We narrowed to 5,933 results for: chia
-
Plasmid#195647PurposeComplementation of MlaC(D165I)DepositorInsertMlaC
ExpressionBacterialMutationMlaC(D165I)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
pBEL2526
Plasmid#195648PurposeComplementation of MlaC(E169R)DepositorInsertMlaC
ExpressionBacterialMutationMlaC(E169R)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
pBEL2527
Plasmid#195649PurposeComplementation of MlaC(G170K)DepositorInsertMlaC
ExpressionBacterialMutationMlaC(G170K)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
pBEL2528
Plasmid#195650PurposeComplementation of MlaC(I130D)DepositorInsertMlaC
ExpressionBacterialMutationMlaC(I130D)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
pBEL2530
Plasmid#195651PurposeComplementation of MlaC(I137D)DepositorInsertMlaC
ExpressionBacterialMutationMlaC(I137D)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
pBEL2531
Plasmid#195652PurposeComplementation of MlaC(L76D)DepositorInsertMlaC
ExpressionBacterialMutationMlaC(L76D)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
pBEL2532
Plasmid#195653PurposeComplementation of MlaC(N155A)DepositorInsertMlaC
ExpressionBacterialMutationMlaC(N155A)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
pBEL2533
Plasmid#195654PurposeComplementation of MlaC(N179E)DepositorInsertMlaC
ExpressionBacterialMutationMlaC(N179E)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
pBEL2534
Plasmid#195655PurposeComplementation of MlaC(Q115A)DepositorInsertMlaC
ExpressionBacterialMutationMlaC(Q115A)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
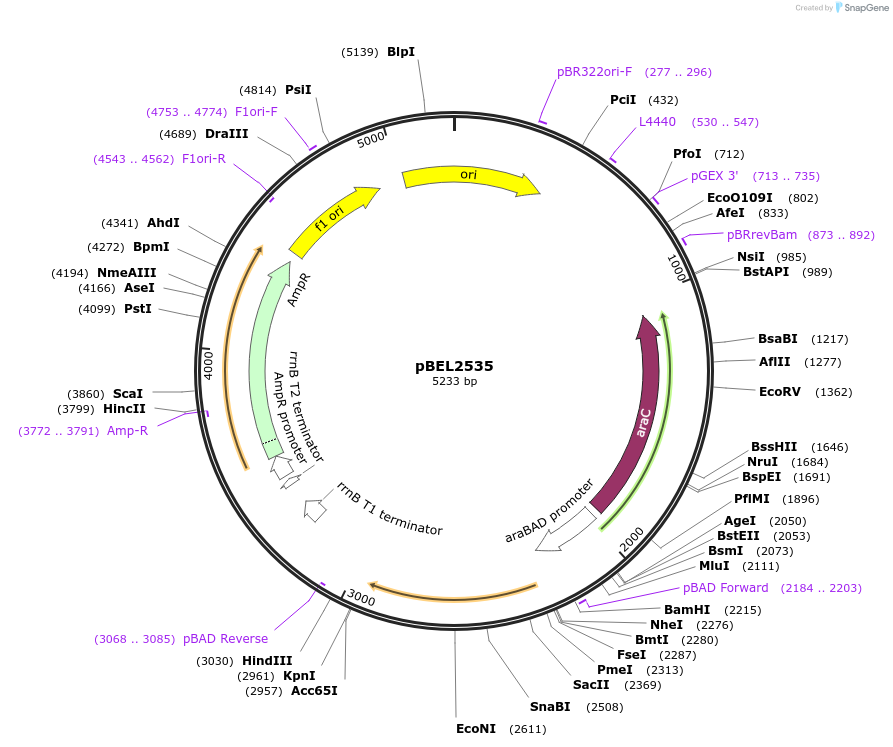
pBEL2535
Plasmid#195656PurposeComplementation of MlaC(R134E)DepositorInsertMlaC
ExpressionBacterialMutationMlaC(R134E)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
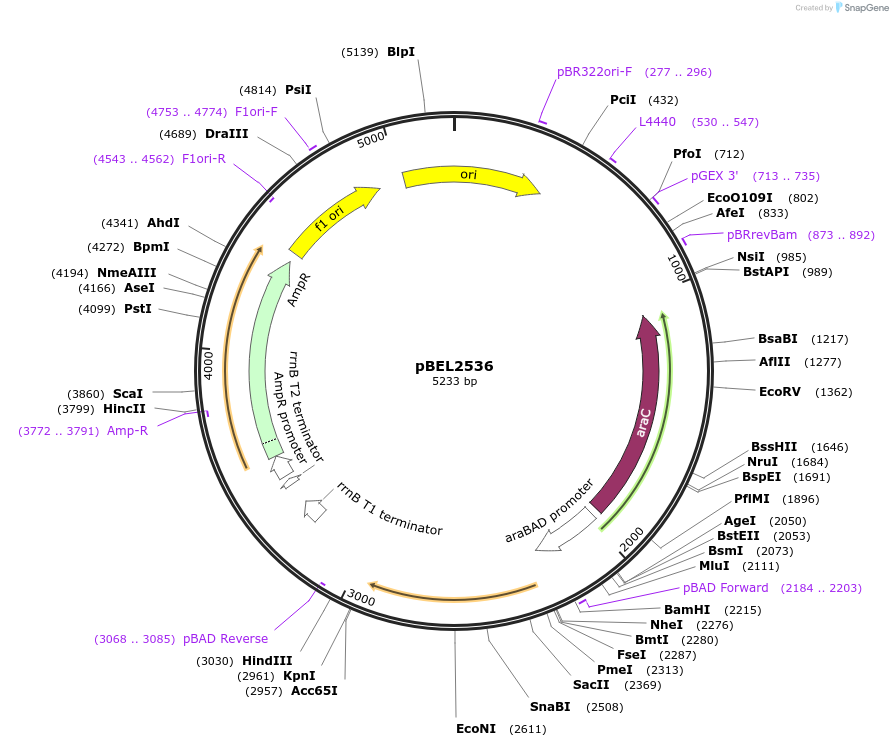
pBEL2536
Plasmid#195657PurposeComplementation of MlaC(R147D)DepositorInsertMlaC
ExpressionBacterialMutationMlaC(R147D)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
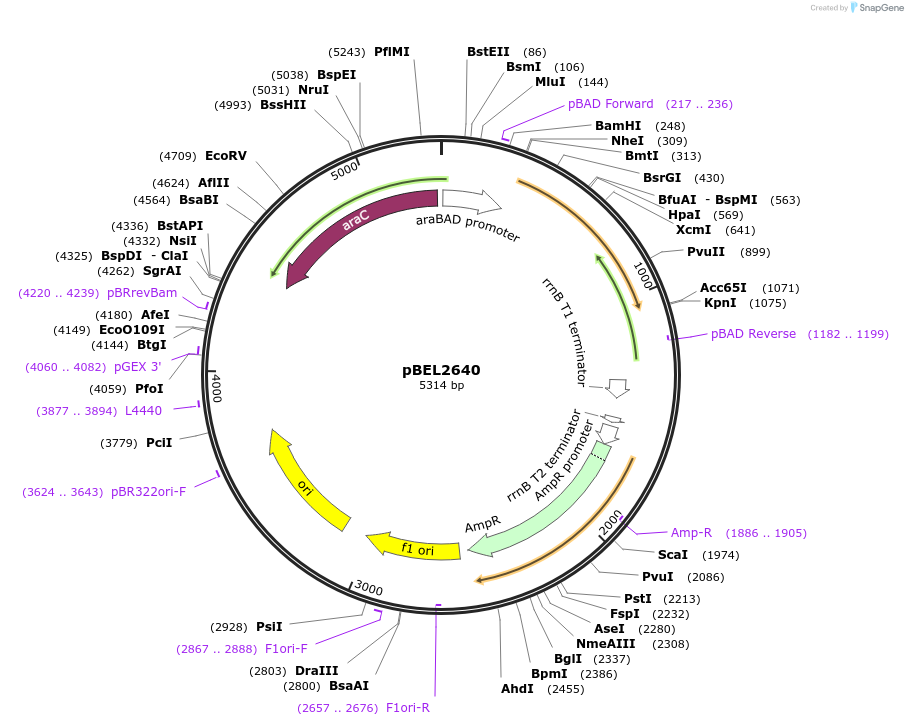
pBEL2640
Plasmid#195629PurposeComplementation of MlaA(L245N)DepositorInsertMlaA
ExpressionBacterialMutationMlaA(L245N)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
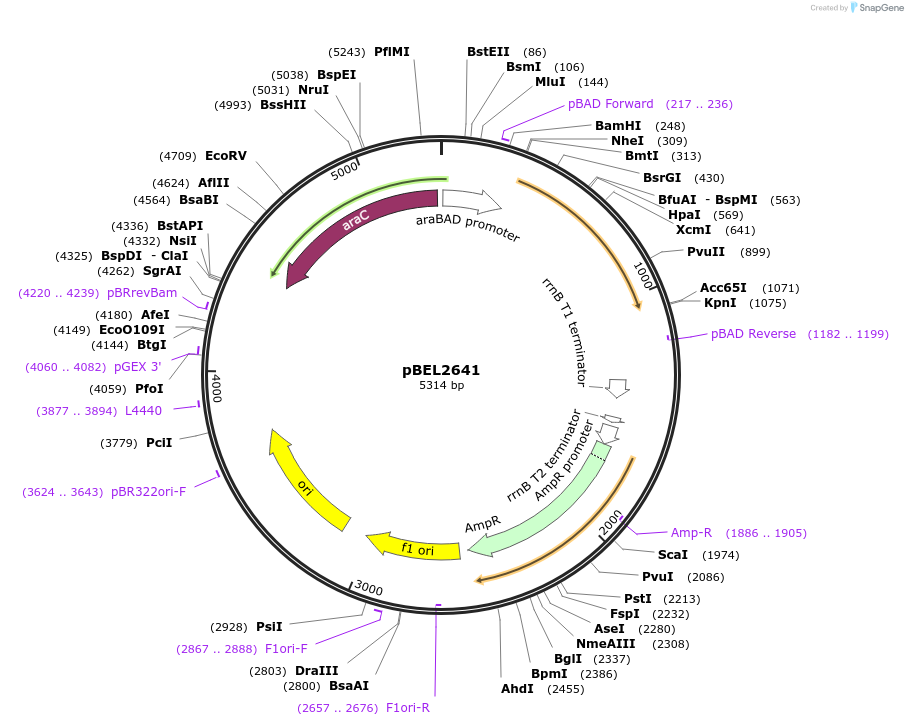
pBEL2641
Plasmid#195630PurposeComplementation of MlaA(I248N)DepositorInsertMlaA
ExpressionBacterialMutationMlaA(I248N)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
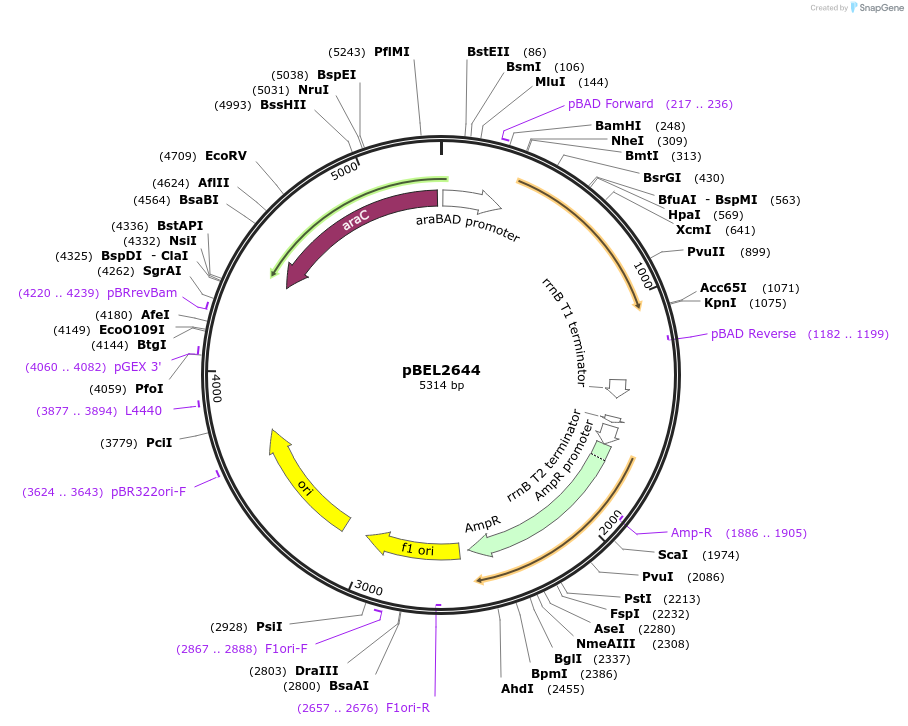
pBEL2644
Plasmid#195631PurposeComplementation of MlaA(D198N)DepositorInsertMlaA
ExpressionBacterialMutationMlaA(D198N)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
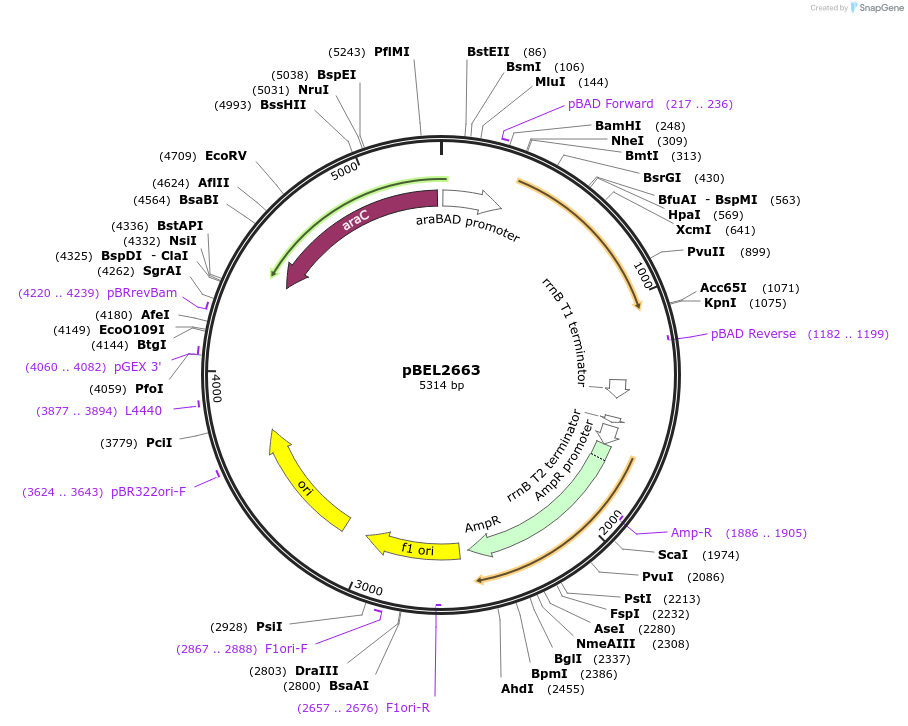
pBEL2663
Plasmid#195632PurposeComplementation of MlaA(F223N)DepositorInsertMlaA
ExpressionBacterialMutationMlaA(F223N)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
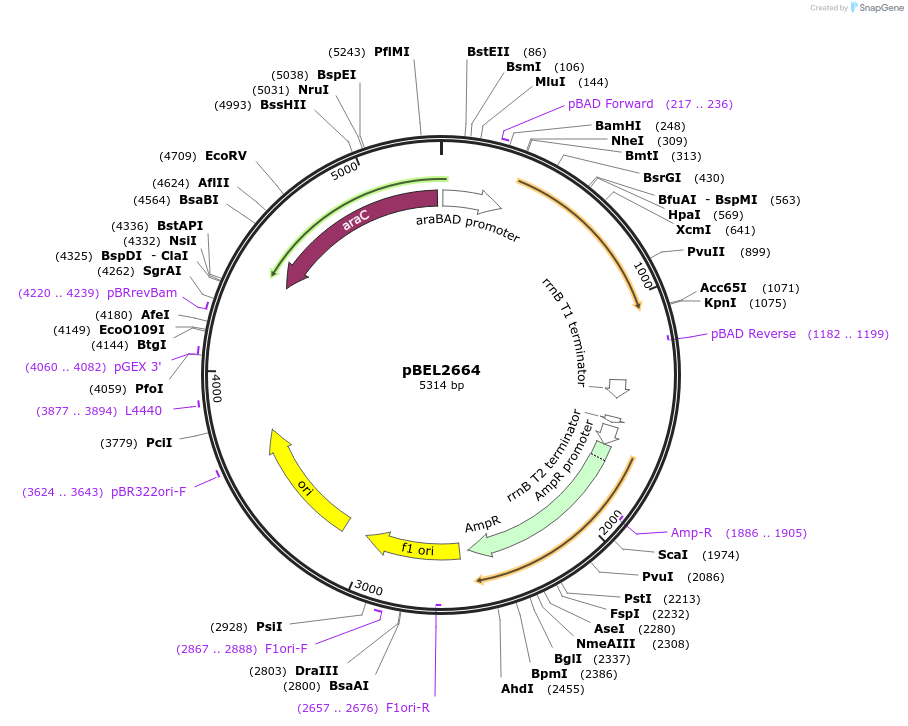
pBEL2664
Plasmid#195633PurposeComplementation of MlaA(L230N)DepositorInsertMlaA
ExpressionBacterialMutationMlaA(L230N)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
pBEL2665
Plasmid#195634PurposeComplementation of MlaA(delta244-251)DepositorInsertMlaA
