Adenoviral Vector Guide
Adenoviruses (AdV) are medium-sized, non-enveloped viruses containing a linear double-stranded DNA (dsDNA) genome. Non-enveloped viruses do not have a lipid bilayer surrounding the viral particle and are only composed of a protein capsid and the viral genetic material inside.
Adenoviral genomes range between 26–45 kb in length and contain multiple heavily-spliced transcripts flanked by two inverted terminal repeats (ITRs). Adenoviral genes are divided into early (E1A, E1B, E2, E3, E4) and late (L1, L2, L3, L4, L5) regions. Early transcripts are expressed shortly after the virus enters the host cell, and encode proteins necessary for viral DNA replication and host cell manipulation. Late transcripts are expressed after DNA replication starts and primarily encode for structural proteins needed to assemble icosahedral capsids and build new virions.

Adenoviruses are part of the Adenoviridae family and can infect dividing and non-dividing cells. AdVs can cause a range of mild diseases in humans and other animals, most commonly respiratory infections, such as the common cold. The immune system recognizes adenoviral elements rapidly and can mount a strong immune response quickly, which has limited their use in therapeutic applications. However, this has also made them the most widely used viral vector platform for vaccine design against a diversity of viruses.
This guide contains many viral vector-specific terms and acronyms, so if you're new to viral vectors or simply need a refresher, we've included a glossary at the end!
Adenovirus versus Adeno-Associated Virus
Adenoviruses fall under the Adenoviridae family, are medium-sized, and contain a double-stranded DNA genome. Adeno-associated viruses (AAV), however, are small single-stranded DNA viruses that belong to the Parvoviridae family. AAVs were discovered as a contaminant of adenovirus preparations, which is how they got their name, but they are not related. AAVs require the presence of adenoviral genes E1, E4, E2a and VA for replication.
For more information about AAVs, read our AAV guide.
Recombinant Adenoviral Vectors
Wild-type adenoviruses have been modified by researchers to create recombinant adenoviral (rAdV) vectors in order to deliver genetic cargo into cells. Most rAdV vectors are based on adenovirus type 5 (Ad5) and have been produced by replacing the early and late adenoviral transcript regions between the two ITRs with the transgenes of interest. The size of the genetic cargo that can be included between the ITRs is limited by the physical space available inside of the adenoviral capsid and directly correlates to the size of the DNA removed from the viral genome. Over time, subsequent generations of rAdV vectors have been developed where more adenoviral genes have been removed. These genes are now supplied in trans, via different plasmids or the packaging cell line, which not only frees up more space for genetic cargo but also improves safety.
In order to produce rAdV vectors, you generally need two plasmids:
- Shuttle plasmid (also known as transfer plasmid) — containing the transgene of interest, the ITRs, and homology regions for recombination with the adenoviral plasmid.
- Adenoviral plasmid — containing the rest of the adenoviral genome necessary for viral production and homology regions for recombination with the shuttle plasmid.
For a summary of all adenoviral plasmid components, see the Adenoviral Plasmid Elements table.
Adenoviral Generations
First-generation rAdV vectors
The first generation of rAdV vectors was developed by removing the E1 region, containing the E1A and E1B transcripts, and the E3 region from the viral genome. This allowed for the inclusion of ~6.5 kb of genetic cargo in first-generation rAdV vectors.
E3 is involved in evading host immunity and is not essential for virus production, whereas the E1 region is essential for viral replication and therefore needs to be supplied through other means. In order to produce rAdV vectors, E1 needs to be expressed by the packaging cell line, normally the human embryonic kidney 293 (HEK293) cell line. There is a rare possibility that recombination between the transfected adenoviral transfer plasmid and the E1 region integrated in the packaging cell’s genome could lead to the production of replication-competent adenoviruses (RCAs) that would be able to infect cells and replicate autonomously. First-generation rAdV vectors remain the most popular choice due to their well-established methods, relative simplicity of production, and higher titers. However, the risk of RCA formation is higher for first-generation rAdV vectors than later generations, which incorporate additional genomic deletions.

