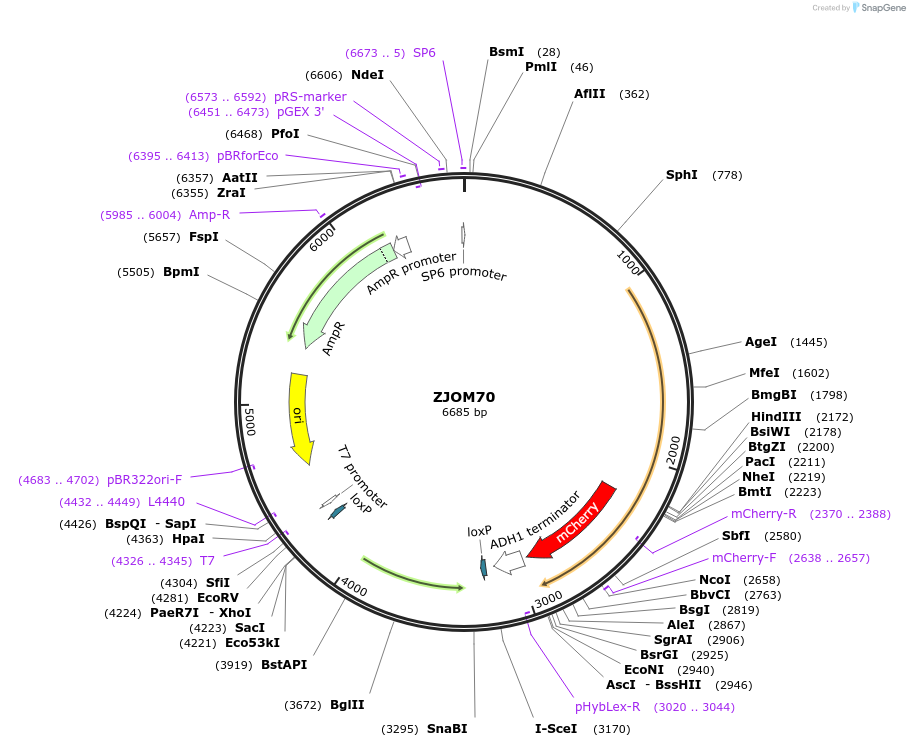
ZJOM70
Plasmid#133658PurposeIntegration of ERG6-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast lipid droplet.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
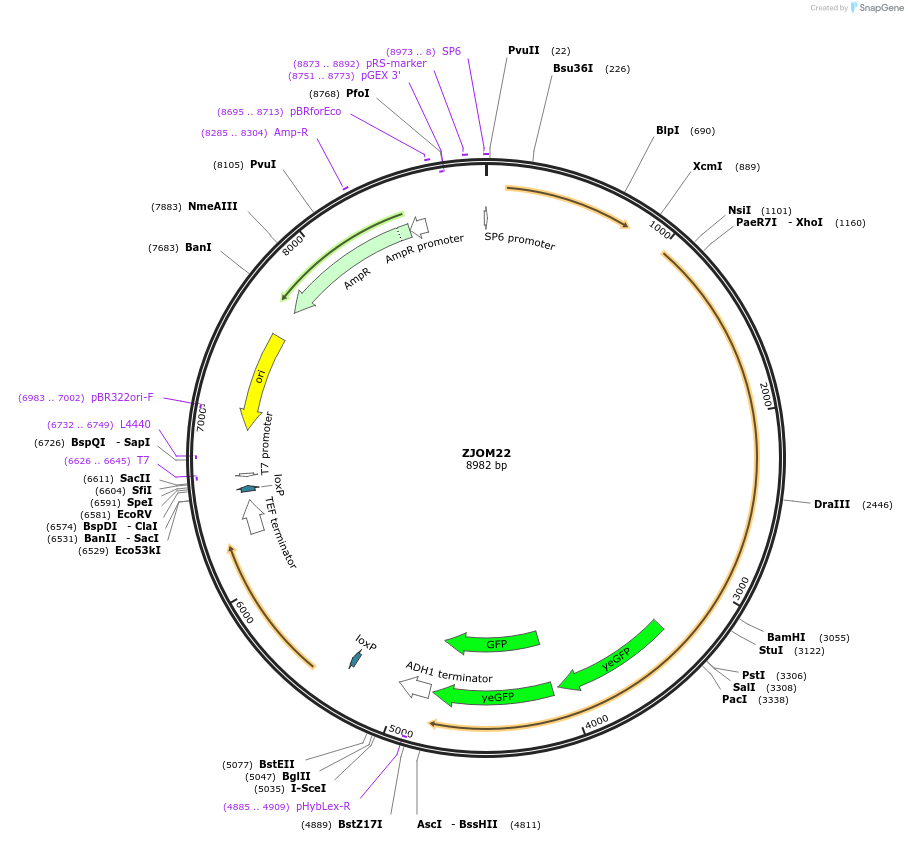
ZJOM22
Plasmid#133629PurposeIntegration of EMC1-2GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Marker for yeast endoplasmic reticulum (ER).DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