ExpressionBacterialMutationMlaA(delta244-251)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
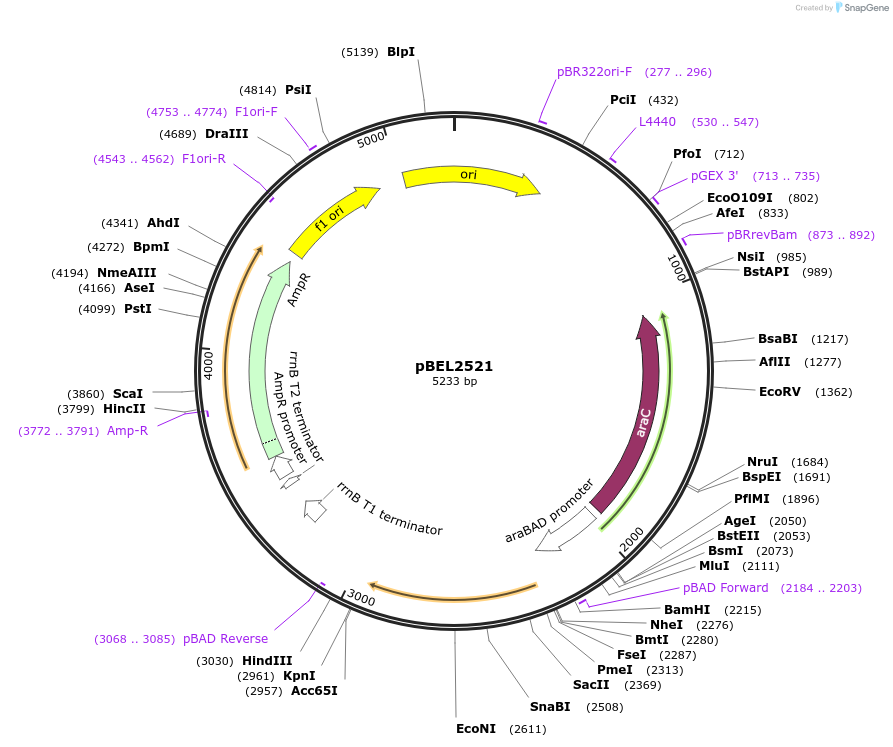
pBEL2521
Plasmid#195643PurposeComplementation of MlaC(A34R)DepositorInsertMlaC
ExpressionBacterialMutationMlaC(A34R)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
pBEL2512
Plasmid#195626PurposeComplementation of MlaA(delta238-251)DepositorInsertMlaA
ExpressionBacterialMutationMlaA(delta238-251)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
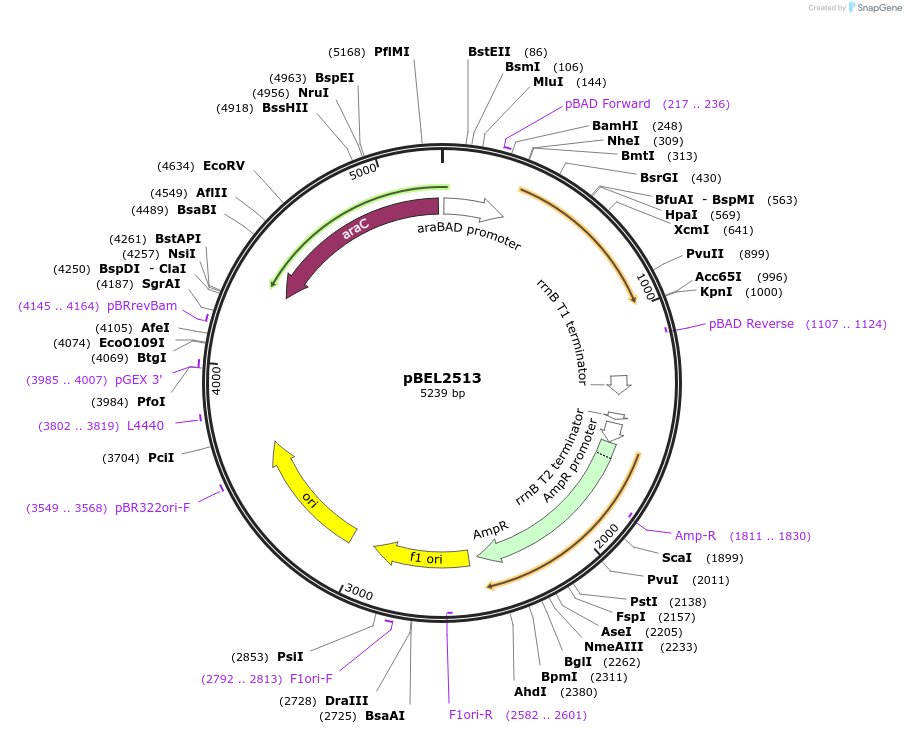
pBEL2513
Plasmid#195627PurposeComplementation of MlaA(delta227-251)DepositorInsertMlaA
ExpressionBacterialMutationMlaA(delta227-251)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
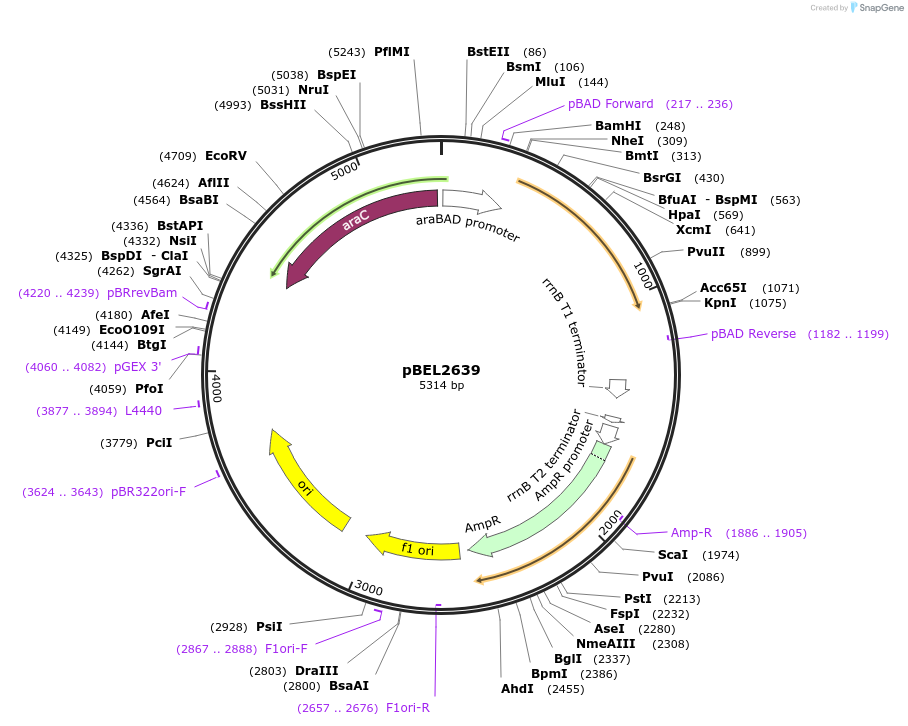
pBEL2639
Plasmid#195628PurposeComplementation of MlaA(I241N)DepositorInsertMlaA
ExpressionBacterialMutationMlaA(I241N)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
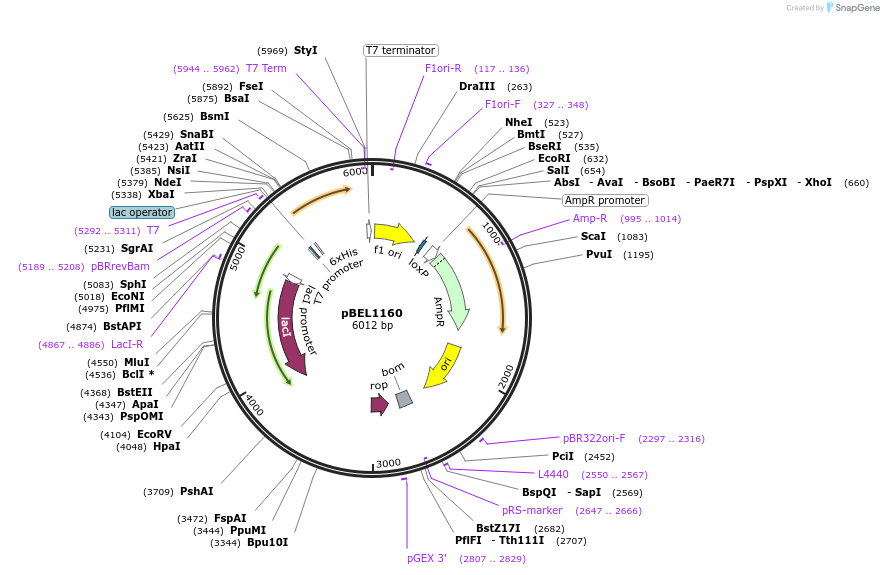
pBEL1160
Plasmid#196025PurposeExpression/purification of 6xHis-TEV-MlaDDepositorInsertMlaD
ExpressionBacterialAvailable SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
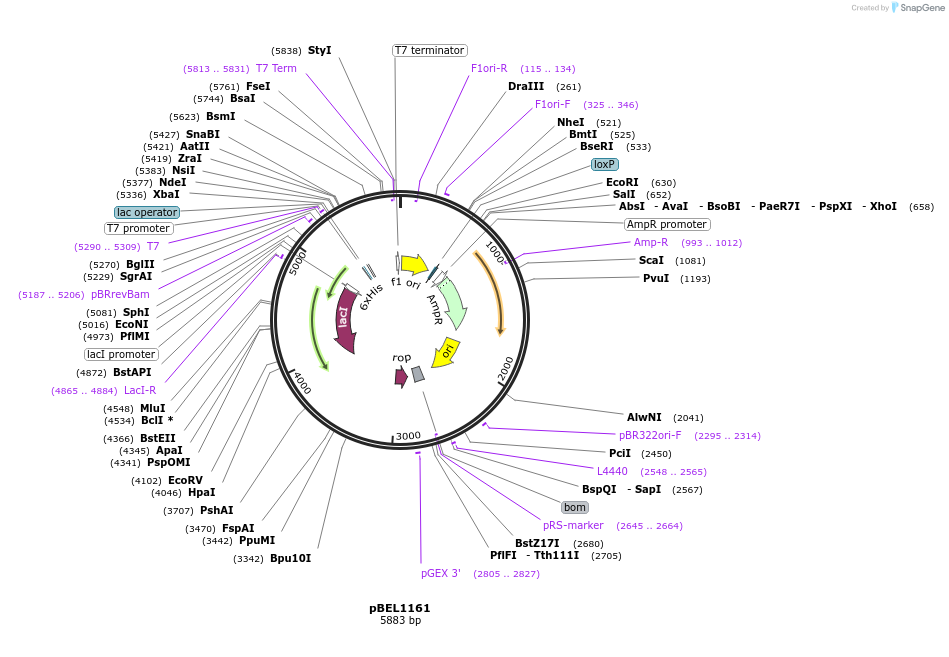
pBEL1161
Plasmid#196026PurposeExpression/purification of 6xHis-TEV-MlaDdelta141-183DepositorInsertMlaD
ExpressionBacterialMutation6xHis-TEV-MlaDdelta141-183Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
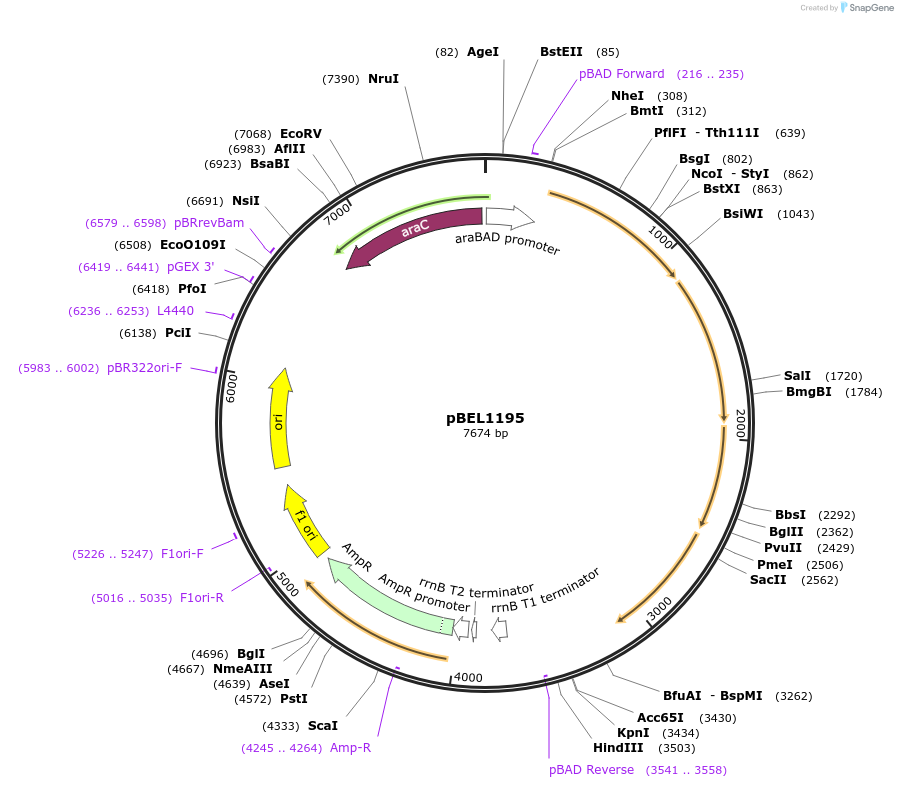
pBEL1195
Plasmid#196027PurposeComplementation of MlaF-MlaE-MlaD-MlaC-MlaBDepositorInsertMlaFEDCB