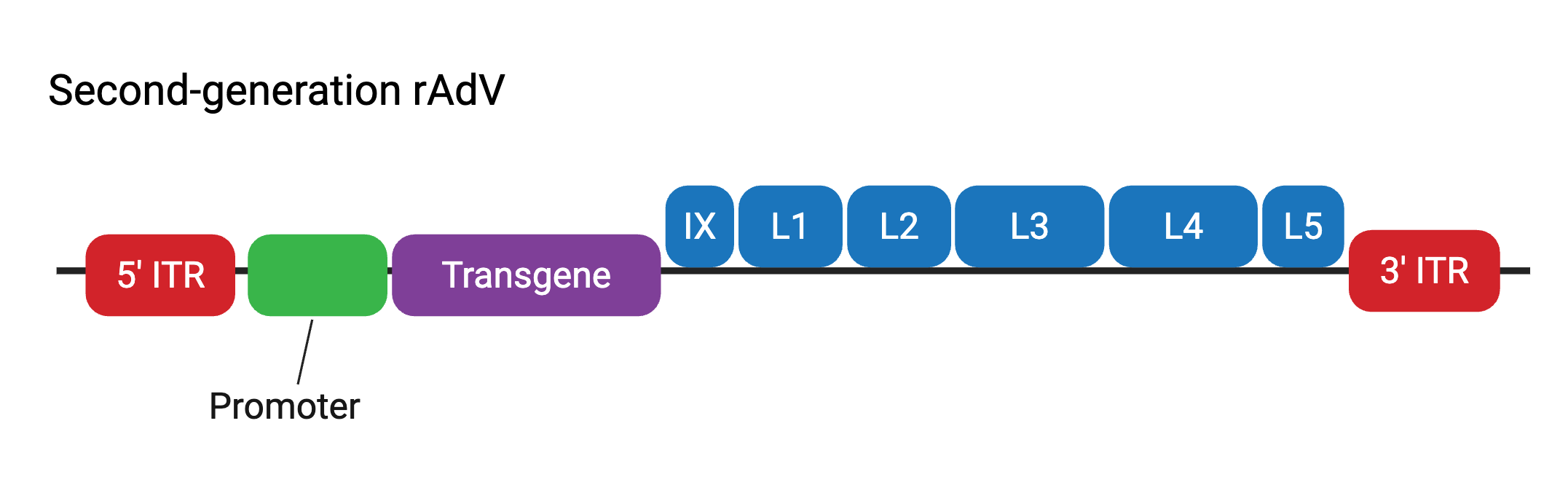
Second-generation rAdV vectors
Second-generation rAdV vectors contain the genomic deletions of first-generation rAdV vectors, as well as the deletion of the other early gene regions, E2 and E4. These modifications aimed to lower immunogenicity issues observed with first-generation rAdV vectors and further increase the transgene packaging capacity up to ~10.5 kb. The deleted E2 and E4 regions are also provided in trans — that is, by engineered packaging HEK293 cell lines. Second-generation rAdV vectors were shown to elicit less immune response when administered in vivo, however deletion of early genes also resulted in a reduction of viral vector amplification and overall lower viral titers. Because of this, along with weaker long-term gene expression patterns that were observed, researchers have tended to favor using first- and third-generation rAdV vectors.
Third-generation rAdV vectors
Third-generation rAdV vectors are also referred to as "gutless" or high capacity AdV (HCAdV) vectors. Here, all viral sequences are deleted, except for the ITRs and the packaging signal, which is located adjacent to the left ITR, resulting in the possibility of accommodating ~36 kb of genetic cargo. Production of HCAdV vectors requires using an additional adenoviral helper virus (HV) to provide the deleted adenoviral genes. The biggest challenge of the HCAdV vector production is to ensure no carryover from the HV in final viral preparations. Several methods to avoid HV contamination have been developed, such as using the Cre-lox and Flp-FRT recombination systems to excise the packaging signal from the HV genome or using other plasmids instead of HVs.

Adenoviral Vector Production
Cloning
Cloning your transgene, gRNA, or shRNA of interest into the transfer plasmid can be done with most standard cloning methods, including restriction enzyme, Gibson Assembly, or Gateway. Some transfer plasmids may have limited restriction sites or may only be compatible with certain cloning methods (such as a Gateway destination vector), so be sure to confirm your chosen plasmid is compatible with cloning methods available in your lab.
When cloning your plasmids, be sure to use recombination deficient bacterial strains, such as NEB Stable cells. These strains reduce the frequency of homologous recombination of unstable regions, like the ITRs found in transfer plasmids. This will ensure that the repeats will be maintained and often results in a greater yield of DNA. However, if the plasmid contains a Gateway cassette containing the ccdB gene, a ccdB-resistant strain is necessary.