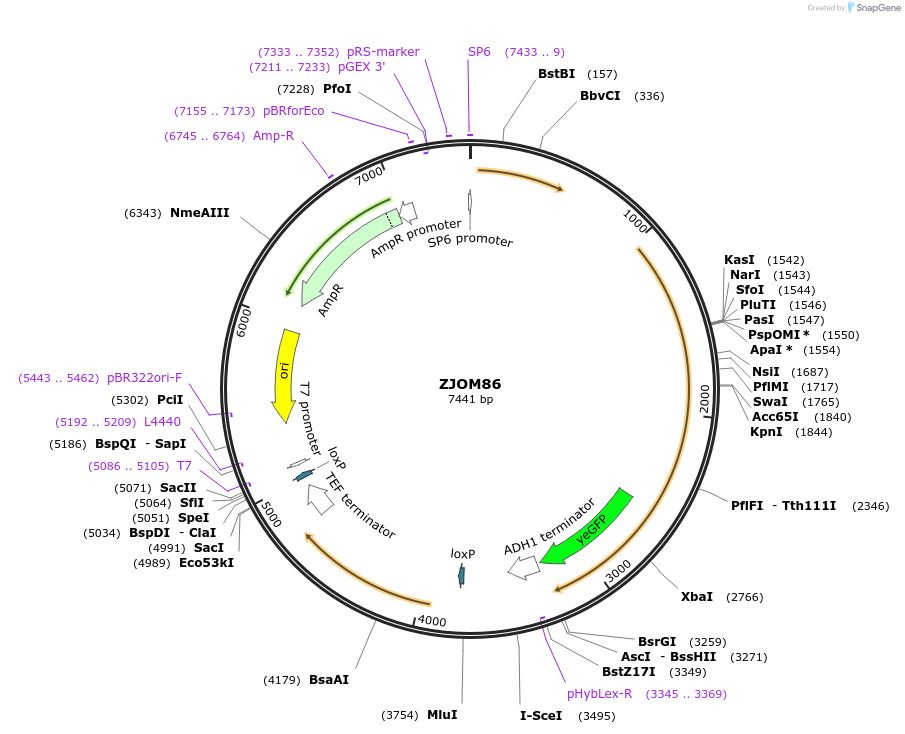
ZJOM86
Plasmid#133634PurposeIntegration of ANP1-GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Marker for yeast early golgi.DepositorAvailable SinceDec. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
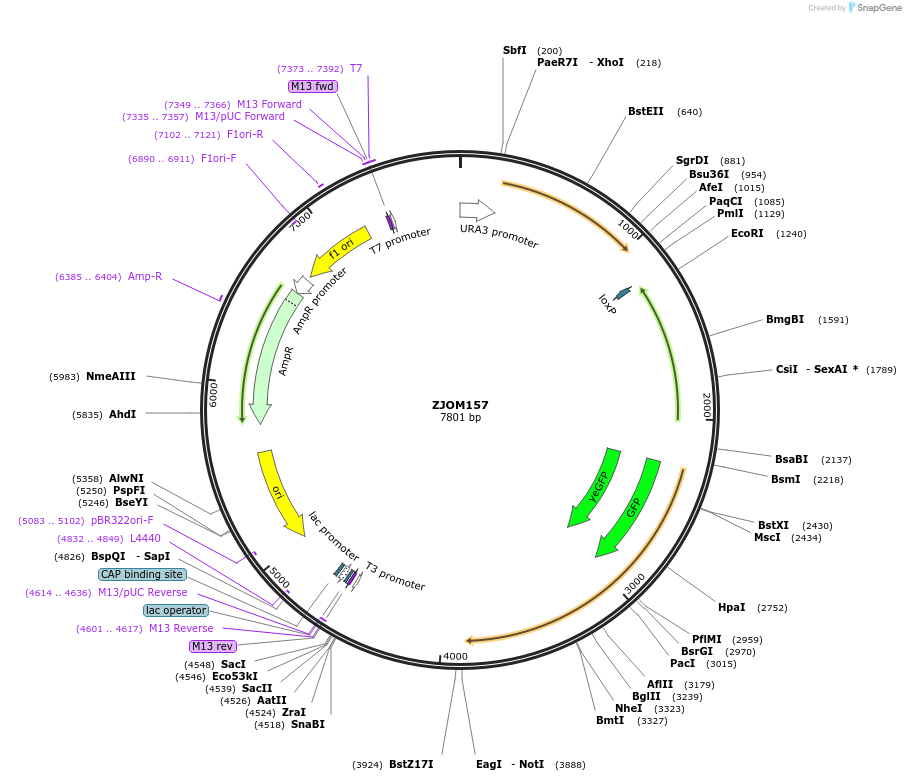
ZJOM157
Plasmid#133640PurposeIntegration of GFP-PEP12 as an additional copy in genome, use auxotrophic marker URA3(Kluyveromyces lactis).Marker for yeast late endosome.DepositorAvailable SinceFeb. 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
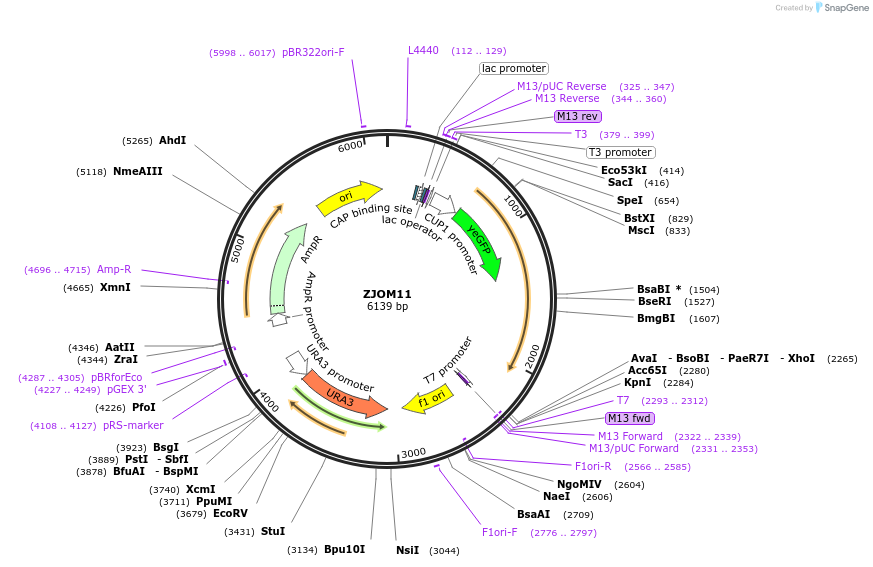
ZJOM11
Plasmid#133637PurposeIntegration of GFP-TLG1 as an additional copy in genome, use auxotrophic marker URA3(Saccharomyces cerevisiae). Marker for yeast late golgi/early endosome.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