ExpressionBacterialAvailable SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
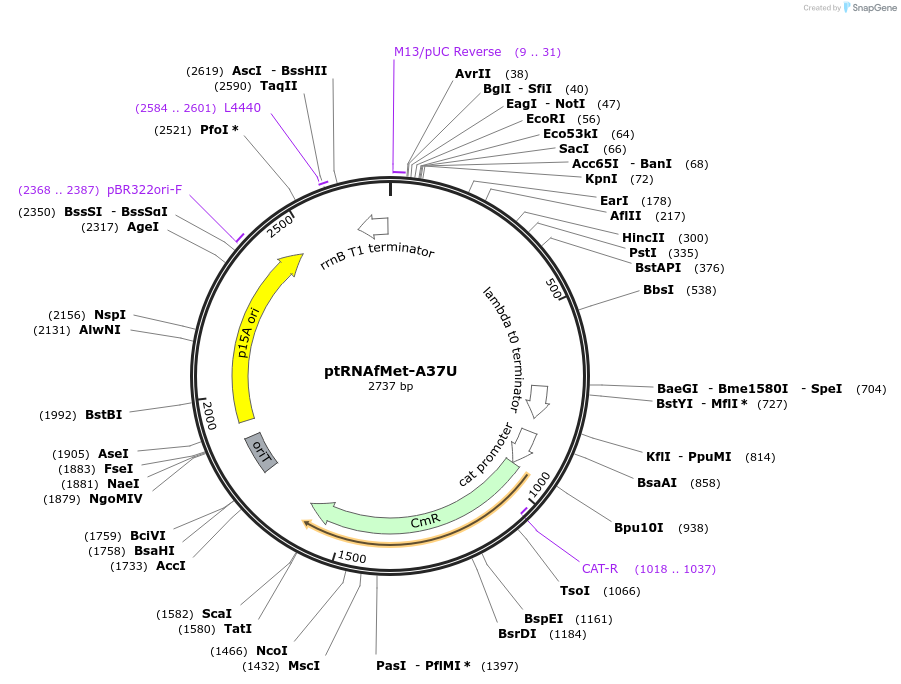
ptRNAfMet-A37U
Plasmid#196659PurposePlasmid for overexpression of mutated initiator tRNA (A37U)DepositorInserttRNAfMet-A37U expression cassette
ExpressionBacterialMutationA37U in tRNAfMetPromotermetY native promoterAvailable SinceMarch 30, 2023AvailabilityAcademic Institutions and Nonprofits only -
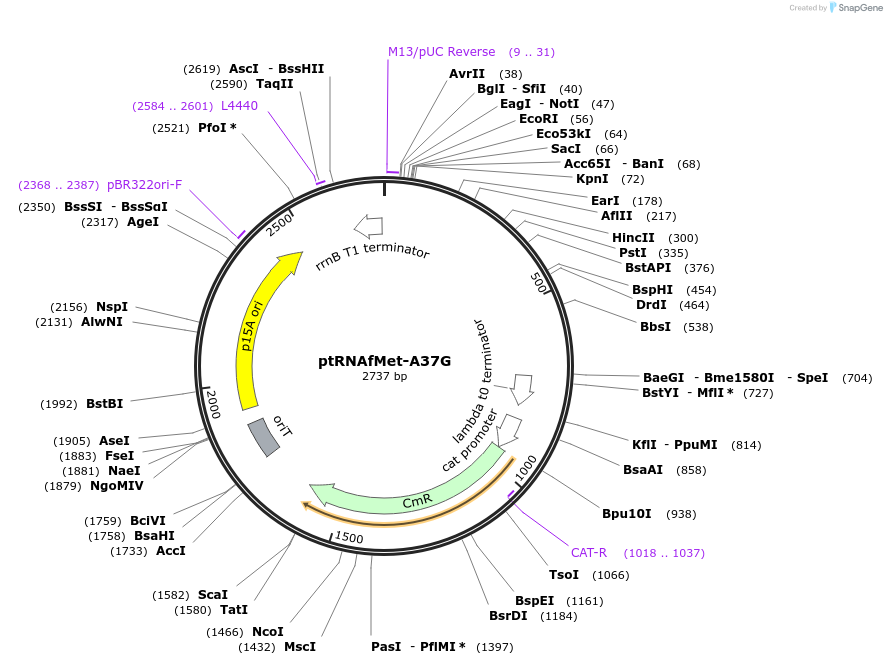
ptRNAfMet-A37G
Plasmid#196658PurposePlasmid for overexpression of mutated initiator tRNA (A37G)DepositorInserttRNAfMet-A37G expression cassette
ExpressionBacterialMutationA37G in tRNAfMetPromotermetY native promoterAvailable SinceMarch 27, 2023AvailabilityAcademic Institutions and Nonprofits only -
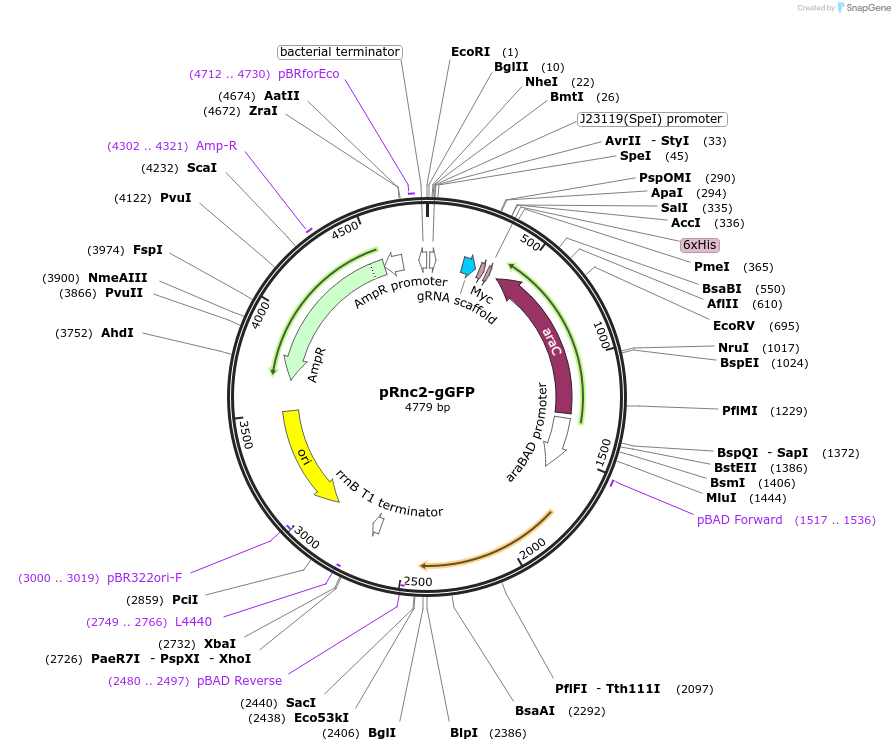
pRnc2-gGFP
Plasmid#189563PurposeExpresses E. Coli RNase III under control of pBAD promoter with native 5' untranslated region and constitutively expresses gUTR-129-GFPDepositorInsertsRNase III
gUTR-129-GFP
ExpressionBacterialMutationMaintains 5' UTR of rncPromoterJ23119 and pBADAvailable SinceMarch 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
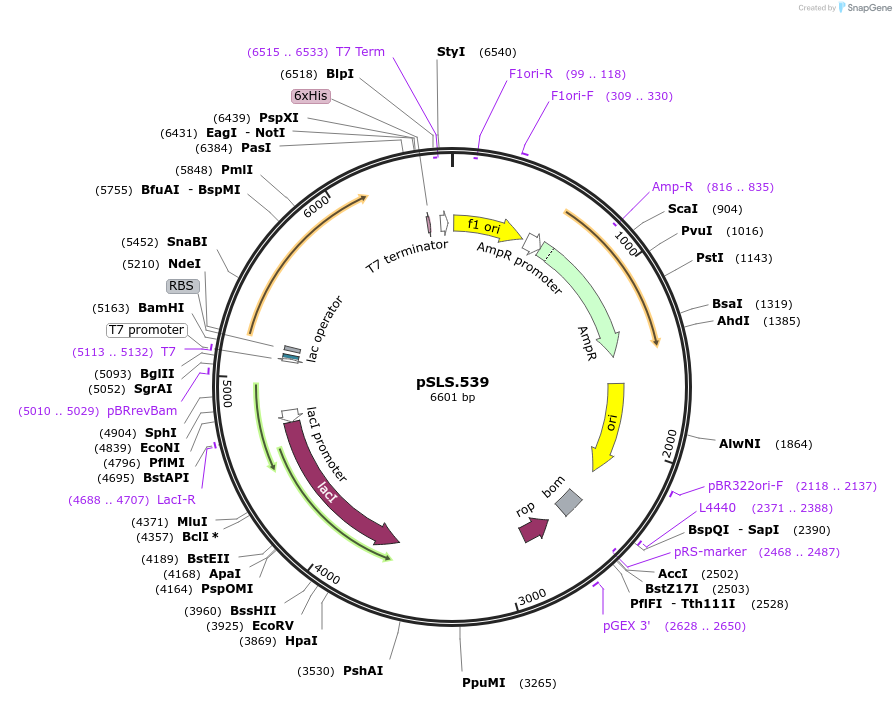
pSLS.539
Plasmid#184958PurposeTest effect of a1/a2 length on editing fabH using retron recombineeringDepositorInsertEco1 RT and recombineering ncRNA, fabH G954A, a1/a2 length: 12
ExpressionBacterialMutationfabH donor G954APromoterT7/lacAvailable SinceJan. 9, 2023AvailabilityAcademic Institutions and Nonprofits only -
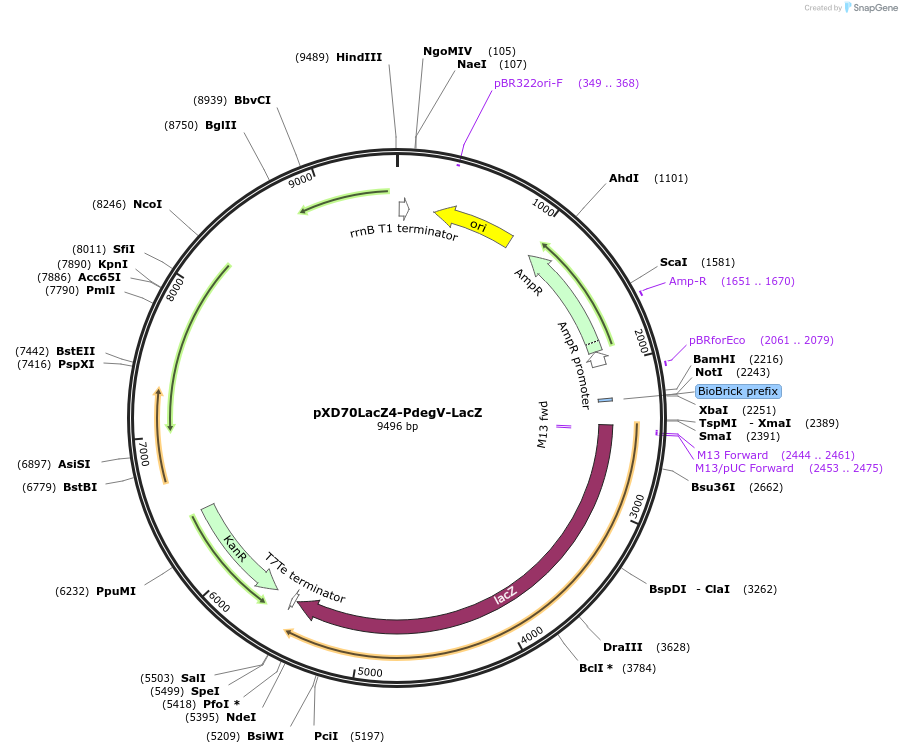