For more information on cloning and working with plasmids, visit Addgene’s Molecular Biology Reference.
AdEasy™
AdEasy™, developed by Bert Vogelstein’s lab, is the most popular method for creating first-generation rAdV vectors, which is still the most commonly used generation for research purposes. The system consists of two plasmids that eventually recombine to form one:
- Shuttle/transfer plasmid (e.g. pAdTrack) — containing the transgene of interest and both ITRs.
- Adenoviral backbone plasmid (e.g. pAdEasy) — containing the adenoviral genes necessary for viral production.
In AdEasy™, the transgene of interest is cloned into the shuttle plasmid, verified, and linearized with the restriction enzyme PmeI. This construct is then transformed into AdEasier-1 cells (Addgene #16399), which are BJ5183 E. coli cells containing pAdEasy. pAdEasy is a ~33 kb adenoviral backbone plasmid containing the adenoviral genes necessary for virus production. The shuttle plasmid and the adenoviral backbone plasmid have matching left and right homology arms which facilitate homologous recombination of the transgene into the adenoviral backbone plasmid. One can also co-transform standard BJ5183 with supercoiled pAdEasy and the shuttle plasmid, but this method results in a higher background of non-recombinant adenoviral plasmids.
Recombinant adenoviral plasmids are then verified for size and proper restriction digest patterns to determine that the transgene has been inserted into the adenoviral backbone plasmid, and that other patterns of recombination have not occurred. Once verified, the recombinant plasmid is linearized with PacI to create a linear dsDNA construct flanked by ITRs. Cells from the packaging HEK293 or HER911 cell lines expressing E1 are transfected with the linearized construct, and rAdV vectors can be harvested about 7–10 days later.
Vogelstein designed multiple shuttle plasmids for different purposes. The pAdTrack series contains an IRES-GFP construct that enables co-expression of GFP with the transgene of interest. With these plasmids, one can track the infection of HEK293/HER911 cells throughout viral vector production. During experiments, GFP can be used to sort cells infected with rAdV vectors, or to verify that the infection rates are equivalent across multiple viruses.
The adenoviral backbone plasmid pAdEasy-1 (Addgene #16400) is suitable for most purposes. For especially long transgenes, the use of pAdEasy-2 (Addgene #16401) can increase the capacity of the rAdV vector. pAdEasy-2 does not contain the viral gene E4, adding 2.7 kb of packaging space. However, these constructs must be transfected into HER911 E4+ cells for rAdV vector production, as HEK293 cells do not contain E4.
Download the Bert Vogelstein’s lab practical guide for using the AdEasy™ system.
Find plasmids for this system on our Adenovirus Plasmid Collection page.

AdMax™
AdMax™ is another well used rAdV vector production platform, which like AdEasy™, relies on the homologous recombination of the shuttle plasmid containing the transgene of interest, and the adenoviral plasmid containing the adenoviral genes necessary for packaging. In the AdMax™ system, recombination between both plasmids occurs directly in the HEK293 packaging cell line, engineered for maximum adenoviral production, and is driven by use of the Cre recombinase.
Genome Integration
Upon infection, wild-type adenovirus as well as rAdV vectors do not integrate into the host genome. Instead, they remain as extrachromosomal elements within the nucleus, normally as double-stranded circular episomes. Due to this, and to the strong host’s immune response against the transduced cells, transgene expression delivered through rAdV vectors is normally transient and short-lived. Second- and third-generation rAdV vectors have been developed to reduce immune response and achieve longer transgene expression.
Adenoviral Types
Serotypes
More than 60 serotypes have been identified within human adenoviruses, which are classified into seven species (A to G). Different serotypes exhibit varying tissue tropisms, meaning they preferentially infect certain tissues, which correlates with the specific diseases they cause. Ad5 and Ad2 are the most commonly used for rAdV vector construction, due to their high gene transfer efficiency. Other serotypes like Ad26 and Ad35 are also of interest for their low seroprevalence and greater ability to avoid vector-neutralizing pre-existing immunity.
Pseudotypes
Pseudotyping is the process of producing viral vectors in combination with foreign capsid proteins. Researchers have further refined the tropism of rAdV vectors by mixing capsids and genomes from different viral serotypes. Pseudotypes are generally denoted using two numbers separated by a slash (e.g. Ad5/35), where the first number indicates the serotype of the genome and the second number the serotype of the capsid. Use of these pseudotyped vectors can modulate viral vector features such as transduction efficiency, tropism and immunogenicity. For example, Ad5/35 and Ad5/26 combine the high transduction efficiency of the Ad5 serotype with the low immunogenicity of the Ad35 and Ad26 serotypes.
