We narrowed to 11,875 results for: Vars
-
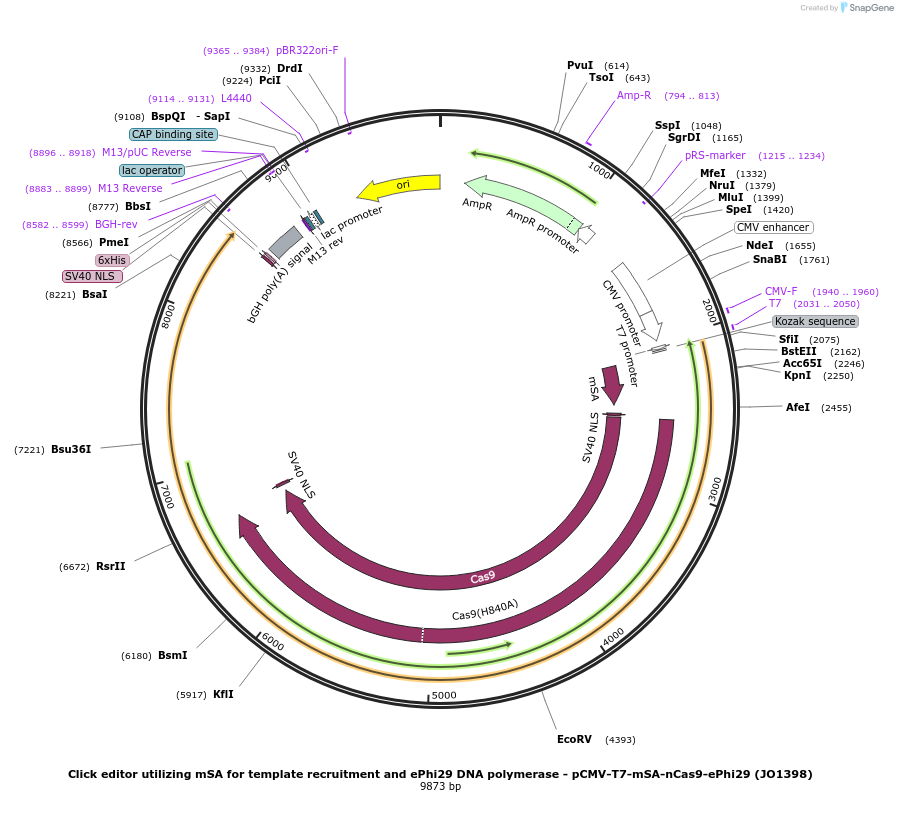
Plasmid#217803PurposeVariant CE1 construct with mSA for template recruitment and ePhi29(D169A) DNA pol, expressed from CMV or T7 promoters.DepositorInsertmSA-linker-SV40NLS-nSpCas9-BPNLS-ePhi29-BPNLS
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLSExpressionMammalianMutationnSpCas9(H840A); Phi29(D169A/M8R/V51A/M97T/G197D/E…PromoterCMV and T7Available SinceSept. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
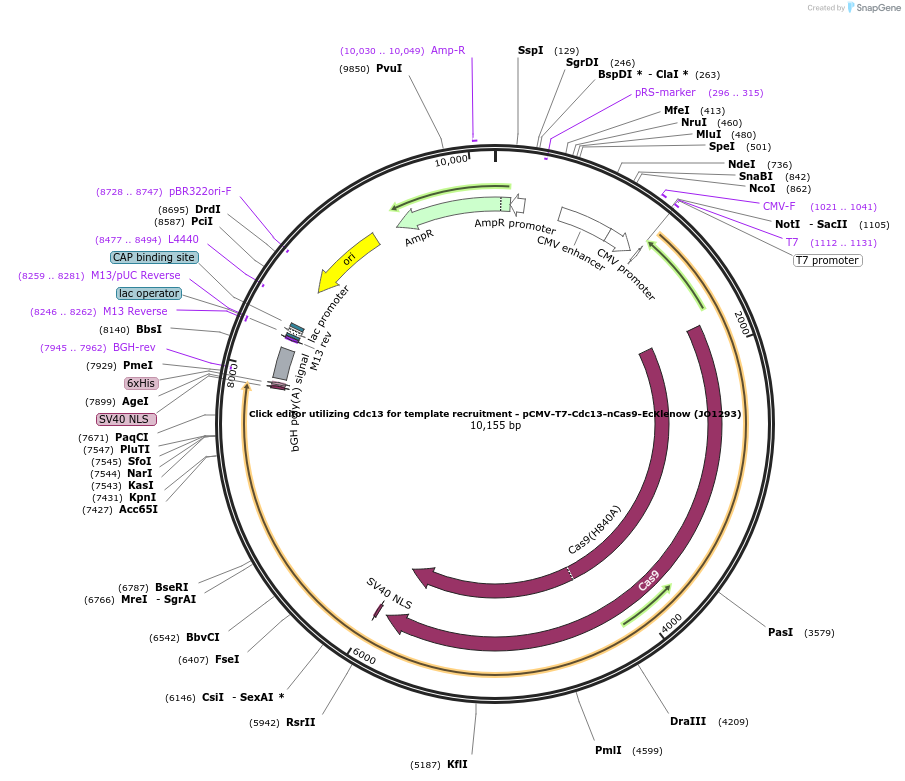
Click editor utilizing Cdc13 for template recruitment - pCMV-T7-Cdc13-nCas9-EcKlenow (JO1293)
Plasmid#217796PurposeVariant CE1 construct with Cdc13 telomere binding protein for template recruitment, expressed from CMV or T7 promoters.DepositorInsertCdc13-XTEN-nSpCas9-BPNLS-EcKlenow(-exo)-BPNLS
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLSExpressionMammalianMutationnSpCas9(H840A); EcKlenow(-exo;D355A/E357A)PromoterCMV and T7Available SinceSept. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
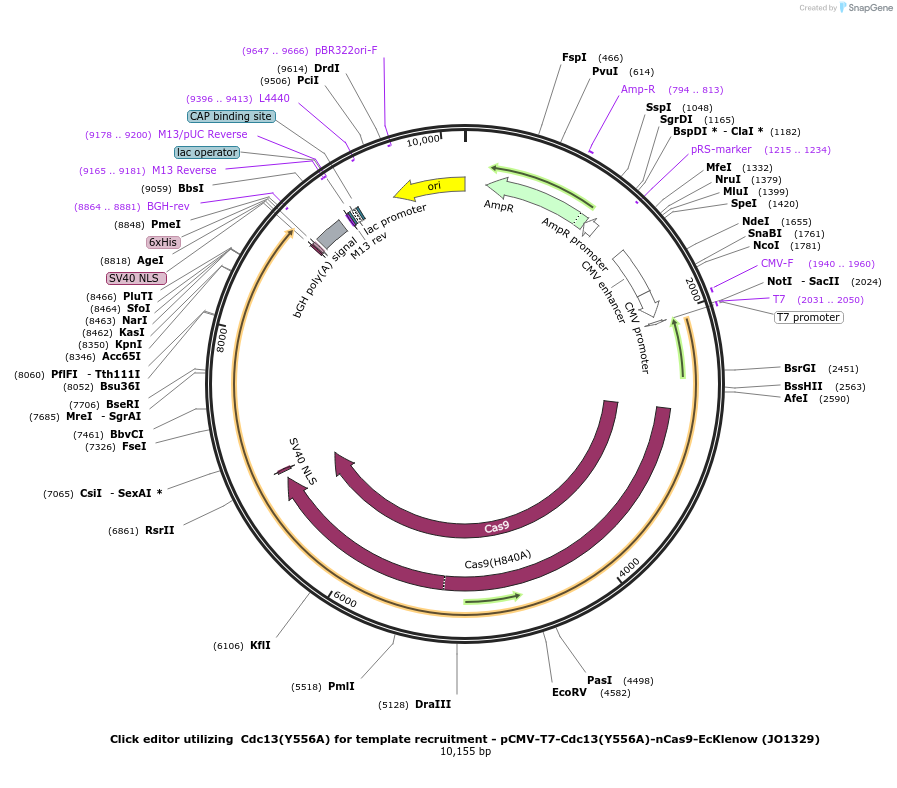
Click editor utilizing Cdc13(Y556A) for template recruitment - pCMV-T7-Cdc13(Y556A)-nCas9-EcKlenow (JO1329)
Plasmid#217797PurposeVariant CE1 construct with Cdc13(Y556A) telomere binding protein for template recruitment, expressed from CMV or T7 promoters.DepositorInsertCdc13(Y556A)-XTEN-nSpCas9-BPNLS-EcKlenow(-exo)-BPNLS
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLSExpressionMammalianMutationCdc13(Y556A); nSpCas9(H840A); EcKlenow(-exo;D355A…PromoterCMV and T7Available SinceSept. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
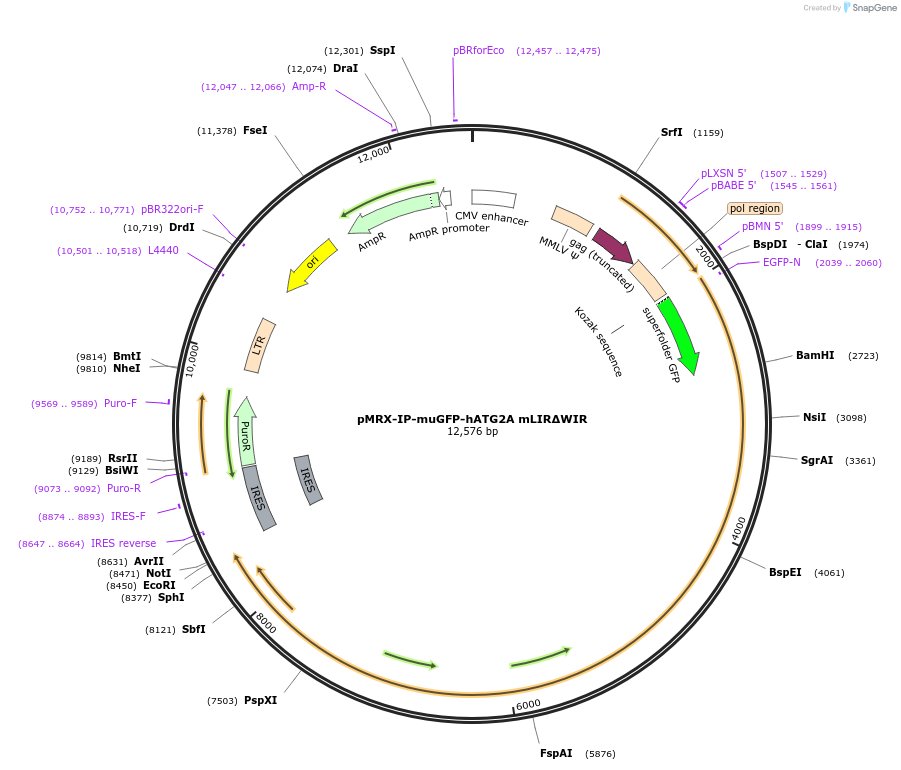
pMRX-IP-muGFP-hATG2A mLIRΔWIR
Plasmid#215508PurposeExpresses GFP tagged human ATG2A which has the mutation in LC3 interacting region and lacks WIPI interacting region.DepositorInsertAutophagy related 2A (ATG2A Human)