pXD70LacZ4-PdegV-LacZ
Plasmid#191620PurposeE. coli - Eggerthella lenta shuttle plasmid (KanR), PdegV promoter-lacZ, E. lenta constitutive promoterDepositorInsertlacZ
ExpressionBacterialAvailable SinceJan. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
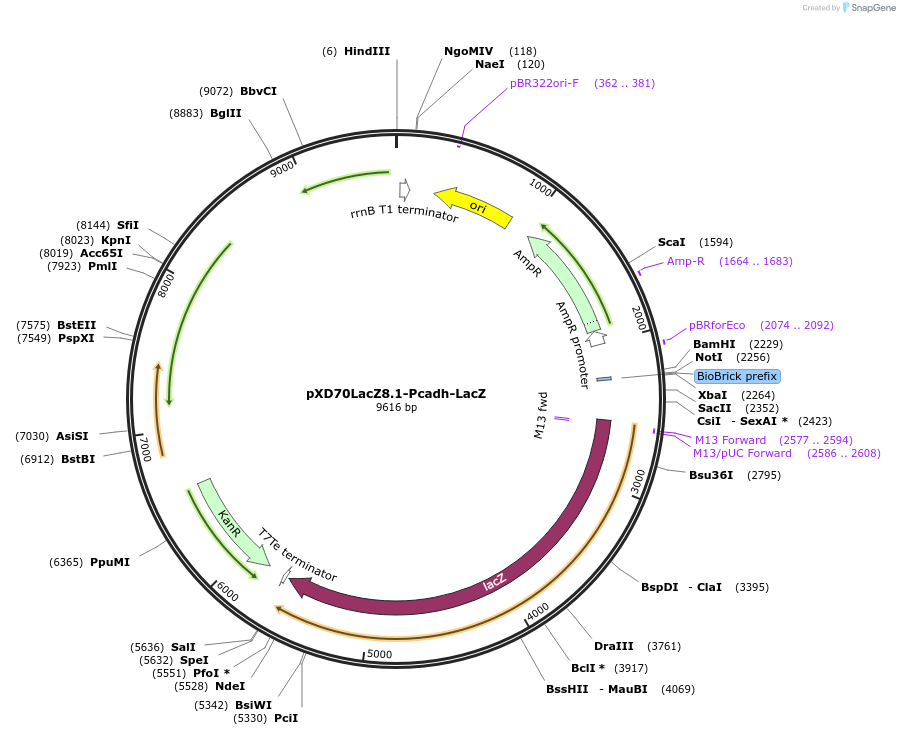
pXD70LacZ8.1-Pcadh-LacZ
Plasmid#191624PurposeE. coli - Eggerthella lenta shuttle plasmid (KanR), Pcadh catechin dehydroxylase promoter-lacZDepositorInsertlacZ
ExpressionBacterialAvailable SinceJan. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
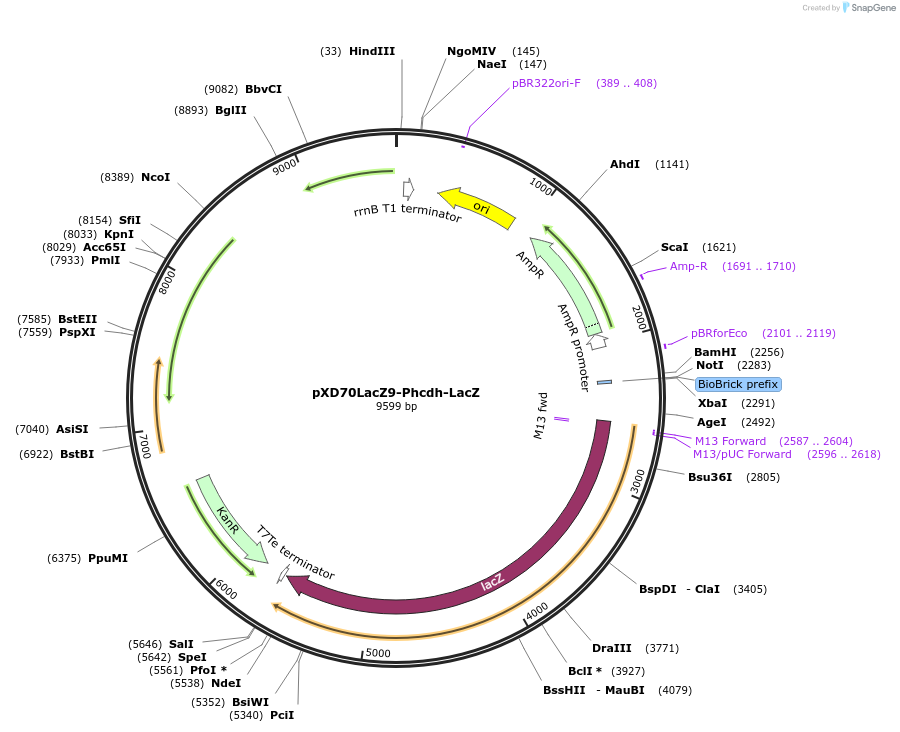
pXD70LacZ9-Phcdh-LacZ
Plasmid#191625PurposeE. coli - Eggerthella lenta shuttle plasmid (KanR), Phcdh hydrocaffeic acid dehydroxylase promoter-lacZDepositorInsertlacZ
ExpressionBacterialAvailable SinceJan. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
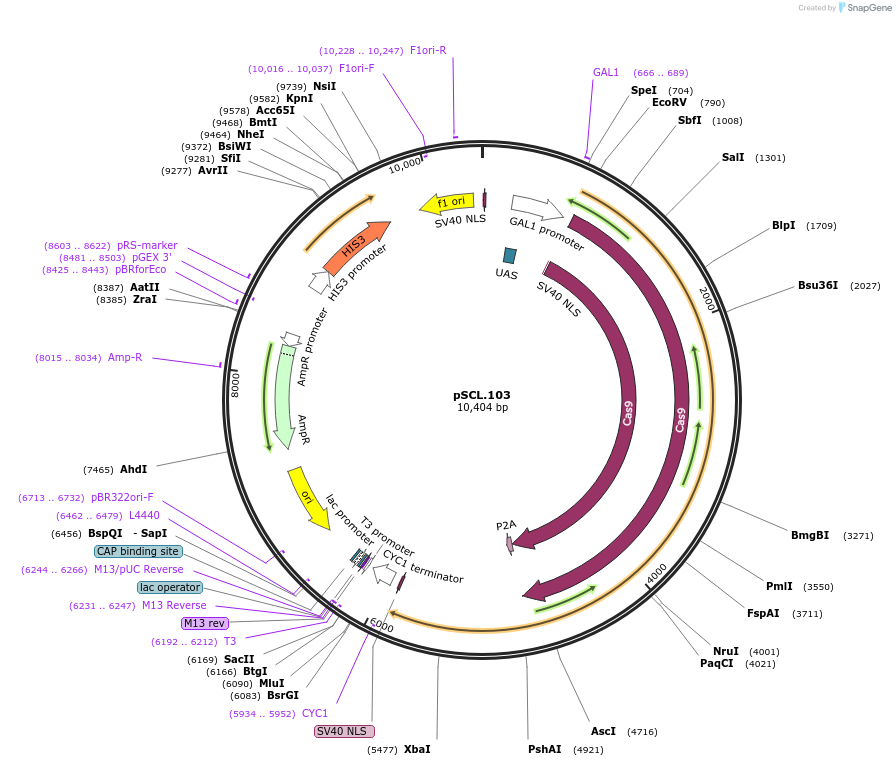
pSCL.103
Plasmid#184988PurposeExpress Cas9-P2A-Eco1RTDepositorInsertCas9-P2A-Eco1RT
TagsSV40NLSExpressionYeastMutationhuman codon optimized RTPromoterGal1-10Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
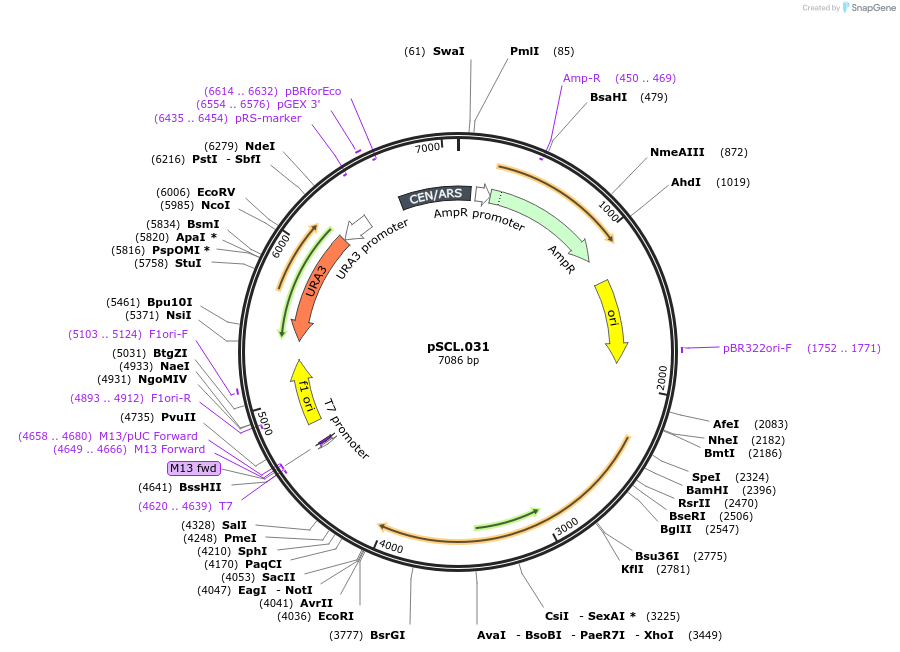
pSCL.031
Plasmid#184968PurposeTest effect of extending -Eco2 a1/a2 in yeastDepositorInsertEco2: RT and ncRNA(extended), a1/a2 length: 29
ExpressionYeastMutationhuman codon optimized RT, a1/a2 length extended t…PromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
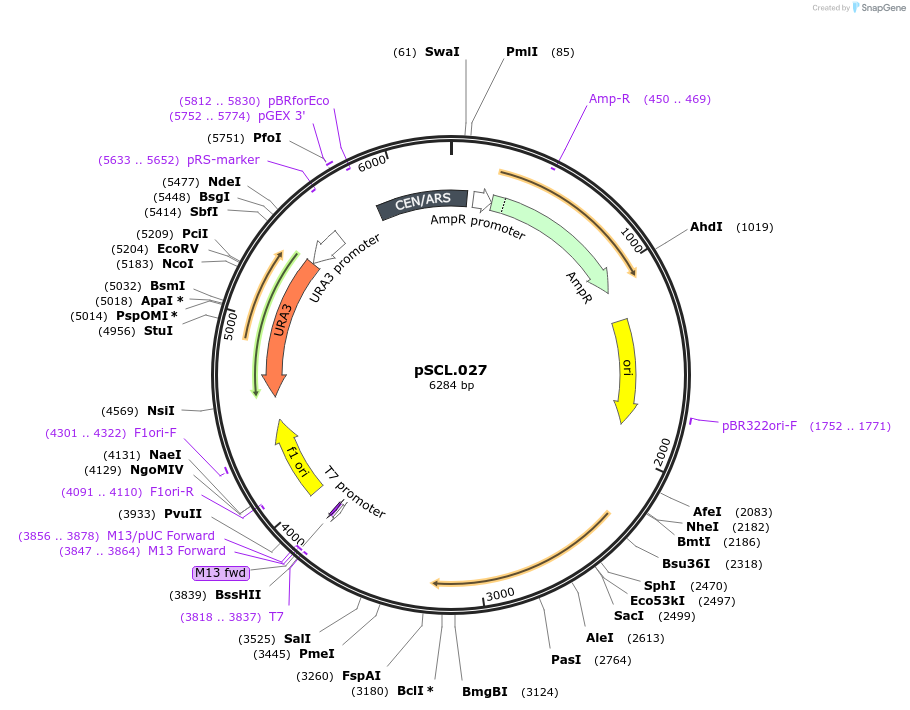
pSCL.027
Plasmid#184964PurposeExpress -Eco1 RT and ncRNA in yeastDepositorInsertEco1: RT and ncRNA(wt), a1/a2 length: 12