Common Uses of Adenoviral Vectors
Although recombinant adeno-associated viral (rAAV) vectors are often the preferred choice among researchers due to their reduced immunogenicity and longer transgene expression, rAdV vectors are used in many research fields, especially when needing transgenes to express for a shorter period of time or when needing to package larger cargo.
CRISPR-Cas9 Delivery
As compared to other viral vectors, rAdV vectors can provide a large transgene packaging capacity. Large protein-coding genes and complex gene expression cassettes are often too big to be packaged in rAAV vectors. This is the case of some commonly used Cas9 homologs, especially when having to express them alongside a guide RNA and other regulatory elements. The packaging capacity of rAdV vectors (~6.5 kb for first-generation rAdV vectors) surpasses that of rAAV vectors (~4.7 kb) and is further increased in second and third-generation vectors. This capacity is large enough to carry the Cas9 gene and a gRNA expression cassette in one single viral particle.
Vaccine Development
rAdV vectors are highly immunogenic because they activate both innate and adaptive immune responses in the host. More than 80% of the human population has been exposed to at least one human AdV serotype and has developed a serotype-specific immune response. Preexisting immunity lowers viral vector uptake and transgene expression. This strong immune response has become a disadvantage for the use of rAdV vectors in gene therapy but has, however, proven to be very useful for designing and producing rAdV-based vaccines. Adenoviral vectors are excellent vectors for delivering target antigens to mammalian hosts and have been used as vaccines against infectious diseases such as COVID-19, Ebola virus, and Zika virus. Chimpanzee adenoviral (ChAdV) vectors have also been developed as vaccine candidates for multiple infectious diseases and prostate cancer.
Cancer Treatment
Recombinant adenoviral vectors are also being used in cancer therapy to stimulate anti-tumor immune response in different ways, such as expressing cytokines and other immune-modulatory molecules. Another strategy has been to use oncolytic rAdV vectors, which are replication-competent adenoviral vectors that can selectively infect, divide in, and lyse tumor cells.
Viral Vector Safety
Recombinant adenoviral vectors, although replication incompetent, are human pathogens and have to be handled at biosafety level two (BSL-2). They are highly immunogenic and can trigger a strong immune response to researchers if exposed. There is a risk of spontaneous reversion to RCAs, although this has been reduced by providing the E1 gene in trans within the packaging cell line.
Recombinant adenoviral vectors typically do not integrate in the host genome and instead are maintained in the cell as episomes. This highly reduces the potential for insertional mutagenesis and the risk of oncogenesis. However, random integration events can occur at a very low frequency.
As with most experiments, infection risks occur with contact to mucous membranes or broken skin. The risk of exposure and reaction severity is increased with rAdV vectors as they are normally produced in high titers for their use in experiments. Needle sticks and ripped gloves are common points of entry. Biosafety should always be considered with respect to the precise nature of experiments being performed. Your biosafety office can provide more information on your institution's best practices with regard to AdV research.
See Addgene’s Biosafety Resource Guide for more information and resources on viral safety.