UseRetroviralTagsmuGFPExpressionMammalianMutationP656R (natural variant with no functional relevan…Available SinceJune 17, 2024AvailabilityAcademic Institutions and Nonprofits only -
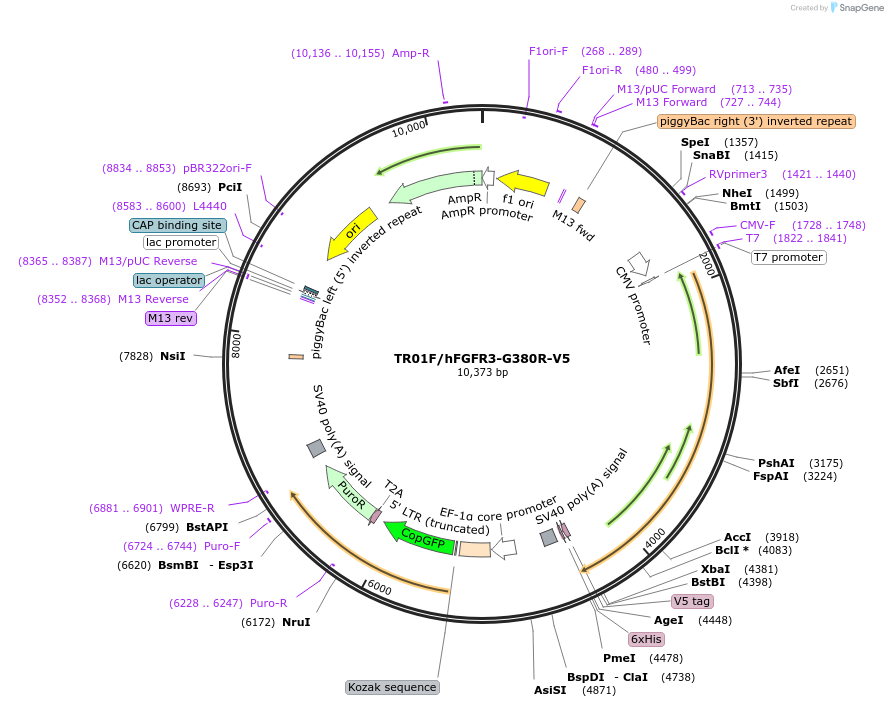
TR01F/hFGFR3-G380R-V5
Plasmid#214908Purposefor PiggyBac mediated integration and stable expression of hFGFR3-G380R variant that is associated with AchondroplasiaDepositorInsertFGFR3 (FGFR3 Human)
TagsV5/HisExpressionMammalianMutationG380R substitutionPromotertruncated CMV promoterAvailable SinceApril 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
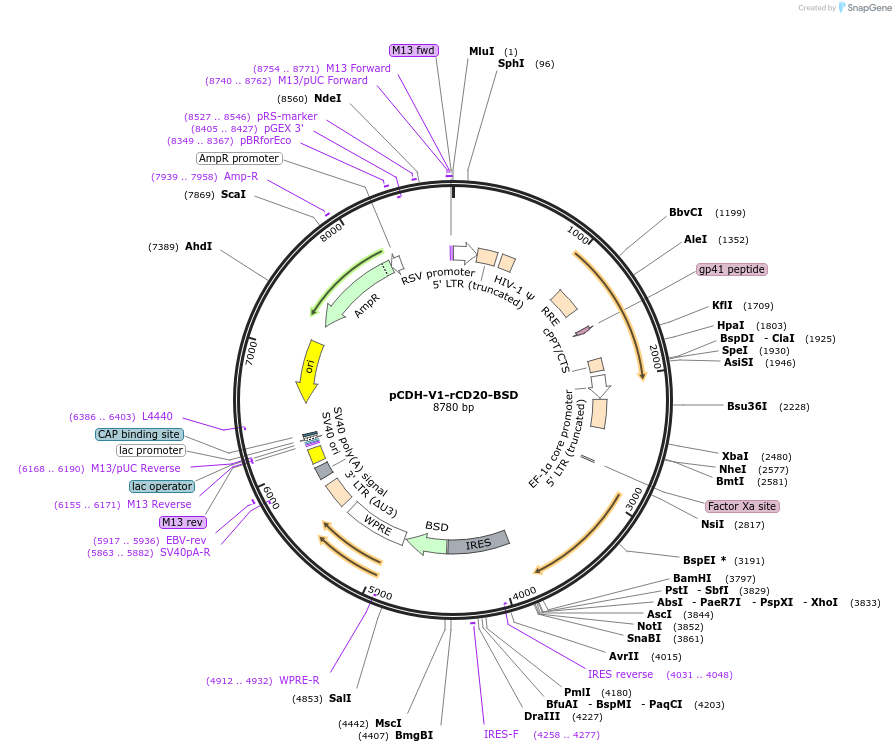
pCDH-V1-rCD20-BSD