ExpressionYeastMutationhuman codon optimized RTPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
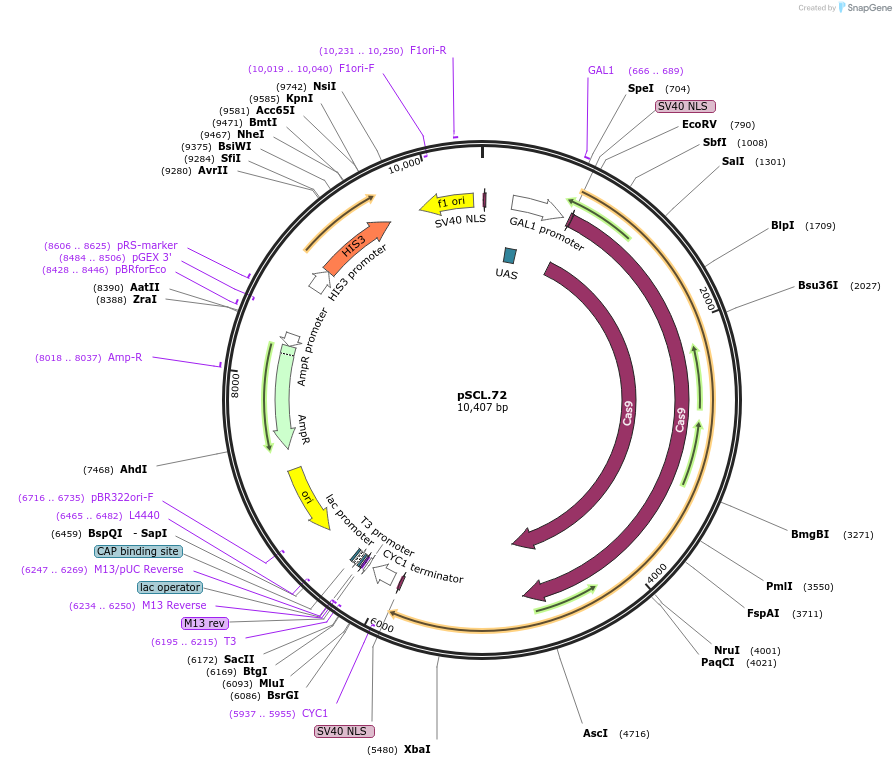
pSCL.72
Plasmid#184984PurposeExpress Cas9/-Eco1 RT fusion, using linker1DepositorInsertCas9-fusion_linker1-Eco1RT
TagsSV40NLSExpressionYeastMutationhuman codon optimized RTPromoterGal1-10Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
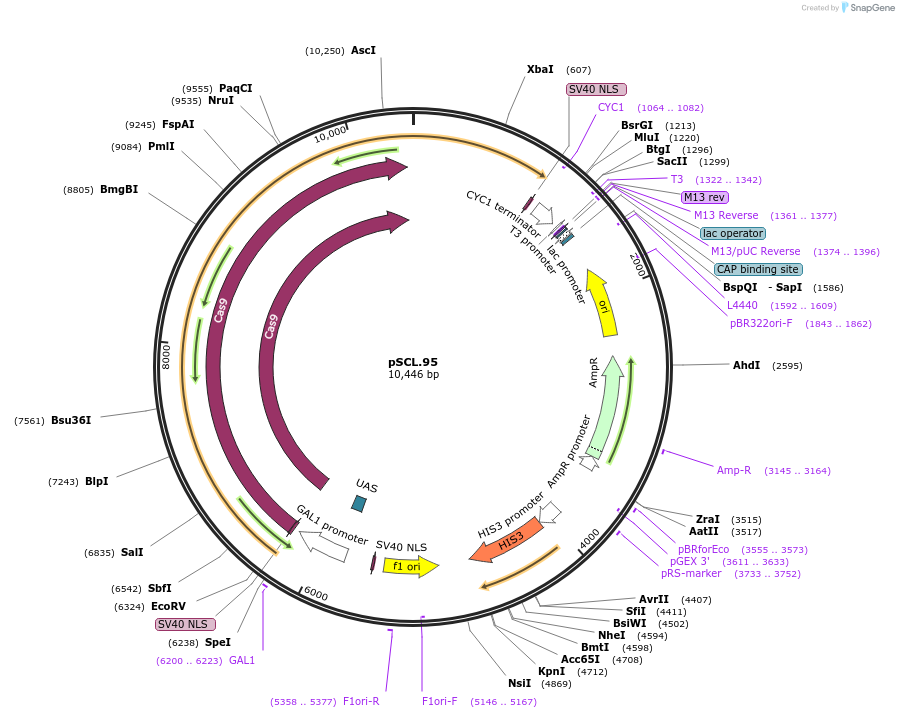
pSCL.95
Plasmid#184986PurposeExpress Cas9/-Eco1 RT fusion, using linker2DepositorInsertCas9-fusion_linker2-Eco1RT
TagsSV40NLSExpressionYeastMutationhuman codon optimized RTPromoterGal1-10Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
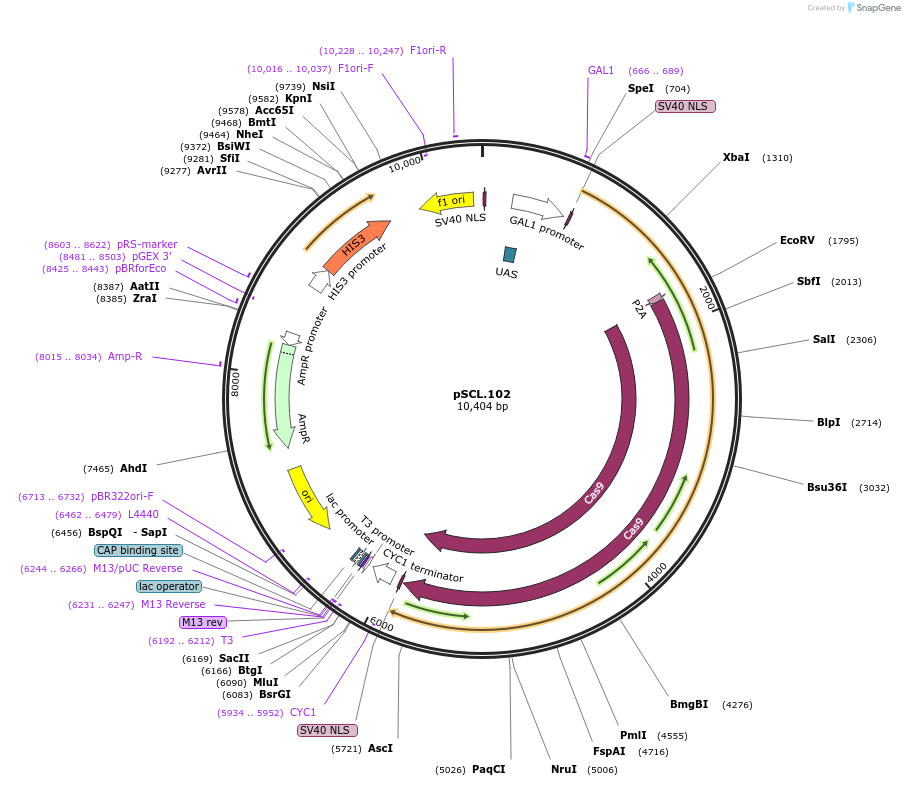
pSCL.102
Plasmid#184987PurposeExpress -Eco1 RT-P2A-Cas9DepositorInsertEco1RT-P2A-Cas9
TagsSV40NLSExpressionYeastMutationhuman codon optimized RTPromoterGal1-10Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
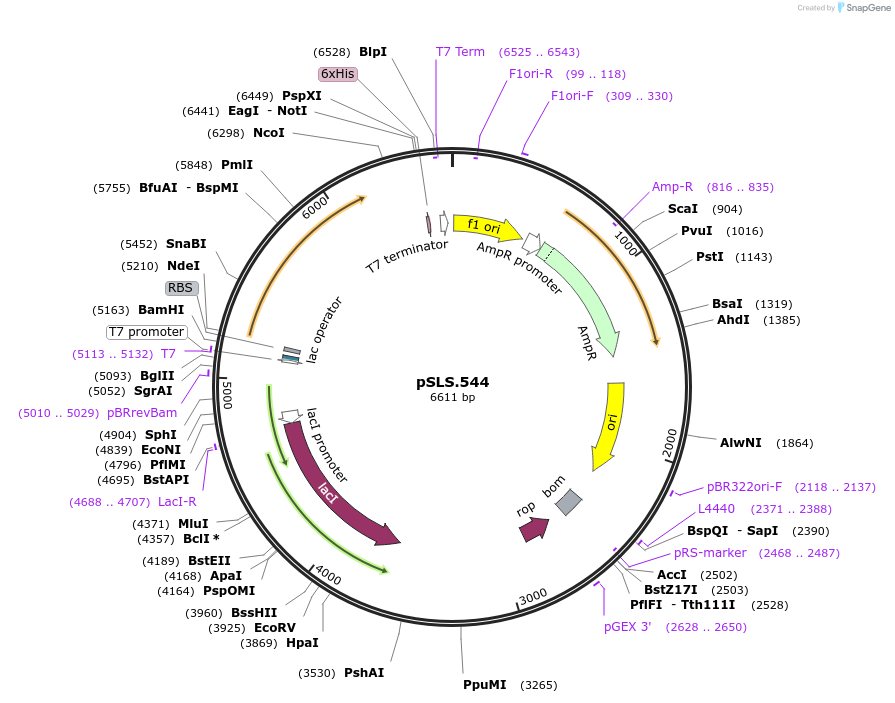
pSLS.544
Plasmid#184961PurposeTest effect of a1/a2 length on editing murF using retron recombineeringDepositorInsertEco1 RT and recombineering ncRNA, murF G1359A, a1/a2 length: 22
ExpressionBacterialMutationmurF donor G1359A, a1/a2 length extended to 22 bpPromoterT7/lacAvailable SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
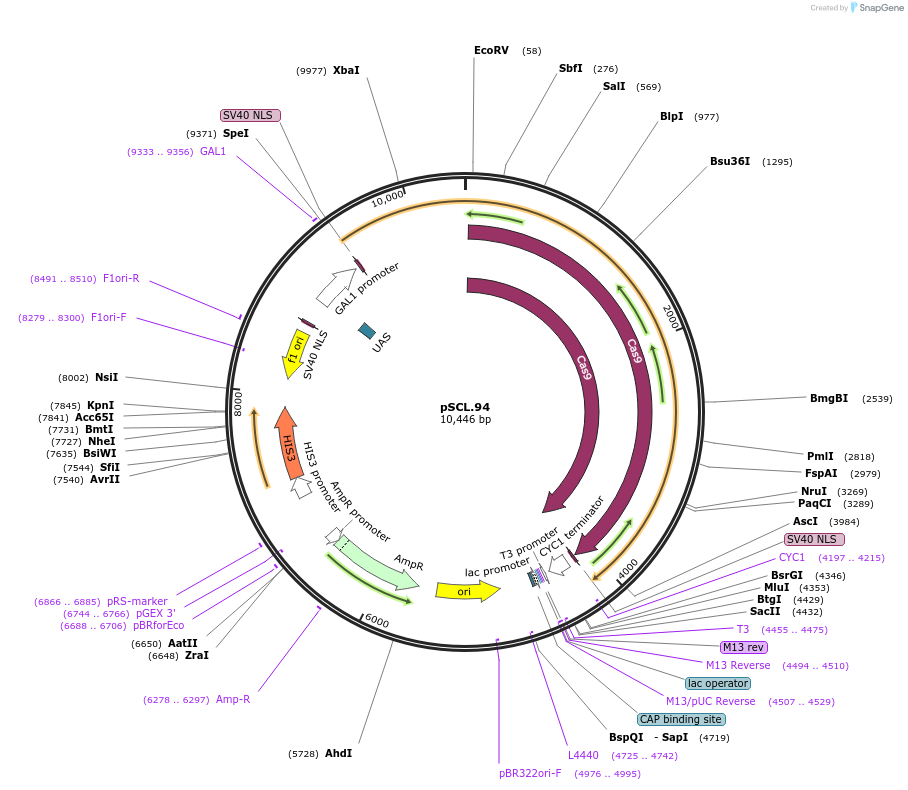
pSCL.94
Plasmid#184985PurposeExpress -Eco1 RT/Cas9 fusion, using linker2DepositorInsertEco1RT-fusion_linker2-Cas9
TagsSV40NLSExpressionYeastMutationhuman codon optimized RTPromoterGal1-10Available SinceNov. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