Resources and References
Resources on addgene.org
Resources on Addgene's Blog
References
Ahi, Y. S., Bangari, D. S., & Mittal, S. K. (2011). Adenoviral vector immunity: its implications and circumvention strategies. Current Gene Therapy, 11(4), 307–320. https://doi.org/10.2174/156652311796150372 (Link opens in a new window) PMID: 21453277 (Link opens in a new window)
Bulcha, J. T., Wang, Y., Ma, H., Tai, P. W. L., & Gao, G. (2021). Viral vector platforms within the gene therapy landscape. Signal Transduction and Targeted Therapy, 6(1), 53. https://doi.org/10.1038/s41392-021-00487-6 (Link opens in a new window) PMID: 33558455 (Link opens in a new window)
Bullard, B. L., Corder, B. N., Gordon, D. N., Pierson, T. C., & Weaver, E. A. (2020). Characterization of a Species E Adenovirus Vector as a Zika virus vaccine. Scientific Reports, 10(1), 3613. https://doi.org/10.1038/s41598-020-60238-5 (Link opens in a new window) PMID: 32107394 (Link opens in a new window)
Ewer, K. J., Sebastian, S., Spencer, A. J., Gilbert, S. C., Hill, A. V. S., & Lambe, T. (2017). Chimpanzee adenoviral vectors as vaccines for outbreak pathogens. Human Vaccines & Immunotherapeutics, 13(12), 3020–3032. https://doi.org/10.1080/21645515.2017.1383575 (Link opens in a new window) PMID: 29083948 (Link opens in a new window)
Geisbert, T. W., Bailey, M., Hensley, L., Asiedu, C., Geisbert, J., Stanley, D., Honko, A., Johnson, J., Mulangu, S., Pau, M. G., Custers, J., Vellinga, J., Hendriks, J., Jahrling, P., Roederer, M., Goudsmit, J., Koup, R., & Sullivan, N. J. (2011). Recombinant adenovirus serotype 26 (Ad26) and Ad35 vaccine vectors bypass immunity to Ad5 and protect nonhuman primates against ebolavirus challenge. Journal of Virology, 85(9), 4222–4233. https://doi.org/10.1128/JVI.02407-10 (Link opens in a new window) PMID: 21325402 (Link opens in a new window)
Gilbert, S. C. (2015). Adenovirus-vectored Ebola vaccines. Expert Review of Vaccines, 14(10), 1347–1357. https://doi.org/10.1586/14760584.2015.1077122 (Link opens in a new window) PMID: 26289977 (Link opens in a new window)
He, T. C., Zhou, S., da Costa, L. T., Yu, J., Kinzler, K. W., & Vogelstein, B. (1998). A simplified system for generating recombinant adenoviruses. Proceedings of the National Academy of Sciences of the United States of America, 95(5), 2509–2514. https://doi.org/10.1073/pnas.95.5.2509 (Link opens in a new window) PMID: 9482916 (Link opens in a new window)
Lee, C. S., Bishop, E. S., Zhang, R., Yu, X., Farina, E. M., Yan, S., Zhao, C., Zheng, Z., Shu, Y., Wu, X., Lei, J., Li, Y., Zhang, W., Yang, C., Wu, K., Wu, Y., Ho, S., Athiviraham, A., Lee, M. J., Wolf, J. M., Reid, R. R., & He, T. C. (2017). Adenovirus-mediated gene delivery: potential applications for gene and cell-based therapies in the new era of personalized medicine. Genes & Diseases, 4(2), 43–63. https://doi.org/10.1016/j.gendis.2017.04.001 (Link opens in a new window) PMID: 28944281 (Link opens in a new window)
Lee, D., Liu, J., Junn, H. J., Lee, E. J., Jeong, K. S., & Seol, D. W. (2019). No more helper adenovirus: production of gutless adenovirus (GLAd) free of adenovirus and replication-competent adenovirus (RCA) contaminants. Experimental & Molecular Medicine, 51(10), 1–18. https://doi.org/10.1038/s12276-019-0334-z (Link opens in a new window) PMID: 31659156 (Link opens in a new window)
Luo, J., Deng, Z. L., Luo, X., Tang, N., Song, W. X., Chen, J., Sharff, K. A., Luu, H. H., Haydon, R. C., Kinzler, K. W., Vogelstein, B., & He, T. C. (2007). A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nature Protocols, 2(5), 1236–1247. https://doi.org/10.1038/nprot.2007.135 (Link opens in a new window) PMID: 17546019 (Link opens in a new window)
Matsunaga, W., & Gotoh, A. (2023). Adenovirus as a vector and oncolytic virus. Current Issues in Molecular Biology, 45(6), 4826–4840. https://doi.org/10.3390/cimb45060307 (Link opens in a new window) PMID: 37367056 (Link opens in a new window)
Mendonça, S. A., Lorincz, R., Boucher, P., & Curiel, D. T. (2021). Adenoviral vector vaccine platforms in the SARS-CoV-2 pandemic. NPJ Vaccines, 6(1), 97. https://doi.org/10.1038/s41541-021-00356-x (Link opens in a new window) PMID: 34354082 (Link opens in a new window)
Rosewell, A., Vetrini, F., & Ng, P. (2011). Helper-dependent adenoviral vectors. Journal of Genetic Syndromes & Gene Therapy, Suppl 5, 001. https://doi.org/10.4172/2157-7412.s5-001 (Link opens in a new window) PMID: 24533227 (Link opens in a new window)
Watanabe, M., Nishikawaji, Y., Kawakami, H., & Kosai, K. I. (2021). Adenovirus biology, recombinant adenovirus, and adenovirus usage in gene therapy. Viruses, 13(12), 2502. https://doi.org/10.3390/v13122502 (Link opens in a new window) PMID: 34960772 (Link opens in a new window)
Adenoviral Plasmid Elements
| Plasmid Type | Element | Delivery relative to transgene | Purpose |
|---|---|---|---|
| Transfer/Shuttle plasmid | 5' ITR (LITR) | in cis | Left Inverted Terminal Repeat, serve as self-priming structures that promote primase-independent DNA replication. |
| 3' ITR (RITR) | in cis | Right Inverted Terminal Repeat, serve as self-priming structures that promote primase-independent DNA replication. | |
| Homology arms (left and right) | in cis | Present also in adenoviral plasmid. Used to combine transgene and ITRs contained in the shuttle plasmid with the adenoviral genes contained in the adenoviral plasmid through homologous recombination. | |
| Psi (Ψ) | in cis | Packaging or encapsidation signal located at the left arm of the genome adjacent to the left ITR that is required for viral genome packaging. In helper viruses, this signal is flanked by loxP sites so they can be excised. | |
| Adenoviral plasmid | E2A, E2B, E3, E4 | in trans | Adenoviral early transcript regions involved in viral DNA replication and host cell manipulation. |
| L1, L2, L3, L4, L5 | in trans | Adenoviral late transcript regions that encode for structural capsid proteins. |
Glossary
| Term | Definition |
|---|---|
| Adaptive immunity | Specific and long-lasting immune response involving specialized immune cells and antibody production that develops after exposure to an antigen. |
| AdEasier-1 cells | BJ5183 E. coli cells containing the pAdEasy-1 packaging plasmid. |
| Adenovirus (AdV) | Wildtype, medium-sized, nonenveloped viruses from the Adenoviridae family. |
| Episome | Genetic element inside the cell that remains as a part of the eukaryotic genome without integration and can replicate independently. |
| Genetic cargo | DNA introduced to cells within the viral vector capsid. |
| Icosahedral | Having the shape of a polyhedron with 20 faces, each being an equilateral triangle. |
| Immunogenicity | The ability of a molecule or substance to induce an immune response in the body. |
| in cis | In the context of viral vector production, in cis refers to genetic elements located in the same plasmid as the gene of interest. |
| in trans | In the context of viral vector production, in trans refers to genetic elements provided outside of the plasmid containing the gene of interest (i.e. other plasmids, packaging cell line). |
| Infection | The natural process of entry and multiplication of a pathogen, like viruses or bacteria, within a host organism, leading to disease. |
| Innate immunity | Non-specific immune response that responds to general signals of infection or damage. |
| Insertional mutagenesis | Random insertion of DNA elements into the genome, often altering gene activity. |
| Oncolytic | Referring to the destruction of cancer cells, generally by lysis. |
| pAdEasy-1 | Adenoviral backbone plasmid that lacks E1 and E3. |
| pAdEasy-2 | Adenoviral backbone plasmid that lacks E1, E3, and E4. Useful for larger inserts. |
| pAdTrack | Class of shuttle/transfer vectors for recombinant adenoviral vector production that contain IRES-GFP. |
| Pseudotyping | The production of viral vectors (or viruses) using viral envelope proteins from different species. Used to alter infectivity and tropism. |
| pShuttle | Class of shuttle/transfer vectors for recombinant adenoviral vector production that do not contain GFP. |
| Recombinant adenoviral vector (rAdV) | Modified form of AdV used to deliver genetic material into cells. |
| Replication-competent adenovirus (RCA) | Mutated adenoviral vector that has the ability to replicate and reproduce within host cells as a result of a spontaneous crossover event between the adenoviral vector and the adenoviral genes provided in trans. |
| Seroprevalence | Proportion of individuals within a population that present antibodies in their blood specific to a given virus or infectious agent at a specific time point. |
| Transduction | The process of artificially introducing foreign DNA into eukaryotic cells using viral methods. |
| Transfection | The process of artificially introducing foreign DNA into eukaryotic cells using non-viral methods. |
| Transformation | The process where bacteria take up DNA from their environment. |
| Tropism | The specific types of cells and tissues that a virus can infect and replicate in. |
Credits
- Contributing Authors
- Written and reviewed by the Scientific Curation team at Addgene.
- Media Credits
- Figures created with BioRender (Link opens in a new window)
- Last Updated
- Content last reviewed on 9 June 2025.


