LbPlant Toolbox Kits
(Kit #
1000000262, 1000000263
)
Depositing Lab: Thorsten Mascher, Georg Fritz
The LbPlant genetic toolbox is a Golden Gate assembly-based collection tailored for Lactiplantibacillus plantarum. It enables the rapid construction of expression vectors in Escherichia coli, followed by transformation into L. plantarum. The collection consists of standardized genetic modules designed for hierarchical plasmid assembly based on iteration of using type IIS restriction enzymes (BsmBI and BsaI).
Kit #1000000262 (LbPlant Toolbox I) made by Thorsten Mascher will be sent as bacterial glycerol stocks in 96-well plate format.
Kit #1000000263 (LbPlant Toolbox II) made by Georg Fritz will be sent as individual bacterial stabs.
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |||
|---|---|---|---|---|---|---|---|
| Kit | 1000000262 | LbPlant Toolbox I | 1 | $440 | Add to Cart | ||
| Kit | 1000000263 | LbPlant Toolbox II | 1 | $473 | Add to Cart | ||
Original Publication
Development of a Golden Gate assembly-based genetic toolbox for Lactiplantibacillus plantarum and its application for engineering monoterpenoid biosynthesis. Li X, Schönberg PY, Wucherpfennig T, Hinze C, Sulaj F, Henle T, Mascher T. ACS Synth Biol. 2024 Sep 20;13(9):2764-2779. doi: 10.1021/acssynbio.4c00075. PubMed (Link opens in a new window) Article (Link opens in a new window)
The Marburg Collection: A Golden Gate DNA assembly framework for synthetic biology applications in Vibrio natriegens. Stukenberg D, Hensel T, Hoff J, Daniel B, Inckemann R, Tedeschi JN, Nousch F, Fritz G. ACS Synth Biol. 2021 Aug 20;10(8):1904-1919. doi: 10.1021/acssynbio.1c00126. PubMed (Link opens in a new window) Article (Link opens in a new window)
Description
Derived from the Marburg collection (Stukenberg et al., 2021), this LbPlant toolbox was adapted for L. plantarum by introducing an additional Level 0 genetic module — the E. coli shuttle part — which enables rapid plasmid assembly and replication in E. coli prior to transformation into L. plantarum. Its modular design, based on the Golden Gate cloning technology, allows for the easy exchange of genetic parts and high-throughput assembly of numerous constructs in a standardized genetic context. This improves the efficiency and predictability of metabolic engineering in L. plantarum. Details about the cloning strategy and characteristics of genetic parts included in this toolbox can be found from our paper published in ACS Synthetic Biology (Li et al., 2024).
Note that when constructing Level 1 plasmids with the broad-host-range origins pWV01 or SH71rep (which are also functional in E. coli), inclusion of a second E. coli origin such as pMB01 should be avoided due to plasmid instability. For the high-throughput screening and characterization of natural promoters, a Level 0 dropout vector is provided in this collection with an sfGFP reporter cassette inserted as a placeholder for promoter + RBS elements. Such modular design can also be adapted by users for other genetic parts of interest such as CDS or terminator.
Although our work focused exclusively on L. plantarum, we find this toolbox to be applicable for other closely related lactic acid bacteria (LAB), including Latilactobacillus sakei and Lacticaseibacillus casei, given that several promoters and replication origins have been experimentally validated in these species.