Plasmid#209752PurposeTo reconstitute CD20 expression with a specific mRNA isoform (variant 1) after CRISPR-Cas9-mediated knockout of CD20.DepositorInsertMS4A1 (MS4A1 Human)
UseLentiviralMutationThe CCTGGGGGGTCTTCTGATGATCC sequence within the C…Available SinceNov. 30, 2023AvailabilityAcademic Institutions and Nonprofits only -
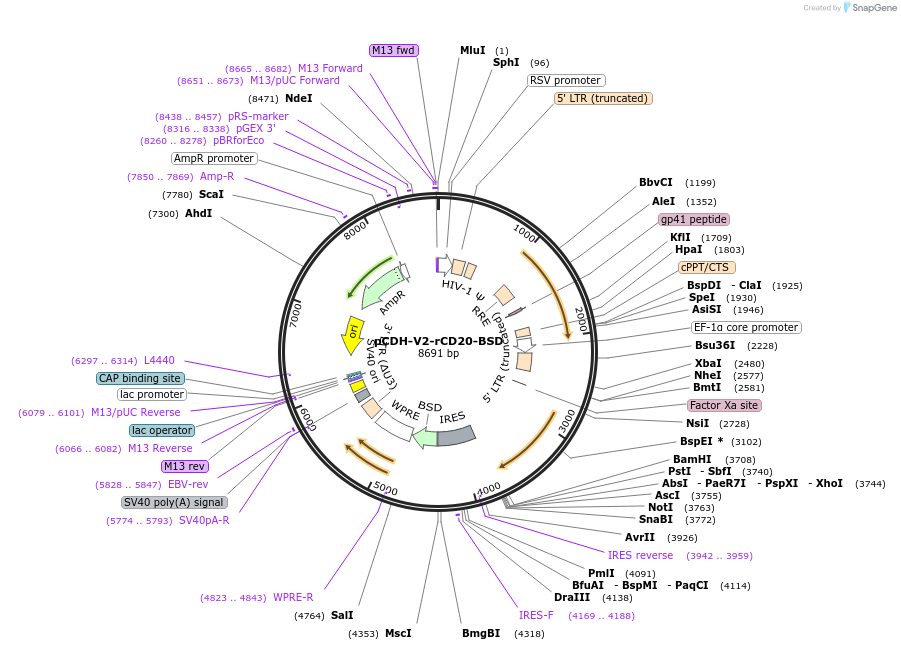
pCDH-V2-rCD20-BSD
Plasmid#209753PurposeTo reconstitute CD20 expression with a specific mRNA isoform (variant 2) after CRISPR-Cas9-mediated knockout of CD20.DepositorInsertMS4A1 (MS4A1 Human)
UseLentiviralMutationThe CCTGGGGGGTCTTCTGATGATCC sequence within the C…Available SinceNov. 30, 2023AvailabilityAcademic Institutions and Nonprofits only -
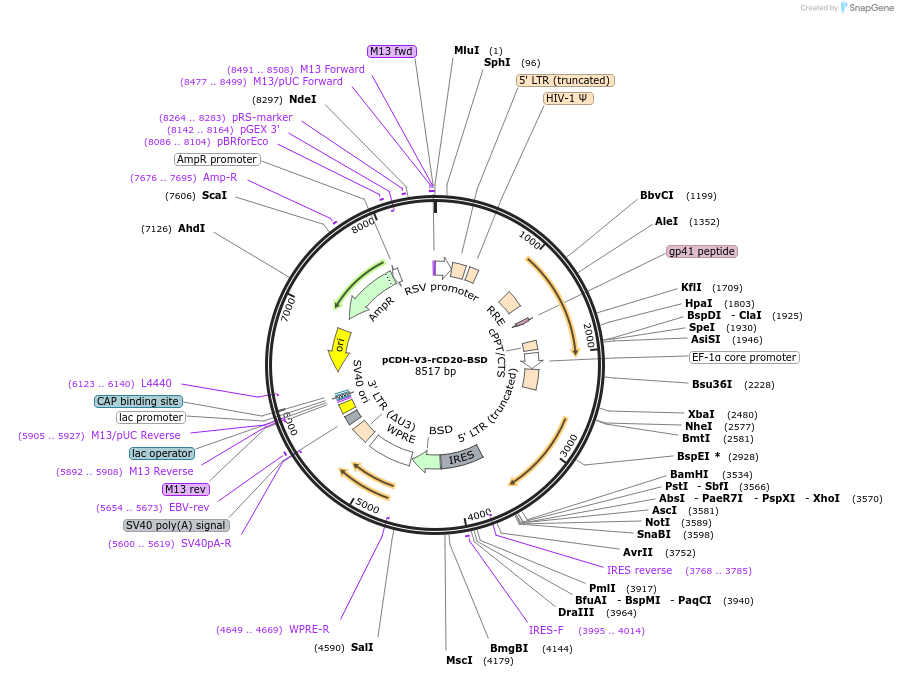
pCDH-V3-rCD20-BSD
Plasmid#209754PurposeTo reconstitute CD20 expression with a specific mRNA isoform (variant 3) after CRISPR-Cas9-mediated knockout of CD20.DepositorInsertMS4A1 (MS4A1 Human)
UseLentiviralMutationThe CCTGGGGGGTCTTCTGATGATCC sequence within the C…Available SinceNov. 30, 2023AvailabilityAcademic Institutions and Nonprofits only -
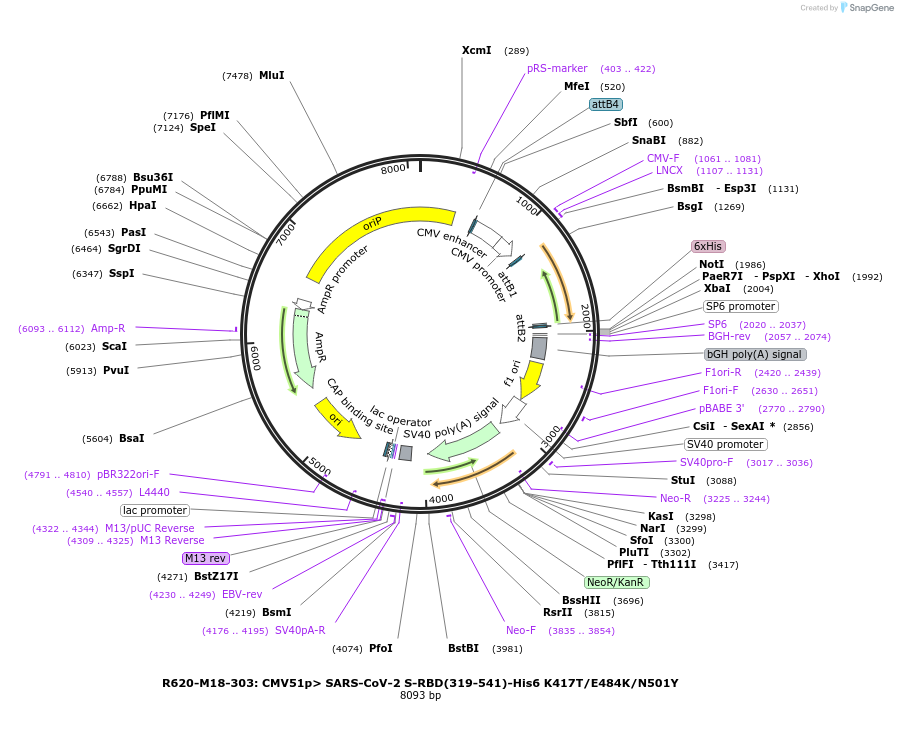
R620-M18-303: CMV51p> SARS-CoV-2 S-RBD(319-541)-His6 K417T/E484K/N501Y
Plasmid#170202PurposeMammalian expression of SARS-CoV-2 spike receptor-binding domain (RBD), N501Y mutant from B.1.1.28 variantDepositorInsertSARS-CoV-2 RBD (S SARS-CoV-2)
TagsHis6ExpressionMammalianMutationK417T, E484K, N501YPromoterCMV51Available SinceMay 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
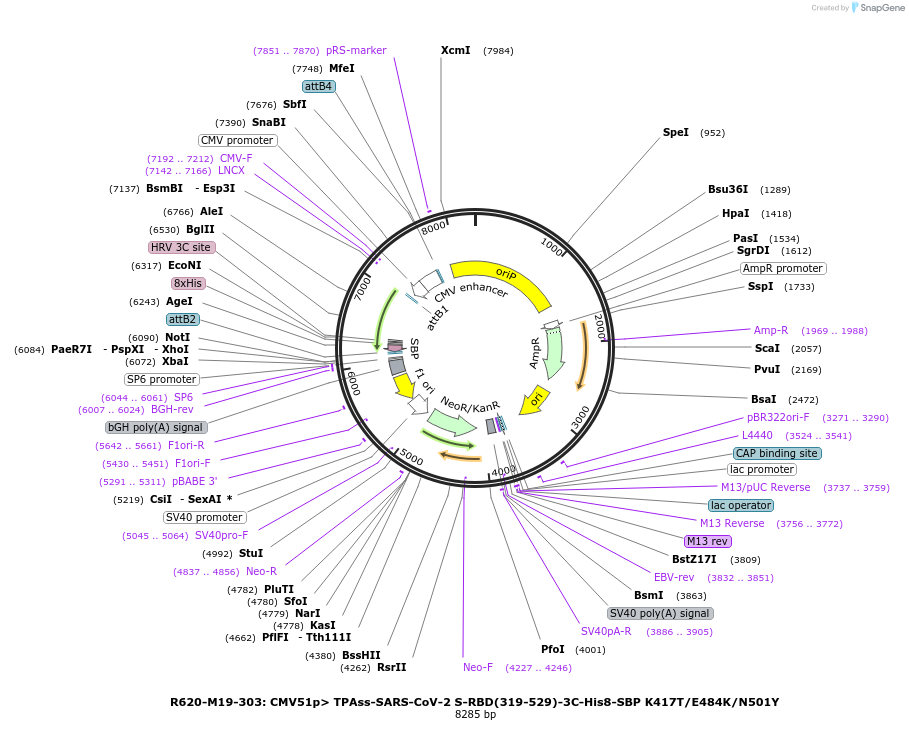
R620-M19-303: CMV51p> TPAss-SARS-CoV-2 S-RBD(319-529)-3C-His8-SBP K417T/E484K/N501Y
Plasmid#170203PurposeMammalian expression of SARS-CoV-2 spike receptor-binding domain (RBD), N501Y mutant from B.1.1.28 variantDepositorInsertSARS-CoV-2 RBD (S SARS-CoV-2)
Tags3C-His8-SBPExpressionMammalianMutationK417T, E484K, N501YPromoterCMV51Available SinceMay 11, 2022AvailabilityAcademic Institutions and Nonprofits only