ENABLE® Gene Editing in planta Toolkit
(Kit #
1000000270
)
Depositing Lab: Kate Creasey Krainer, Vladimir Nekrasov, Nicola Patron
The ENABLE® Gene Editing in planta toolkit provides a streamlined set of molecular tools for constructing CRISPR gene-editing vectors in just two simple cloning steps. Compatible with both monocot and dicot species, the kit supports transient and stable transformation approaches and is designed for use by researchers with any level of CRISPR experience.
This kit will be sent as individual bacterial stabs at room temperature.
Original Publication
Developing a Molecular Toolkit to ENABLE all to apply CRISPR/Cas9-based Gene Editing in planta. Abate BA, Hahn F, Chirivì D, Betti C, Fornara F, Molloy JC, Creasey Krainer KM. bioRxiv. 2025 November 13. doi: 10.1101/2025.11.09.687425. Article (Link opens in a new window)
Description
The Grow More Foundation (Link opens in a new window) is a nonprofit organization dedicated to ensuring equitable access to plant biotechnology in low- and middle-income countries. Through its ENABLE® program, the foundation works to turn advanced plant science into practical solutions for underutilized staple crops, empowering scientists in the Global South to address the agricultural challenges most relevant to their communities.
Our goal is to reduce technical and financial barriers to gene editing, allowing students, professors, researchers, and others to use genome-editing tools in local research projects. We see ENABLE® as a foundation for hands-on training, innovation, and creating new traits in neglected and underused crops — and as a step toward making crop research and development more accessible worldwide.