For complete implementation of the toolbox, we recommend users to acquire both kits — toolbox I and the complementary toolbox II originally developed by Dr. Georg Fritz (Stukenberg et al., 2021), which has been made available on Addgene with their permission.
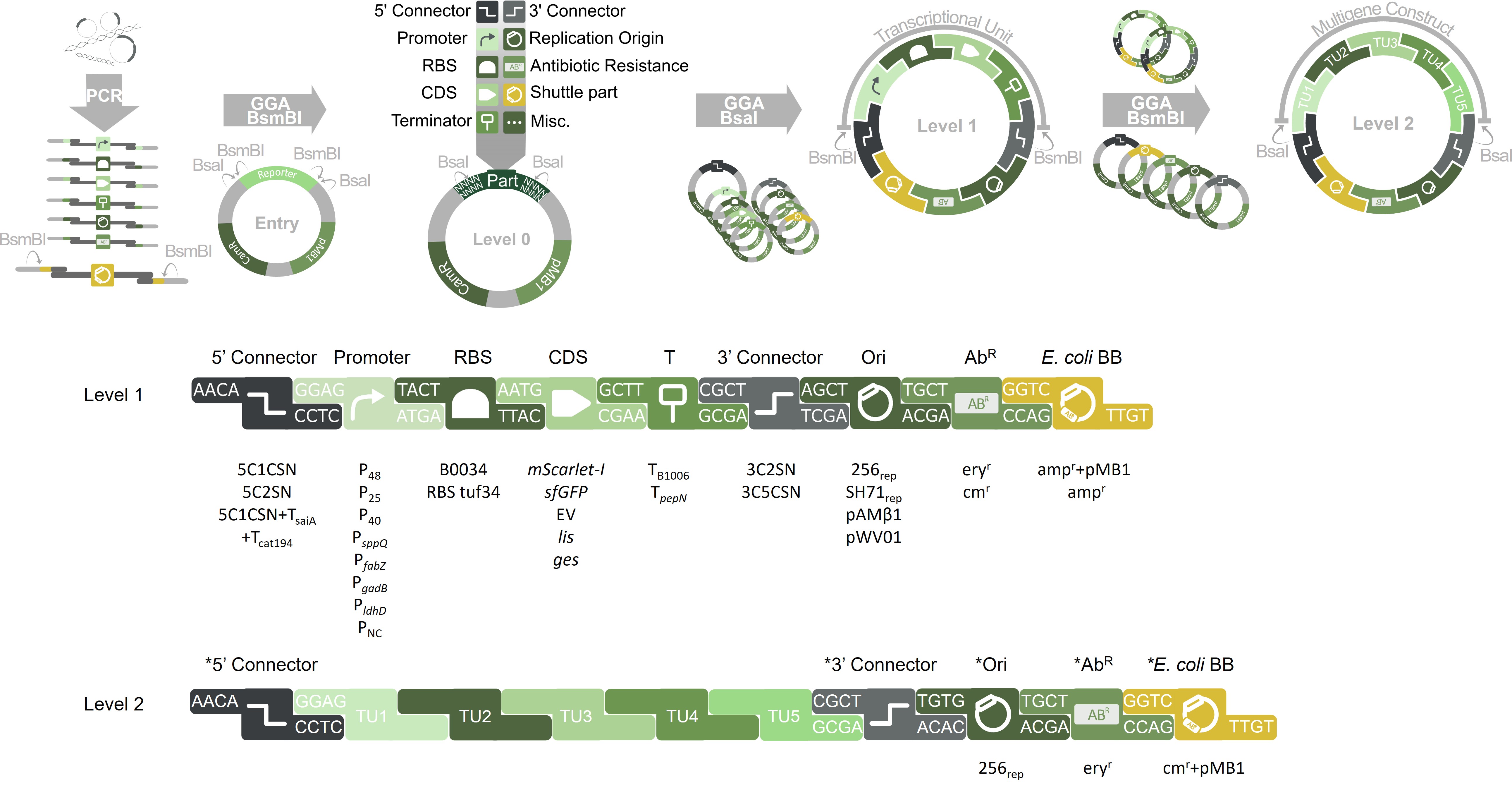
Figure 1: Schematic drawing of LbPlant toolbox for L. plantarum. This collection contains a set of standardized genetic modules that can be used for hierarchical assembly based on the Golden Gate Assembly technique. Level 0 parts from each standard class can be cut out of their vector with the type IIS restriction enzyme BsaI. The resulting fragments have predesigned overhangs and can be ligated into Level 1 plasmids. The obtained transcriptional units can be further directionally assembled into multigene constructs (Level 2) using BsmBI. RBS: ribosome binding site; CDS: coding sequence; T: terminator; Ori: L. plantarum origin of replication; AbR: L. plantarum antibiotic resistance cassette; E. coli BB: genetic parts for E. coli shuttling.
How to Cite this Kit
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which they were created, and include Addgene in the Materials and Methods of your future publications.
For your Materials and Methods section:
"The LbPlant Toolbox I Kit was a gift from Thorsten Mascher (Addgene kit #1000000262)."
"The LbPlant Toolbox II Kit was a gift from Georg Fritz (Addgene kit #1000000263)."
For your Reference section:
Development of a Golden Gate assembly-based genetic toolbox for Lactiplantibacillus plantarum and its application for engineering monoterpenoid biosynthesis. Li X, Schönberg PY, Wucherpfennig T, Hinze C, Sulaj F, Henle T, Mascher T. ACS Synth Biol. 2024 Sep 20;13(9):2764-2779. doi: 10.1021/acssynbio.4c00075. PubMed (Link opens in a new window) Article (Link opens in a new window)
The Marburg Collection: A Golden Gate DNA assembly framework for synthetic biology applications in Vibrio natriegens. Stukenberg D, Hensel T, Hoff J, Daniel B, Inckemann R, Tedeschi JN, Nousch F, Fritz G. ACS Synth Biol. 2021 Aug 20;10(8):1904-1919. doi: 10.1021/acssynbio.1c00126. PubMed (Link opens in a new window) Article (Link opens in a new window)
LbPlant toolbox I - #1000000262
- Resistance Color Key
Each circle corresponds to a specific antibiotic resistance in the kit plate map wells.
- Inventory
Searchable and sortable table of all plasmids in kit. The Well column lists the plasmid well location in its plate. The Plasmid column links to a plasmid's individual web page.
- Kit Plate Map
96-well plate map for plasmid layout. Hovering over a well reveals the plasmid name, while clicking on a well opens the plasmid page.
Resistance Color Key
| Chloramphenicol | |
| Ampicillin | |
| Erythromycin |
Inventory
| Well | Plasmid | Resistance |
|---|---|---|
| A / 1 | pLP3 |
|
| A / 2 | pLPC53 |
|
| A / 3 | pLPP1 |
|
| A / 4 | pLPP2 |
|
| A / 5 | pLPP3 |
|
| A / 6 | pLPP4 |
|
| A / 7 | pLPP5 |
|
| A / 8 | pLPP6 |
|
| A / 9 | pLPP7 |
|
| A / 10 | pLPP8 |
|
| A / 11 | pLPP9 |
|
| A / 12 | pLPS2 |
|
| B / 1 | pLP-EV |
|
| B / 2 | pLPT2 |
|
| B / 3 | pLPO1 |
|
| B / 4 | pLPO2 |
|
| B / 5 | pLPO3 |
|
| B / 7 | pLPR2 |
|
| B / 8 | pLPE3 |
|
| B / 9 | pLP4 |
|
| B / 10 | pLP5 |
|
| B / 11 | pLP6 |
|
| B / 12 | pLPE1 |
|
| C / 1 | pLPE2 |
|
| C / 2 | pLPC54 |
|
| C / 3 | pLPC33 |
|
| C / 4 | pLPO5 |
|
| C / 5 | pLPR1 |
|
| C / 6 | pLPR3 |
|