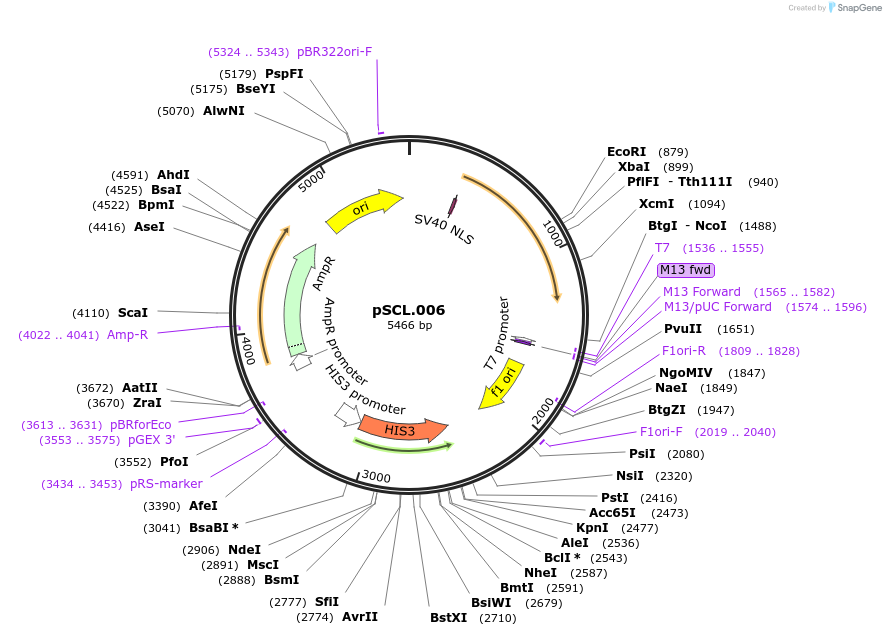
pSCL.006
Plasmid#184970PurposeExpress -Eco1 RT in yeastDepositorInsertIntegrating inducible cassette: Eco1 RT
TagsSV40NLSExpressionYeastMutationhuman codon optimized RTPromoterGal1-10Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
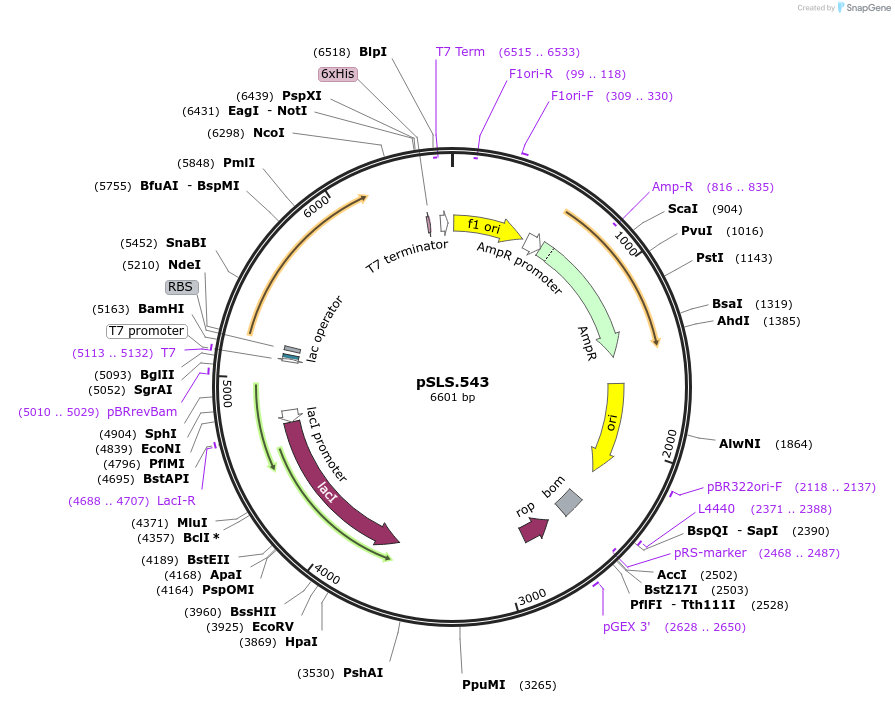
pSLS.543
Plasmid#184960PurposeTest effect of a1/a2 length on editing murF using retron recombineeringDepositorInsertEco1 RT and recombineering ncRNA, murF G1359A, a1/a2 length: 12
ExpressionBacterialMutationmurF donor G1359APromoterT7/lacAvailable SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
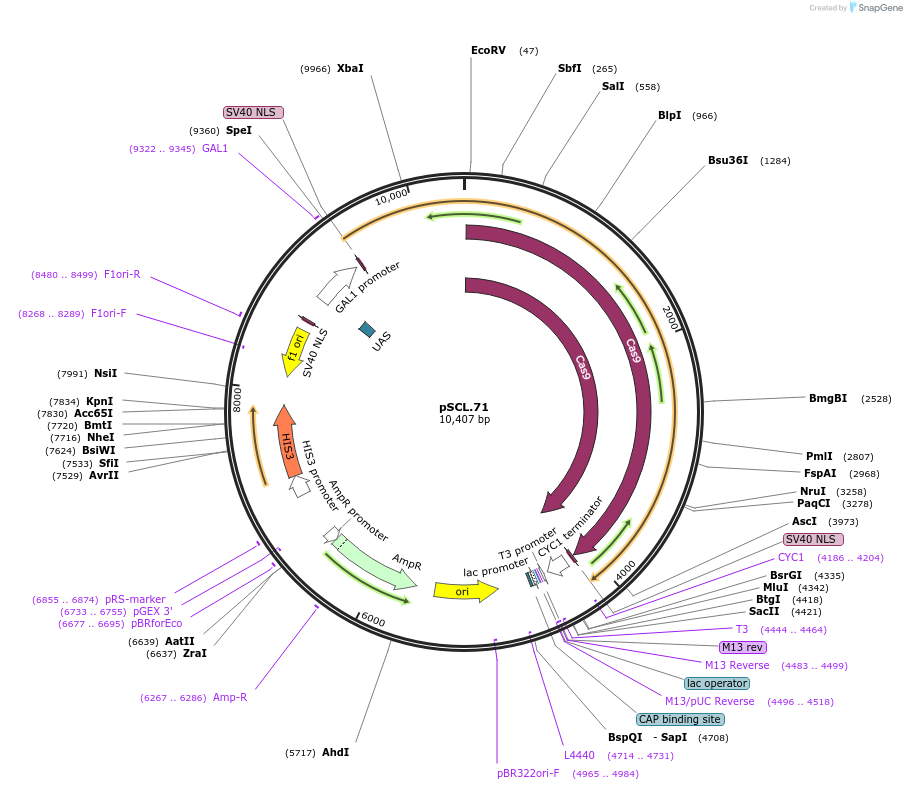
pSCL.71
Plasmid#184983PurposeExpress -Eco1 RT/Cas9 fusion, using linker1DepositorInsertEco1RT-fusion_linker1-Cas9
TagsSV40NLSExpressionYeastMutationhuman codon optimized RTPromoterGal1-10Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
pSLS.545
Plasmid#184962PurposeTest effect of a1/a2 length on editing priB using retron recombineeringDepositorInsertEco1 RT and recombineering ncRNA, priB G1069A, a1/a2 length: 12
ExpressionBacterialMutationpriB donor G1069APromoterT7/lacAvailable SinceNov. 1, 2022AvailabilityAcademic Institutions and Nonprofits only -
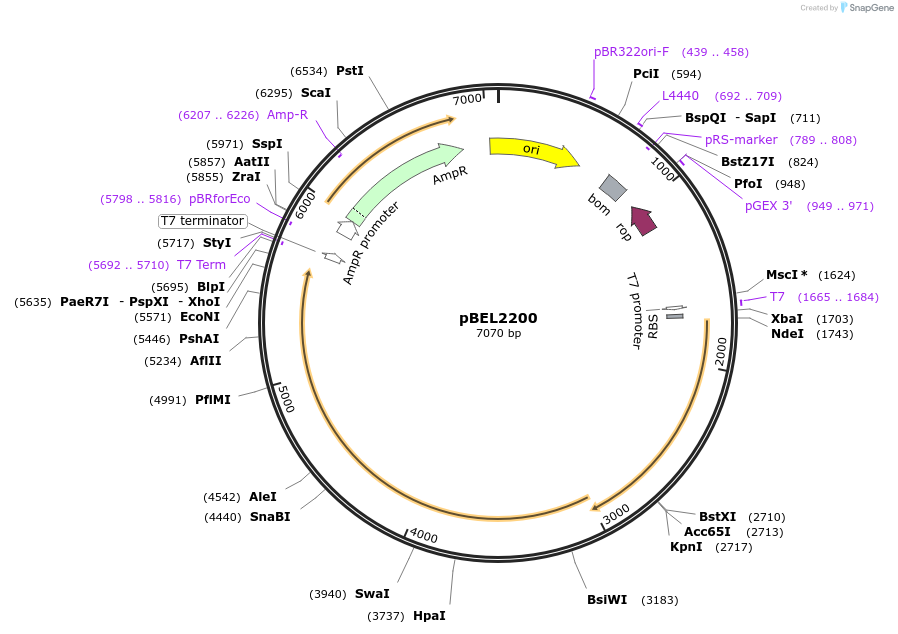
pBEL2200
Plasmid#170407PurposeGenetic complementation plasmid of LetA-[LetB PLL6->3] mutantDepositorExpressionBacterialMutationLetA-LetB(Δ702-716, replace with 347-360)Available SinceOct. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
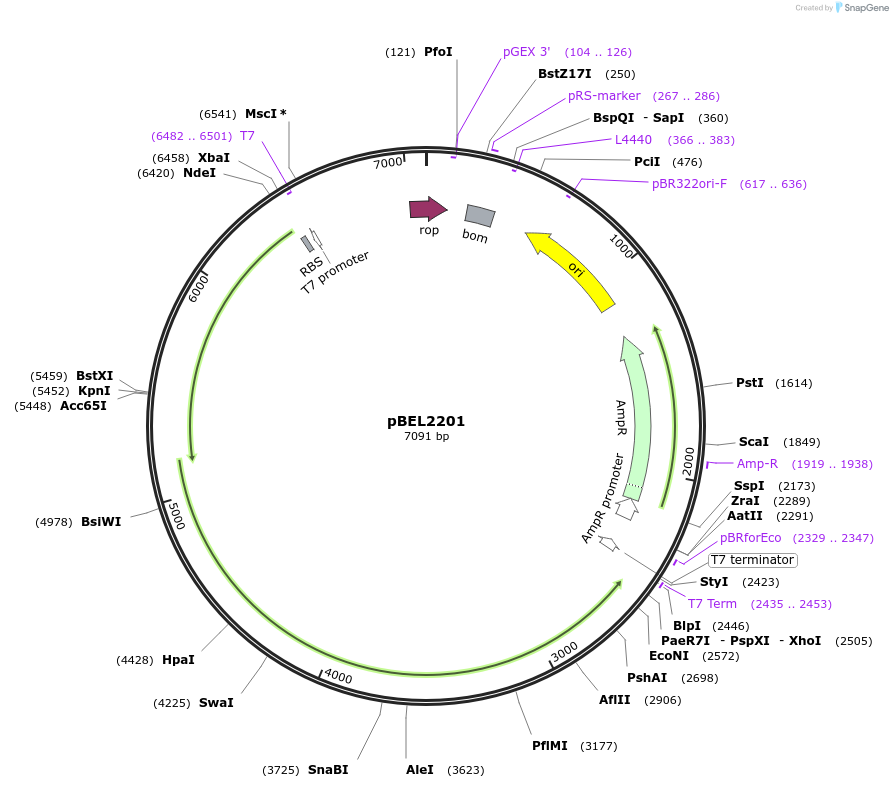
pBEL2201
Plasmid#170408PurposeGenetic complementation plasmid of LetA-[LetB PLL6->5] mutantDepositorExpressionBacterialMutationLetA-LetB(Δ702-716, replace with 580-600)Available SinceOct. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
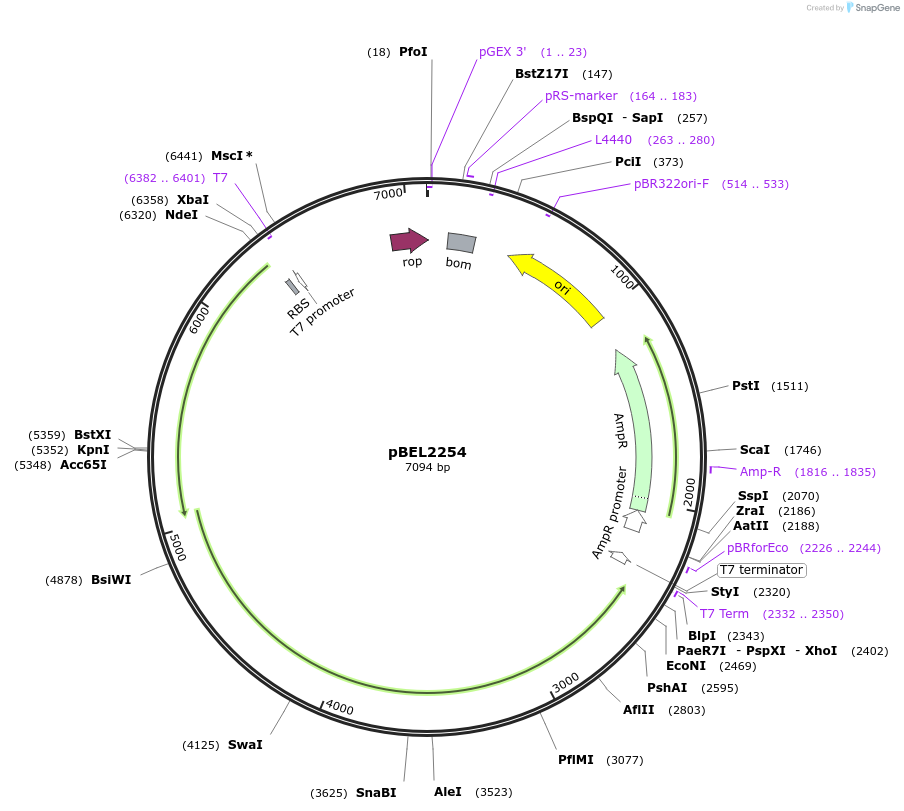
pBEL2254
Plasmid#170409PurposeGenetic complementation plasmid of LetA-[LetB PLL6->2] mutantDepositorExpressionBacterialMutationLetA-LetB(Δ700-717, replace with 225-249)Available SinceOct. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
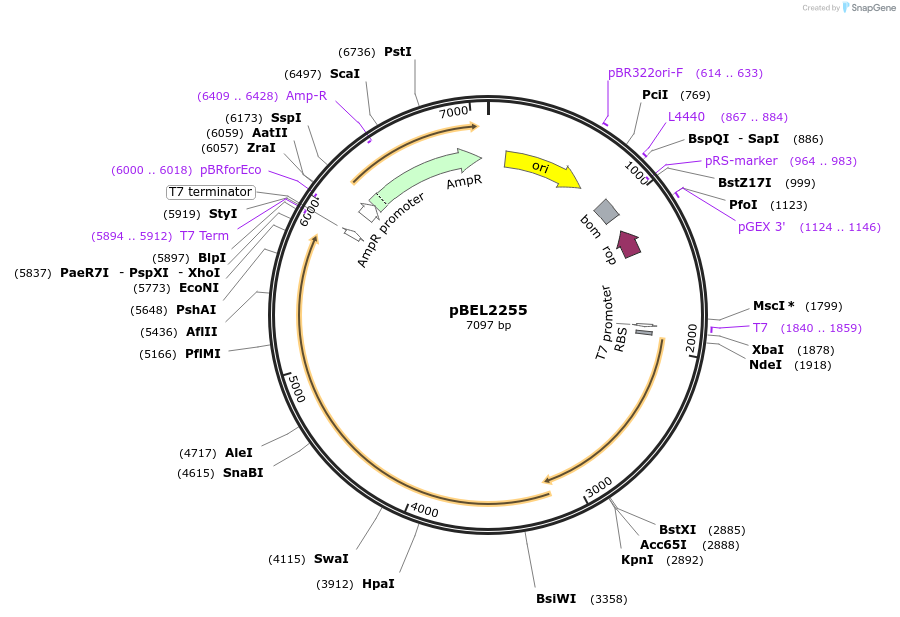
pBEL2255
Plasmid#170410PurposeGenetic complementation plasmid of LetA-[LetB PLL6->4] mutantDepositorExpressionBacterialMutationLetA-LetB(Δ700-716, replace with 445-479)Available SinceOct. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
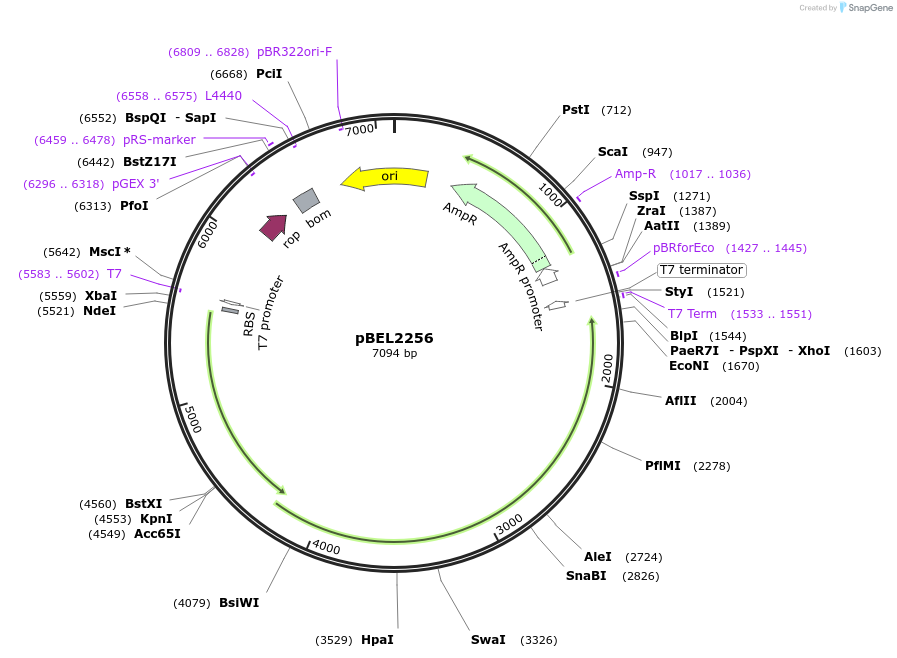
pBEL2256
Plasmid#170411PurposeGenetic complementation plasmid of LetA-[LetB PLL6->7] mutantDepositorExpressionBacterialMutationLetA-LetB(Δ700-717, replace with 812-836)Available SinceOct. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
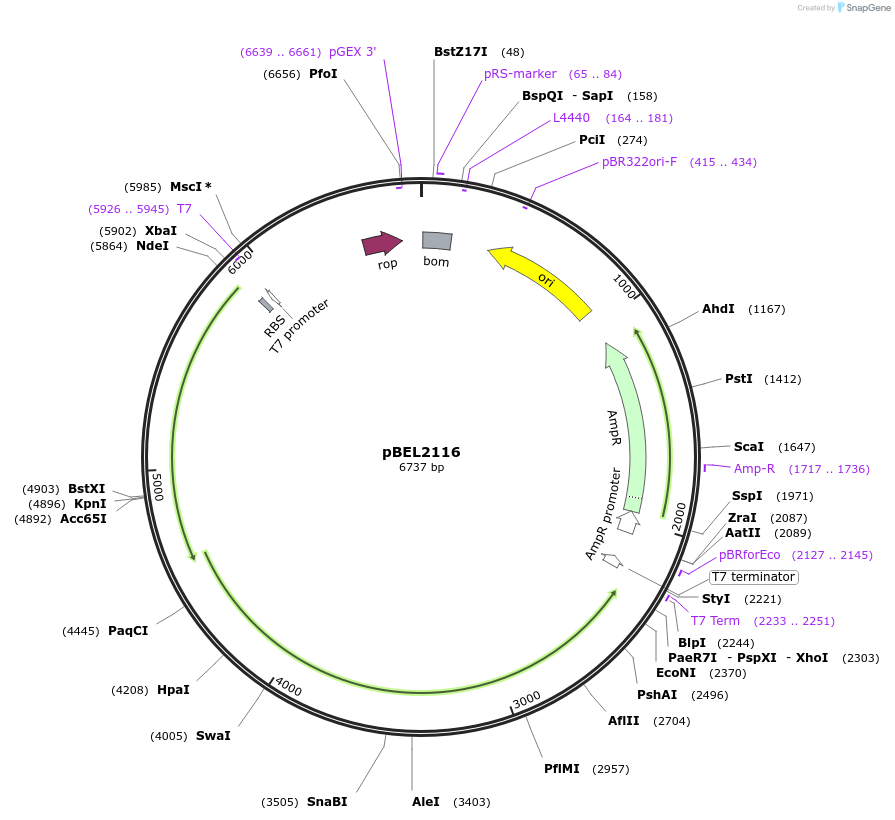
pBEL2116
Plasmid#170393PurposePlasmid for genetic complementation of LetAB with ΔRing 1 LetB mutantDepositorAvailable SinceOct. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
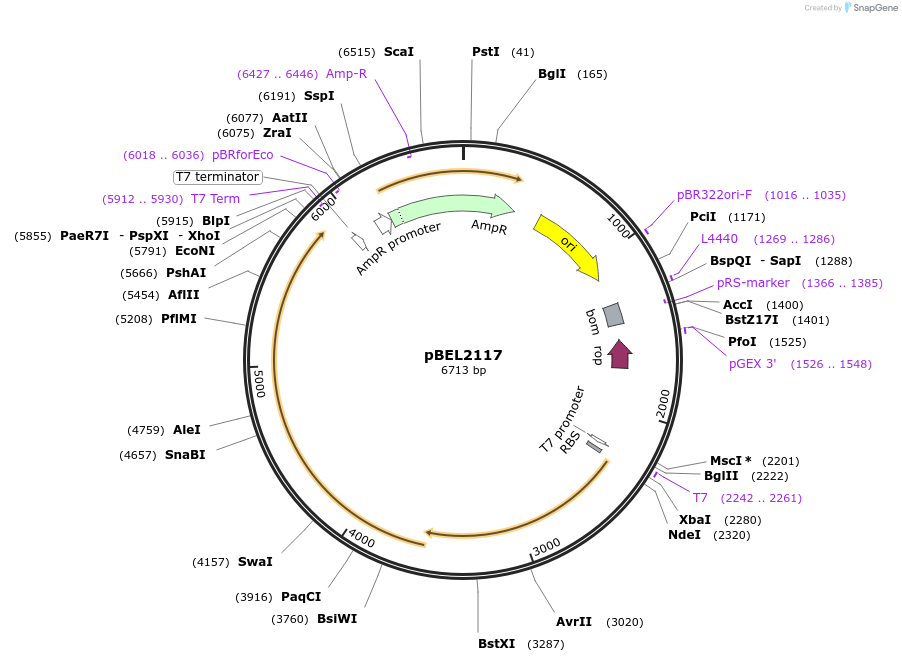
pBEL2117
Plasmid#170394PurposePlasmid for genetic complementation of LetAB with ΔRing 2 LetB mutantDepositorAvailable SinceOct. 3, 2022AvailabilityAcademic Institutions and Nonprofits only