We narrowed to 4,598 results for: golden gate
-
Plasmid#245592PurposeLevel 0 Golden Gate part C-Terminal Tag with overhangs TTCG - GCTTDepositorInsertC-Terminal tag eGFP
UseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutation, also c…Available SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
pICSL01008_CP
Plasmid#245690PurposeLevel 0 Golden Gate Promoter + 5' UTR acceptor with AmilCP negative selection cassette GGAG - CCATDepositorTypeEmpty backboneUseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
pICSL01002_CP
Plasmid#245687PurposeLevel 0 Golden Gate N-terminal tag acceptor with AmilCP negative selection cassette CCAT - AATGDepositorTypeEmpty backboneUseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
pICSL50005
Plasmid#245578PurposeLevel 0 Golden Gate part C-Terminal Tag with overhangs TTCG - GCTTDepositorInsertC-Terminal tag YFP tag (yellow variant of GFP)
UseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
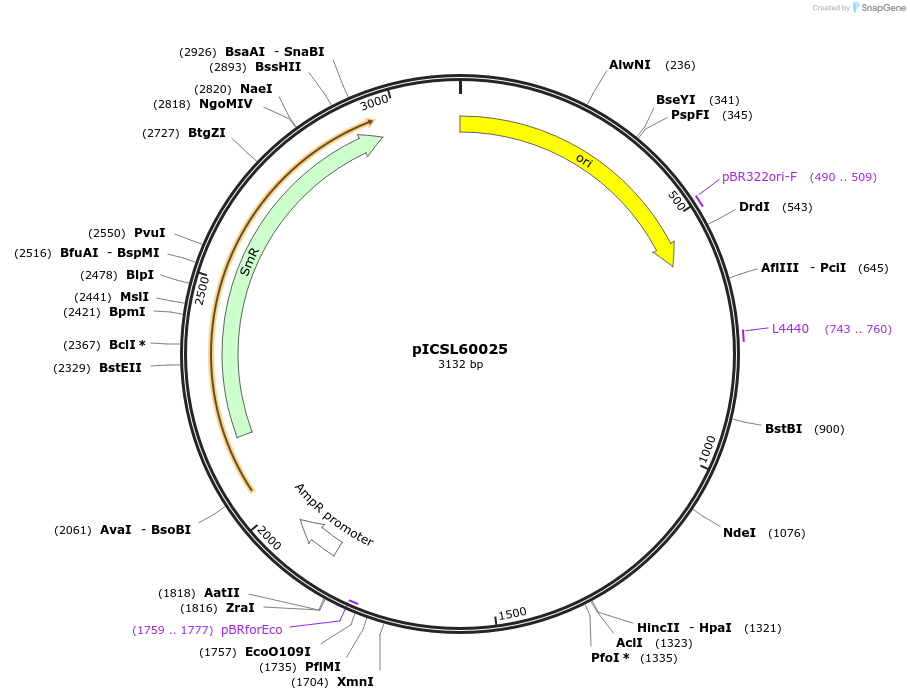
pICSL60025
Plasmid#245673PurposeLevel 0 Golden Gate part Rb7MAR (D144-437 variant) terminator, GCTT - CGCTDepositorInsertRb7MAR (D144-437 variant) terminator
UseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
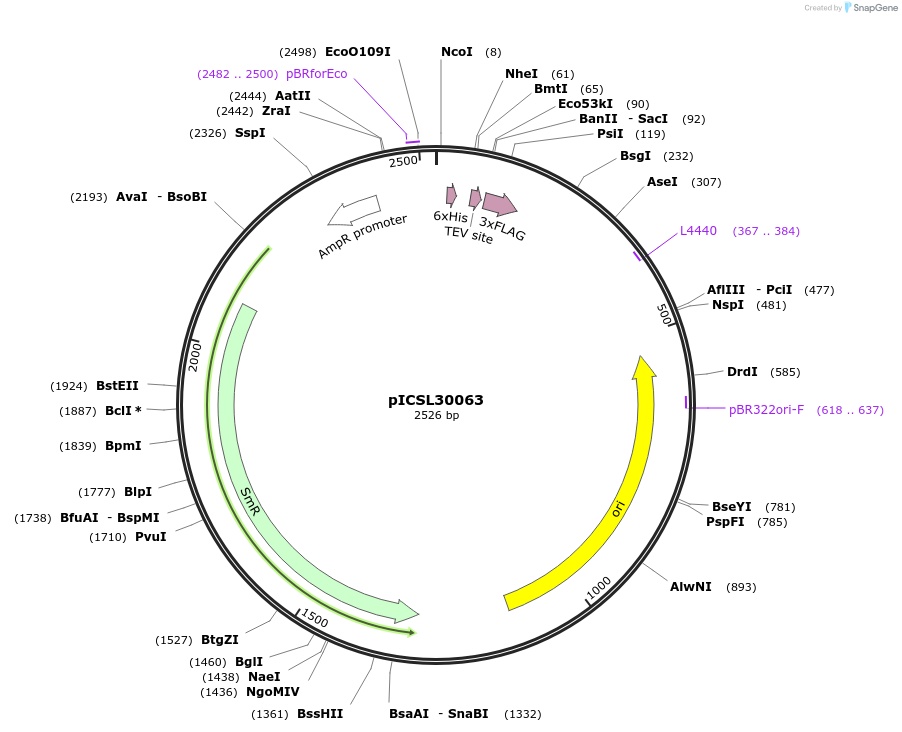
pICSL30063
Plasmid#245566PurposeLevel 0 Golden Gate part N-Terminal Tag with overhangs CCAT - AATGDepositorInsertN-Terminal tag Self-Cleaving HF tag compatible with ProteinSelect purification system
UseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
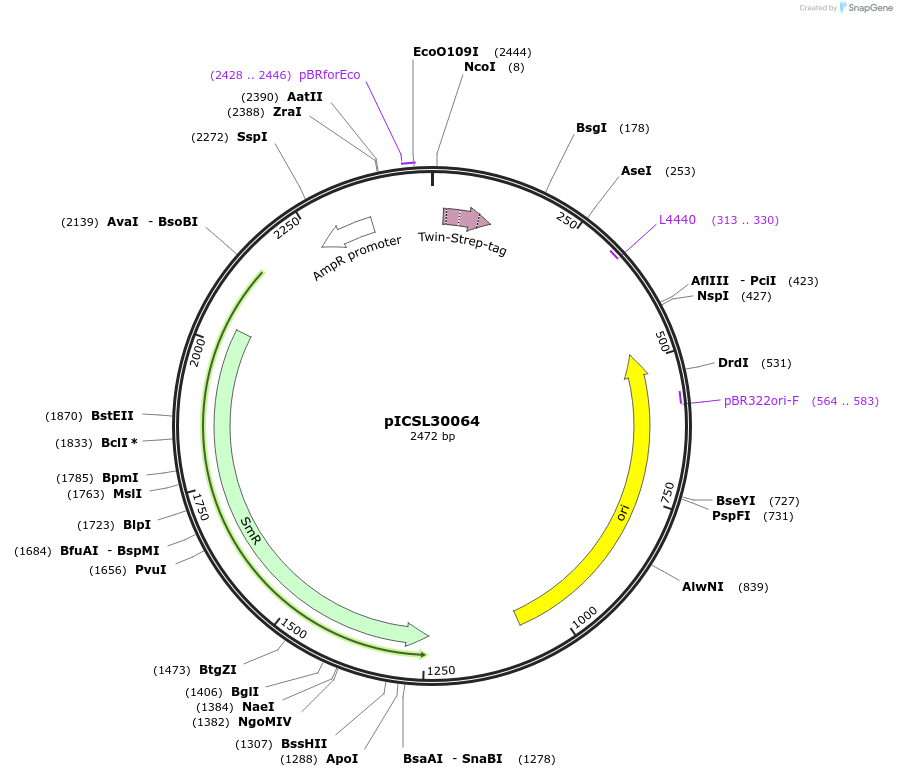
pICSL30064
Plasmid#245567PurposeLevel 0 Golden Gate part N-Terminal Tag with overhangs CCAT - AATGDepositorInsertN-Terminal tag Self-Cleaving STREPII tag compatible with ProteinSelect purification system
UseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
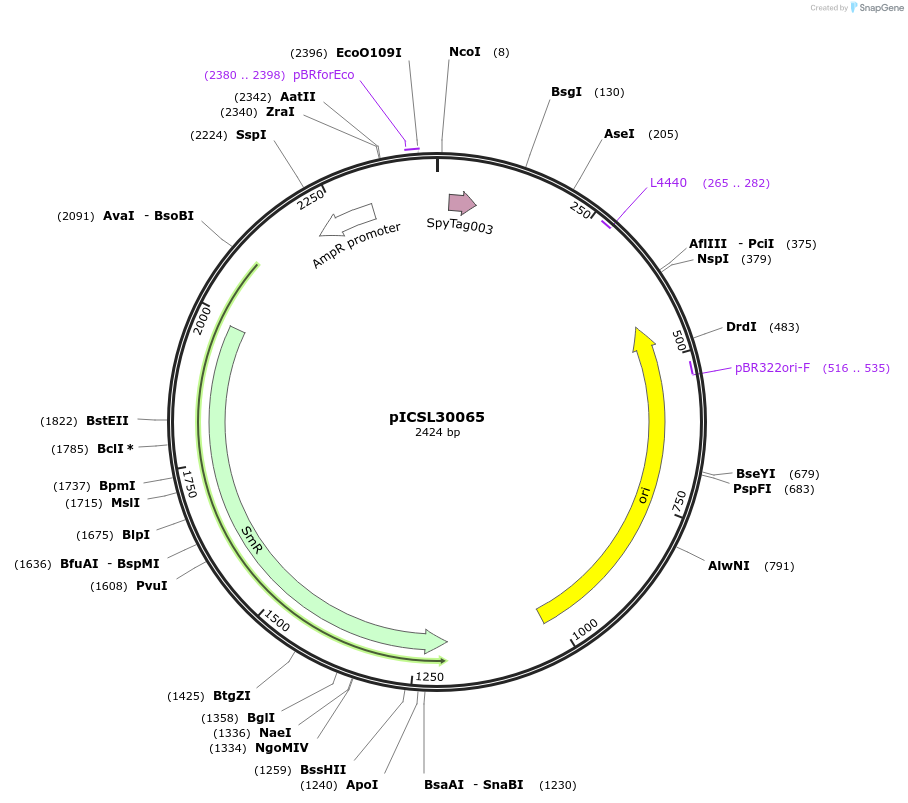
pICSL30065
Plasmid#245568PurposeLevel 0 Golden Gate part N-Terminal Tag with overhangs CCAT - AATGDepositorInsertN-Terminal tag Self-Cleaving SPY tag compatible with ProteinSelect purification system
UseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
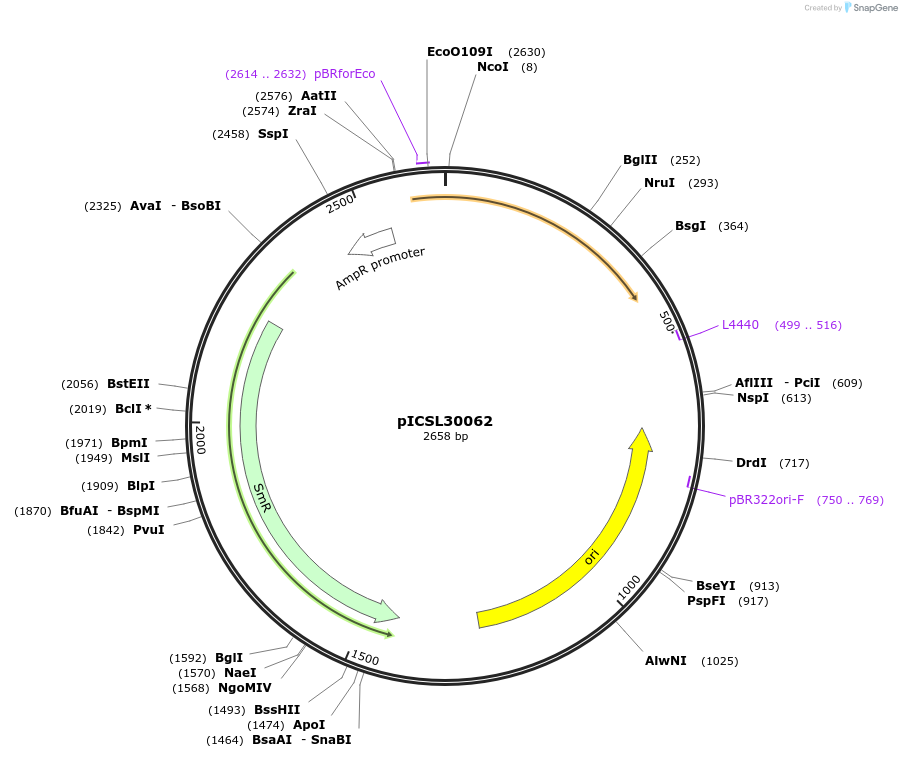
pICSL30062
Plasmid#245565PurposeLevel 0 Golden Gate part N-Terminal Tag with overhangs CCAT - AATGDepositorInsertN-Terminal tag Self-Cleaving SUMO tag compatible with ProteinSelect purification system
UseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
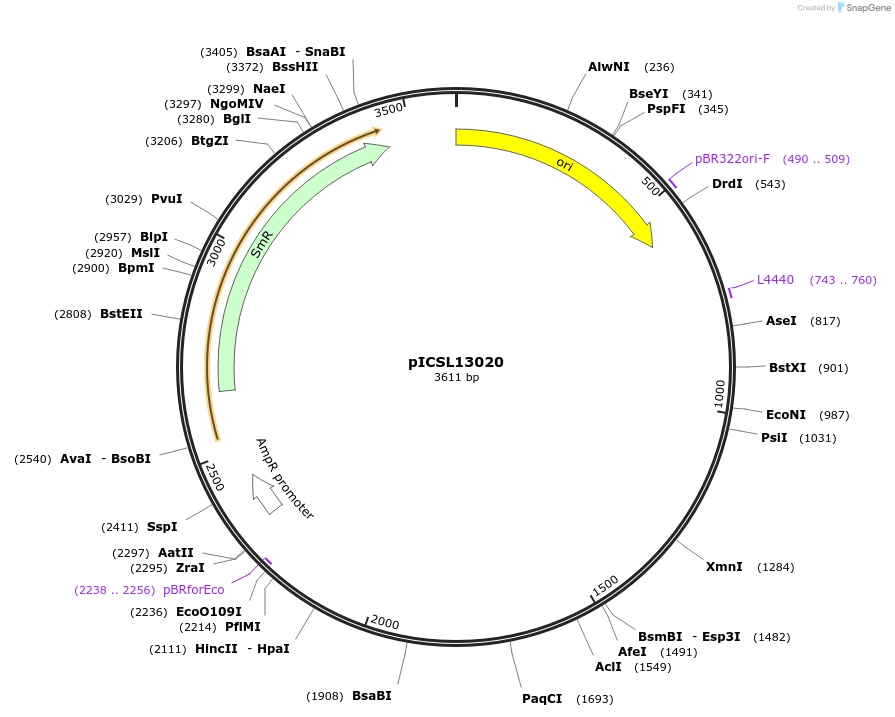
pICSL13020
Plasmid#245664PurposeLevel 0 Golden Gate part EC1.2-EN-EC1.1 Enhancer Fusion promoter, GGAG - CCATDepositorInsertEC1.2-EN-EC1.1 Enhancer Fusion promoter
UseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
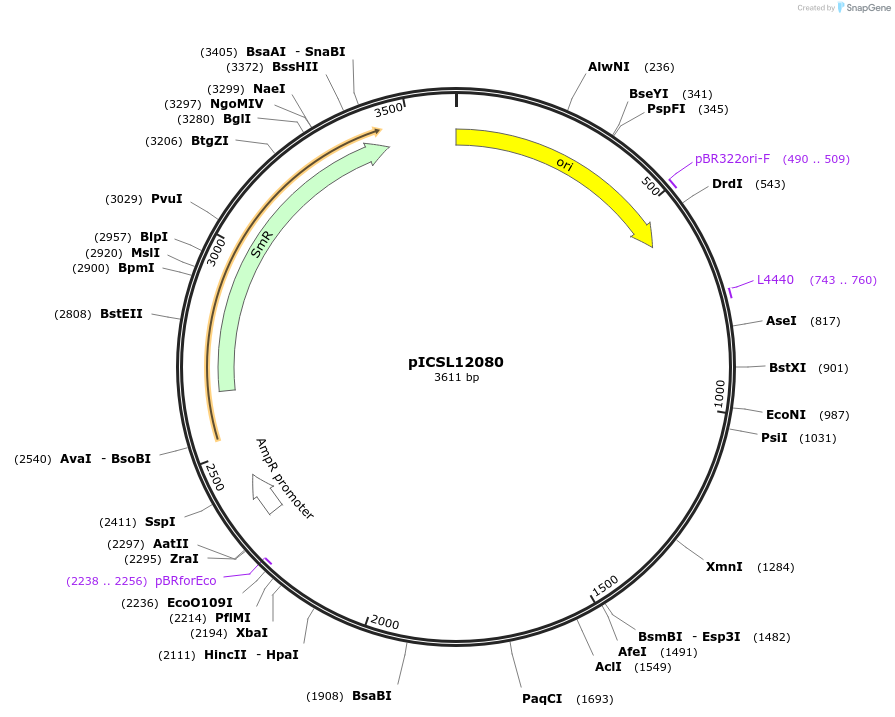
pICSL12080
Plasmid#245650PurposeLevel 0 Golden Gate part EC1.2-EN-EC1.1 Enhancer Fusion promoter, GGAG - AATGDepositorInsertEC1.2-EN-EC1.1 Enhancer Fusion promoter
UseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
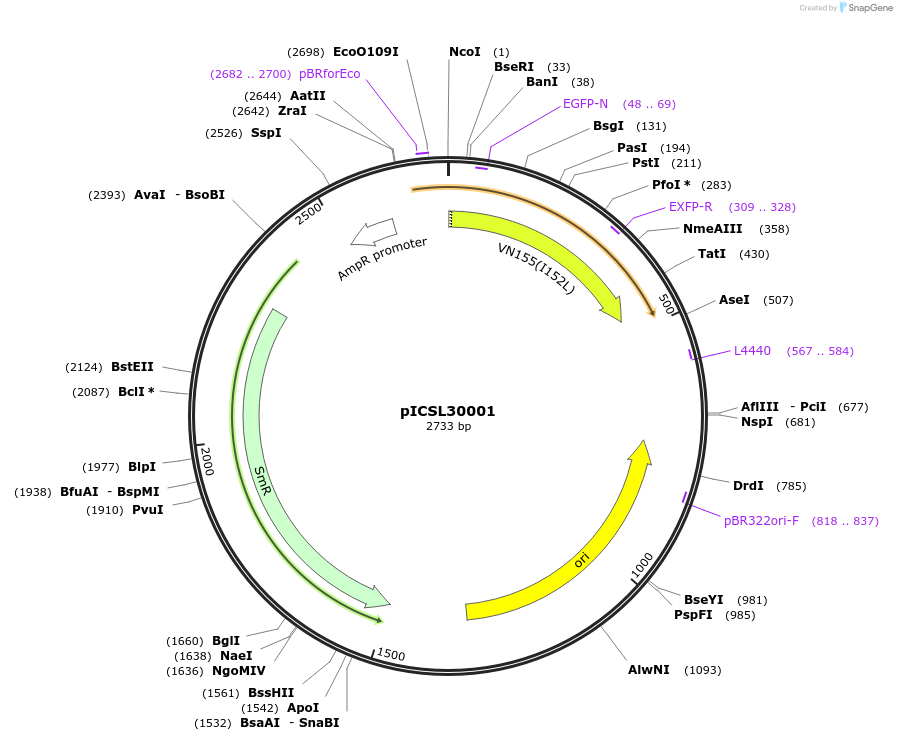
pICSL30001
Plasmid#245529PurposeLevel 0 Golden Gate part N-Terminal Tag with overhangs CCAT - AATGDepositorInsertN-Terminal tag SplitYFP tag- N terminus of YFP
UseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
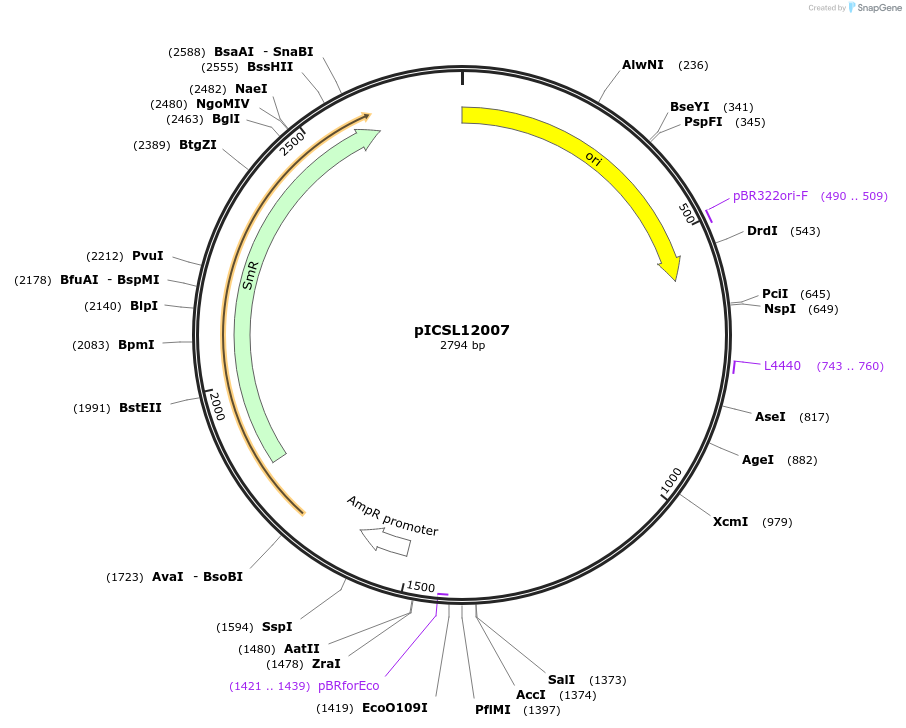
pICSL12007
Plasmid#245628PurposeLevel 0 Golden Gate part pAt3: Cysteine Synthase (At3g61440) promoter, GGAG - AATGDepositorInsertpAt3: Cysteine Synthase (At3g61440) promoter
UseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
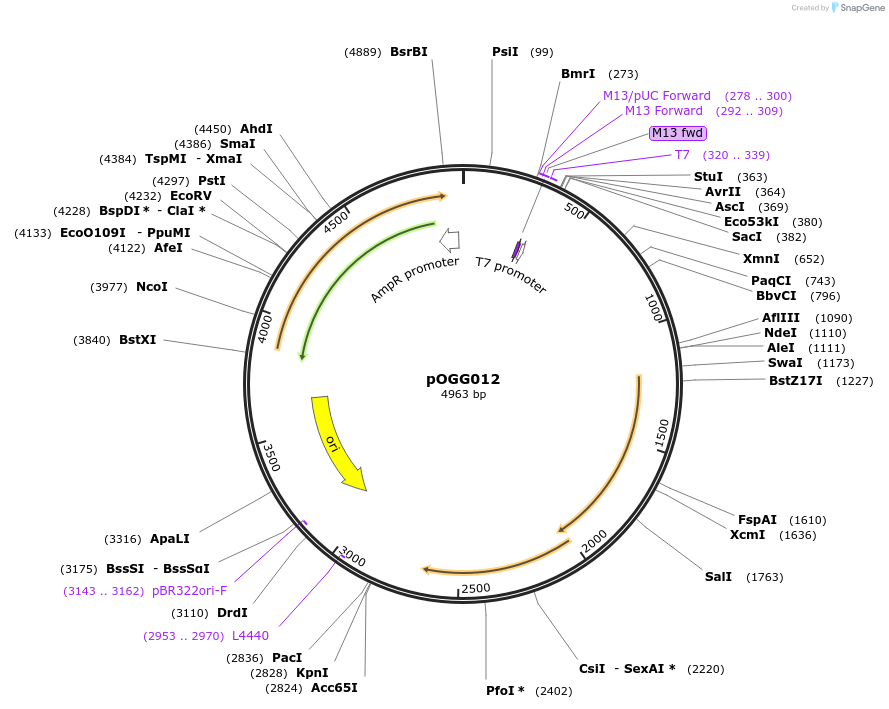
pOGG012
Plasmid#113986PurposeVector construction module. Partition genes (parABCDE) for plasmid stability in absence of antibiotic selection for construction of Golden Gate cloning vectorsDepositorInsertPartition genes (parABCDE)
ExpressionBacterialMutationDomesticated for Golden Gate cloningAvailable SinceJan. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
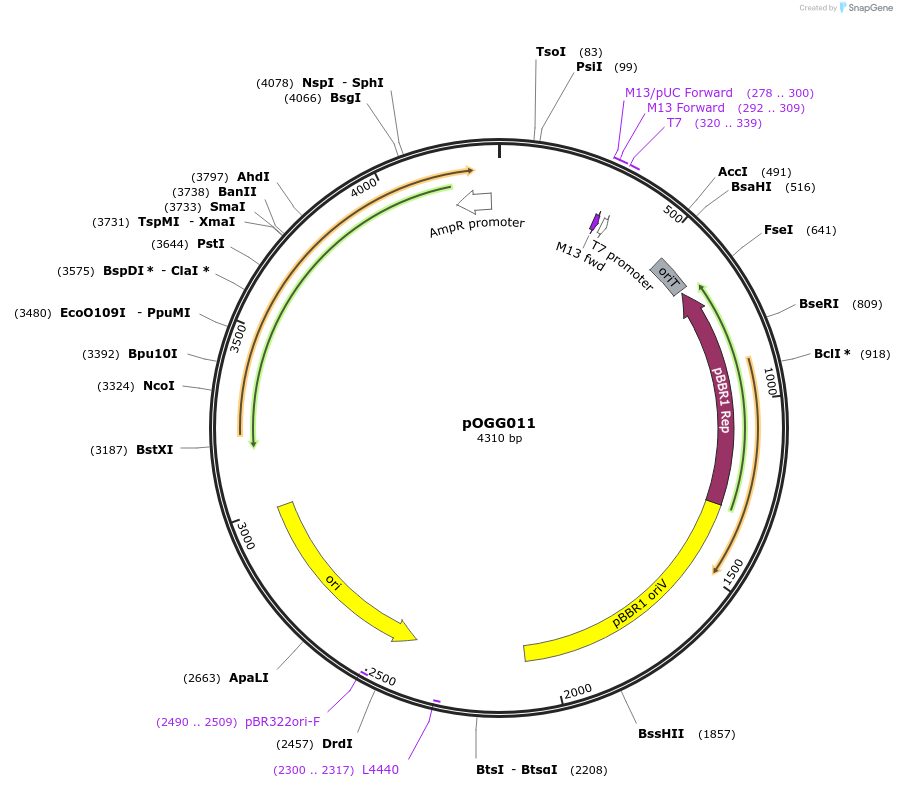
pOGG010
Plasmid#113984PurposeVector construction module. RK2 (IncP) low-copy number origin of replication for construction of Golden Gate cloning vectorsDepositorInsertRK2 (IncP)
ExpressionBacterialMutationDomesticated for Golden Gate cloningAvailable SinceJan. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
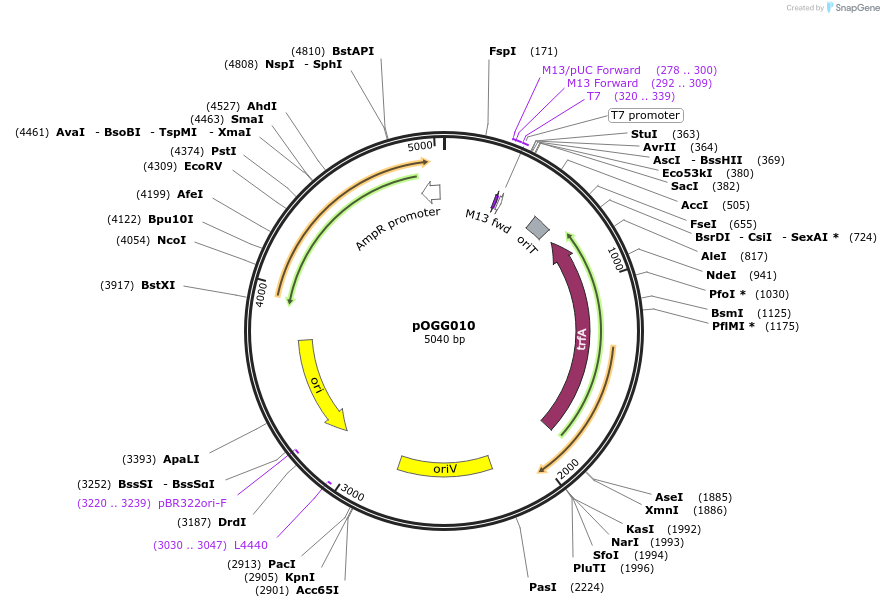
pOGG011
Plasmid#113985PurposeVector construction module. pBBR1 medium-copy number origin of replication for construction of Golden Gate cloning vectorsDepositorInsertpBBR1
ExpressionBacterialMutationDomesticated for Golden Gate cloningAvailable SinceJan. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
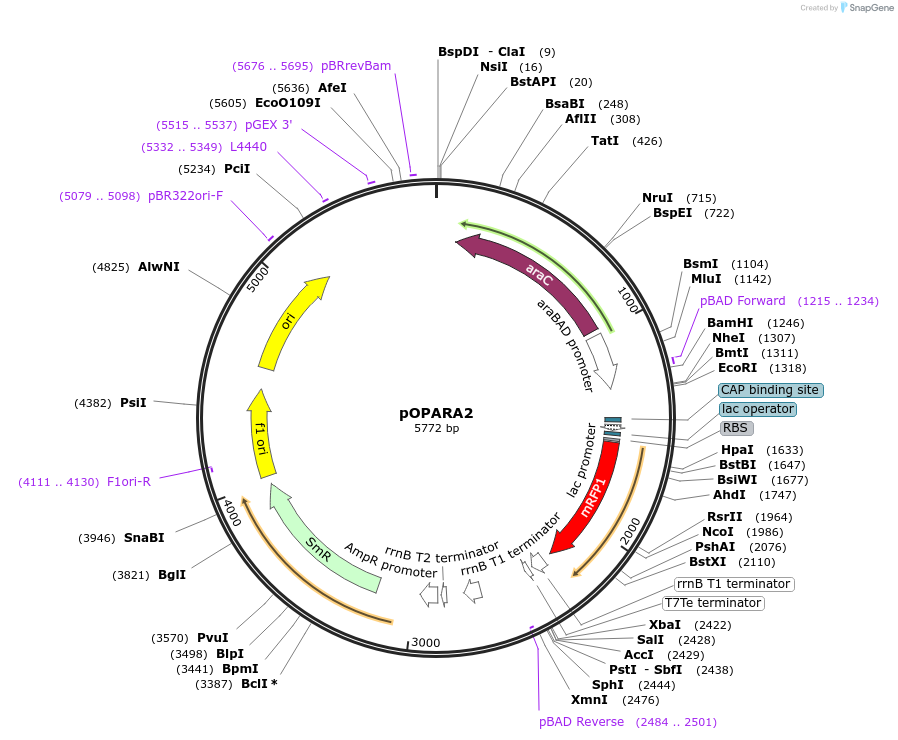
pOPARA2
Plasmid#200417PurposeGolden Gate assembly of expression constructs containing pBAD promoter (spectinomycin resistance)DepositorArticleTypeEmpty backboneExpressionBacterialPromoterpBADAvailable SinceMay 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
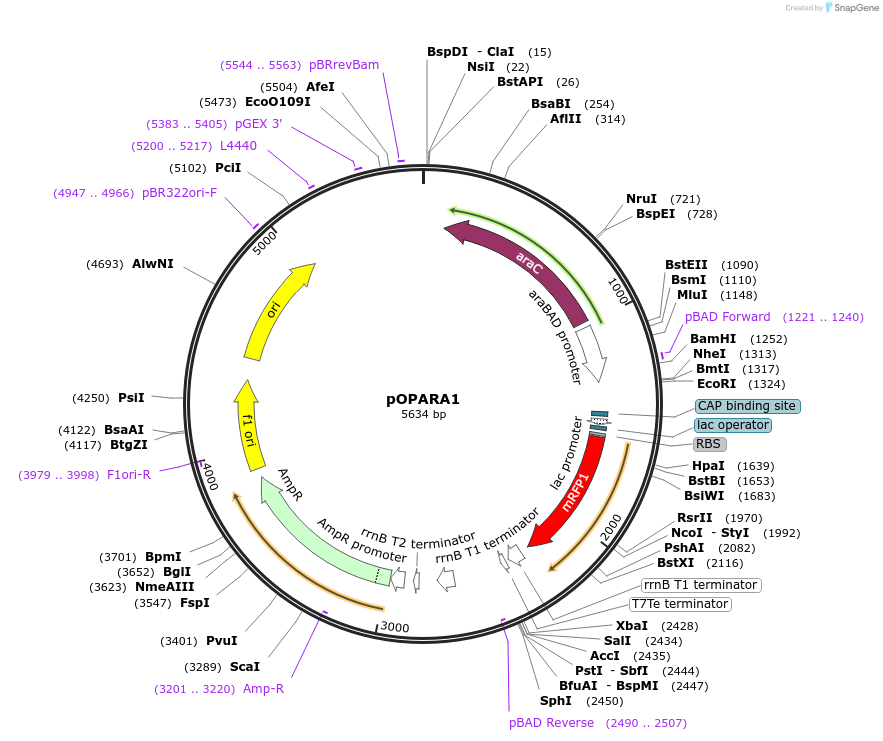
pOPARA1
Plasmid#200416PurposeGolden Gate assembly of expression constructs containing pBAD promoter (carbenicillin resistance)DepositorArticleTypeEmpty backboneExpressionBacterialPromoterpBADAvailable SinceMay 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
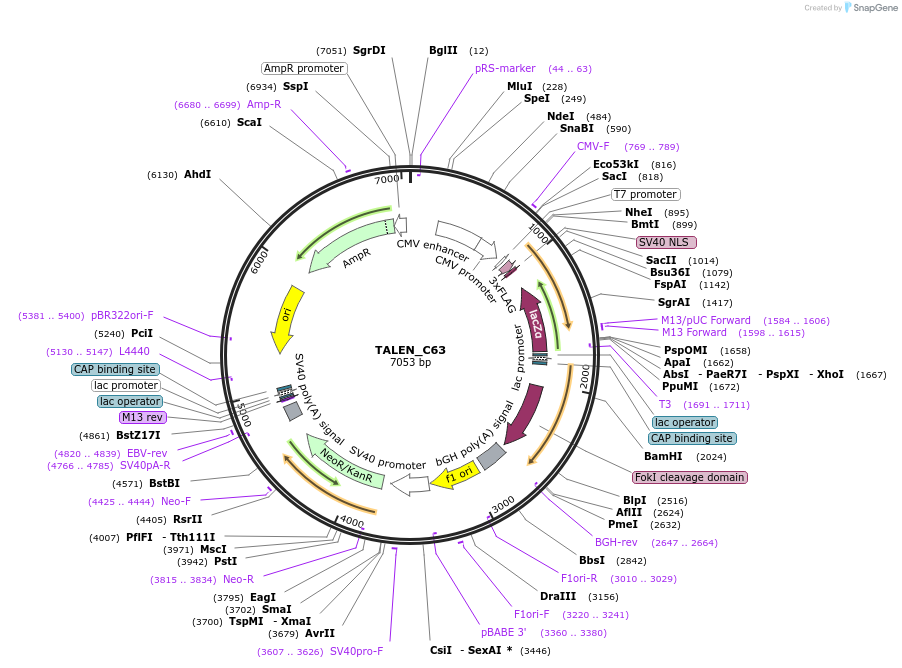
TALEN_C63
Plasmid#40788PurposeGolden Gate Compatible TALEN Construct for site specific modification of genome, +63 C Terminus, Codon Optimized FokIDepositorInsertTranscription Activator Like Effector Nuclease
UseTALENExpressionMammalianMutation+63 C Terminus, Codon Optimized FokI EndonucleasePromoterCMVAvailable SinceDec. 12, 2013AvailabilityIndustry, Academic Institutions, and Nonprofits -
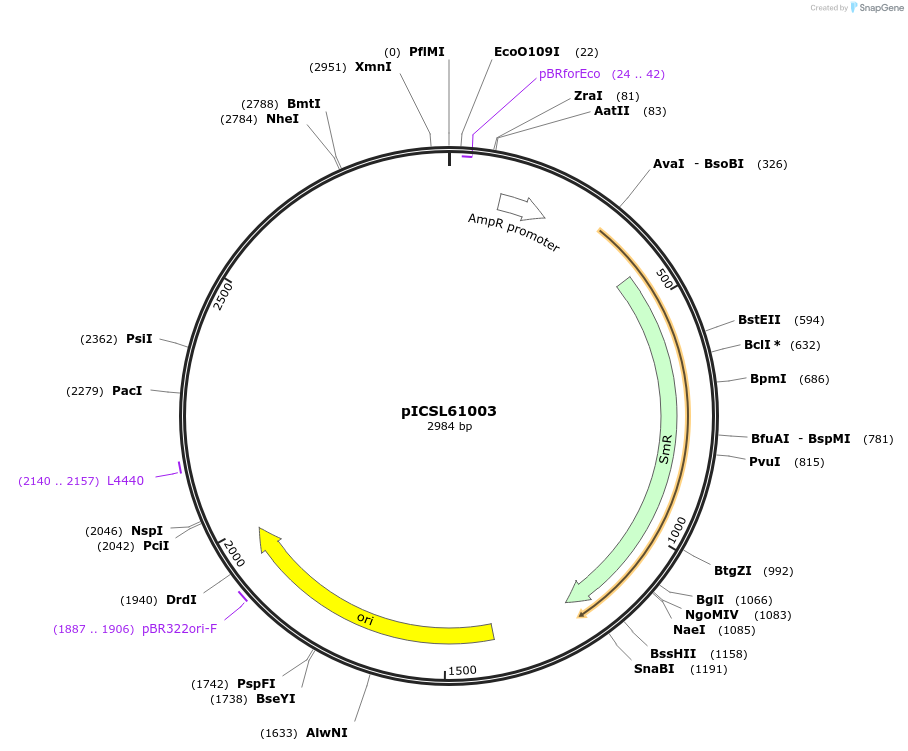
pICSL61003
Plasmid#245680PurposeLevel 0 Golden Gate part 3' Extensin-Full length-IEU (TerP1) terminator P1, GCTT - TAGADepositorInsert3' Extensin-Full length-IEU (TerP1) terminator P1
UseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceDec. 18, 2025AvailabilityAcademic Institutions and Nonprofits only