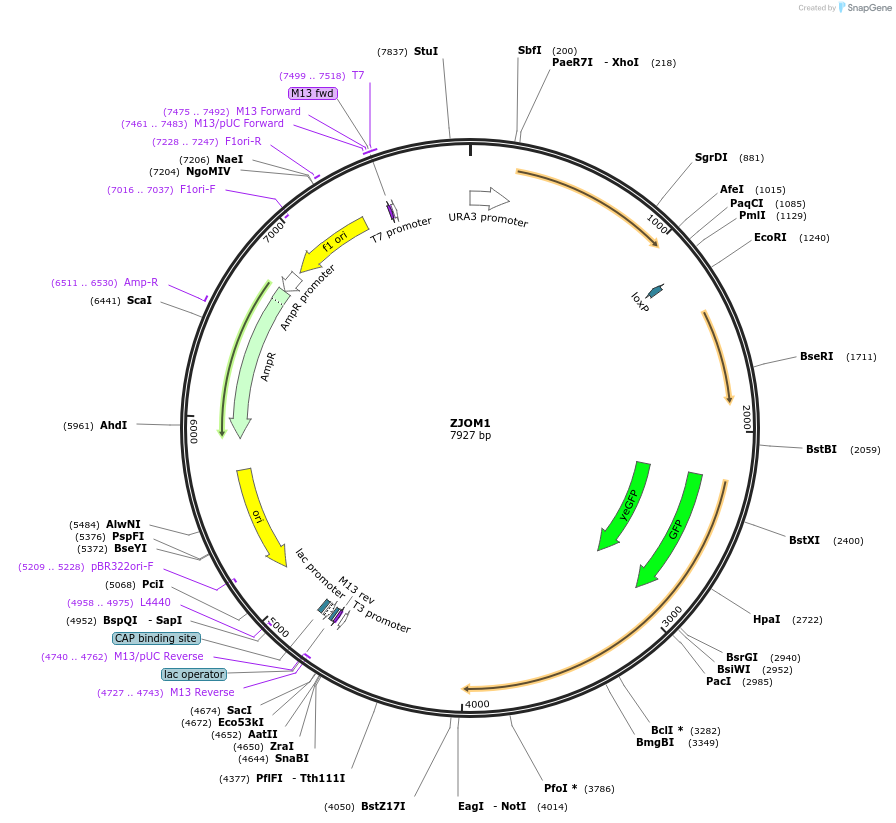
ZJOM1
Plasmid#133632PurposeIntegration of GFP-SED5 as an additional copy in genome, use auxotrophic marker URA3(Kluyveromyces lactis). Marker for yeast early golgi.DepositorAvailable SinceApril 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
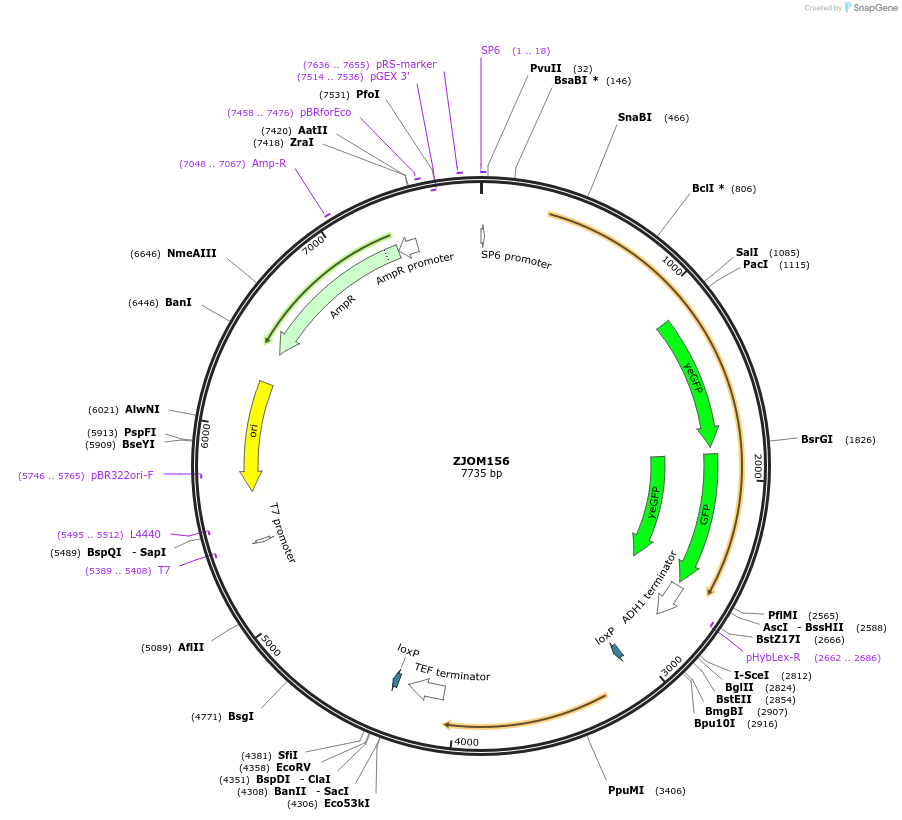
ZJOM156
Plasmid#133636PurposeIntegration of 2xGFP tag at SEC7 C terminus, use auxotrophic marker URA3(Candida albicans). Marker for yeast late golgi/early endosome.DepositorAvailable SinceFeb. 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
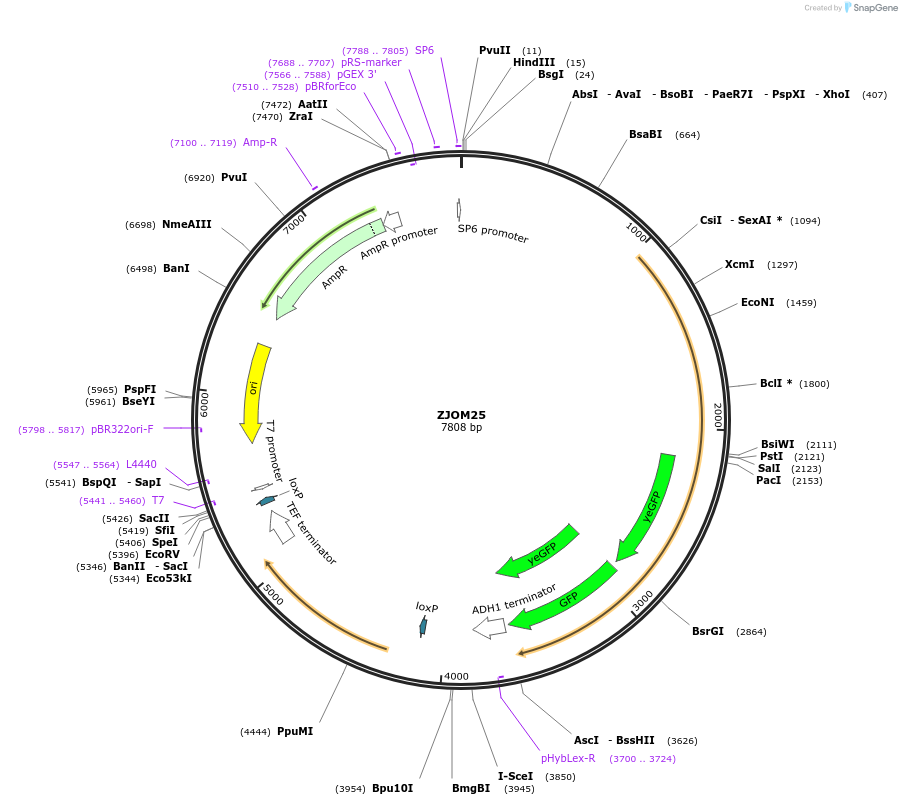
ZJOM25
Plasmid#133672PurposeIntegration of MSP1-2GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Present on mitochondria and peroxisomes.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only