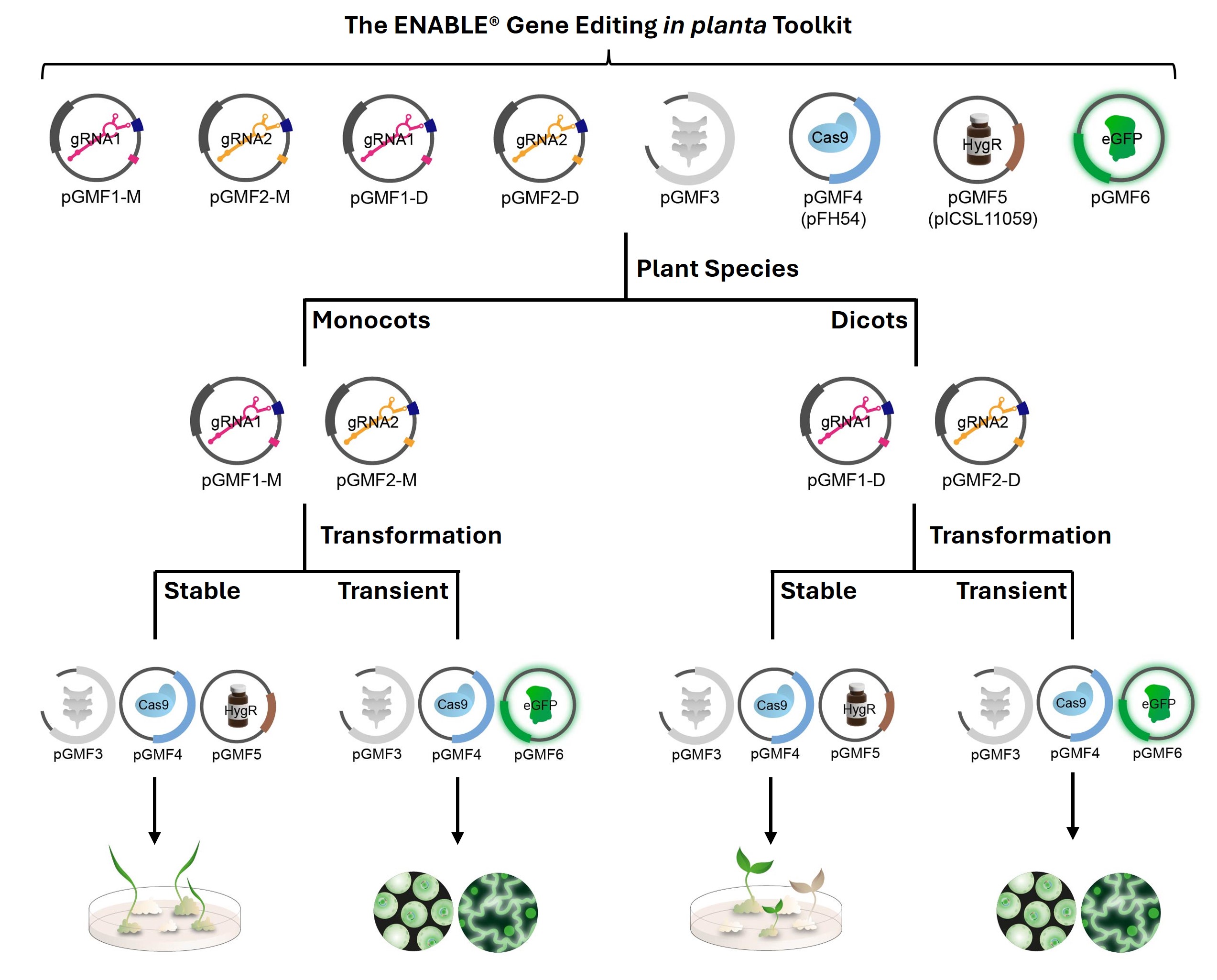
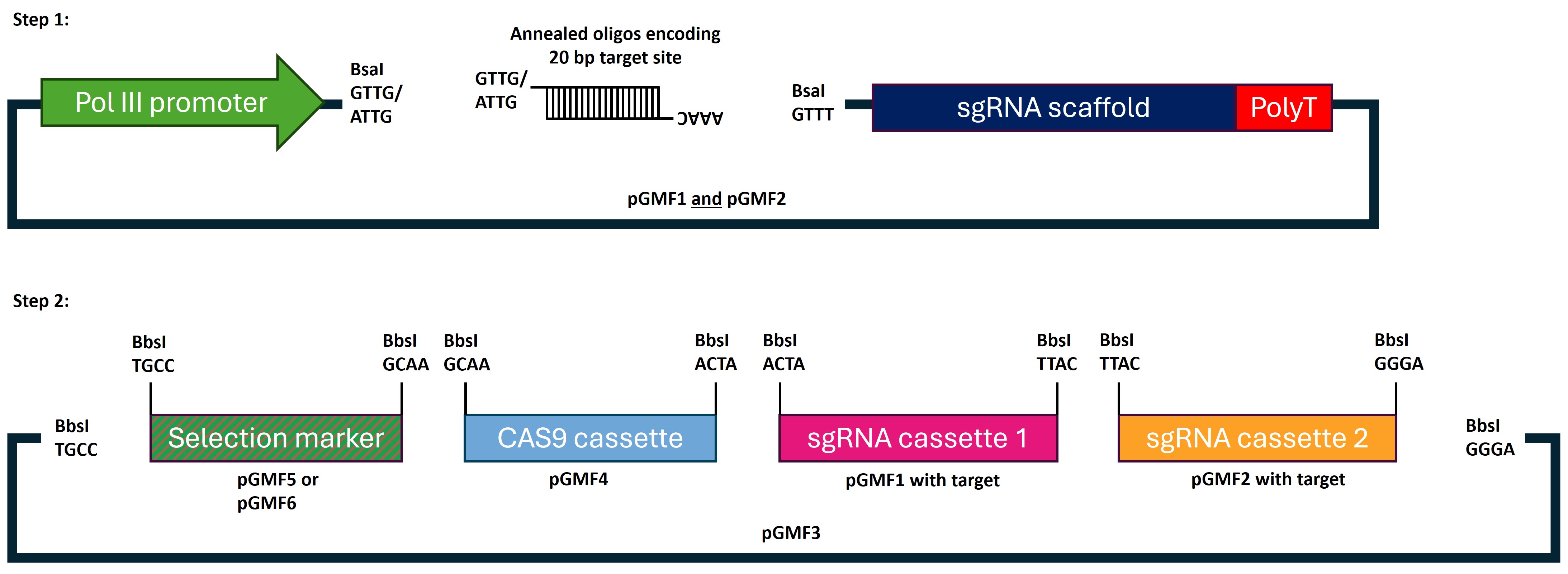
We created the ENABLE® Gene Editing in planta toolkit, a straightforward molecular toolkit that includes eight CRISPR-related plasmids along with comprehensive documentation. Our toolkit enables gene editing in both monocot and dicot plants and is suitable for stable or transient plant transformation.
For more detail, read the accompanying publication (Abate et al., 2025 (Link opens in a new window)) and protocols linked in the Protocols & Resources tab, or visit the Grow More Foundation's resource page (Link opens in a new window).
Kit Documentation
How to Cite this Kit
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which they were created, and include Addgene in the Materials and Methods of your future publications.
For your Materials and Methods section:
"The ENABLE® Gene Editing in planta toolkit was a gift from Kate Creasey Krainer, Vladimir Nekrasov, and Nicola Patron (Addgene kit #1000000270)."
For your Reference section:
Developing a Molecular Toolkit to ENABLE all to apply CRISPR/Cas9-based Gene Editing in planta. Abate BA, Hahn F, Chirivì D, Betti C, Fornara F, Molloy JC, Creasey Krainer KM. bioRxiv. 2025 November 13. doi: 10.1101/2025.11.09.687425. Article (Link opens in a new window)
The ENABLE® Gene Editing in planta Toolkit contains eight plasmids. Please refer to the individual plasmid pages below for more details on each plasmid in this kit:


