We narrowed to 23,252 results for: Sele
-
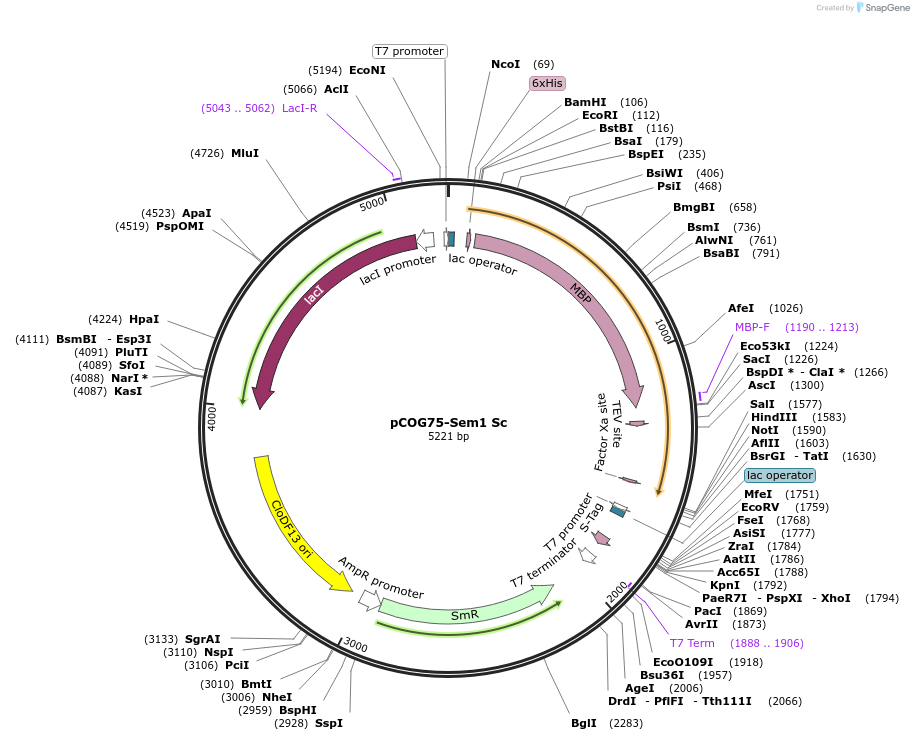
Plasmid#83233PurposepCDF-His6-MBP-Sem1 ScDepositorAvailable SinceNov. 22, 2016AvailabilityAcademic Institutions and Nonprofits only
-
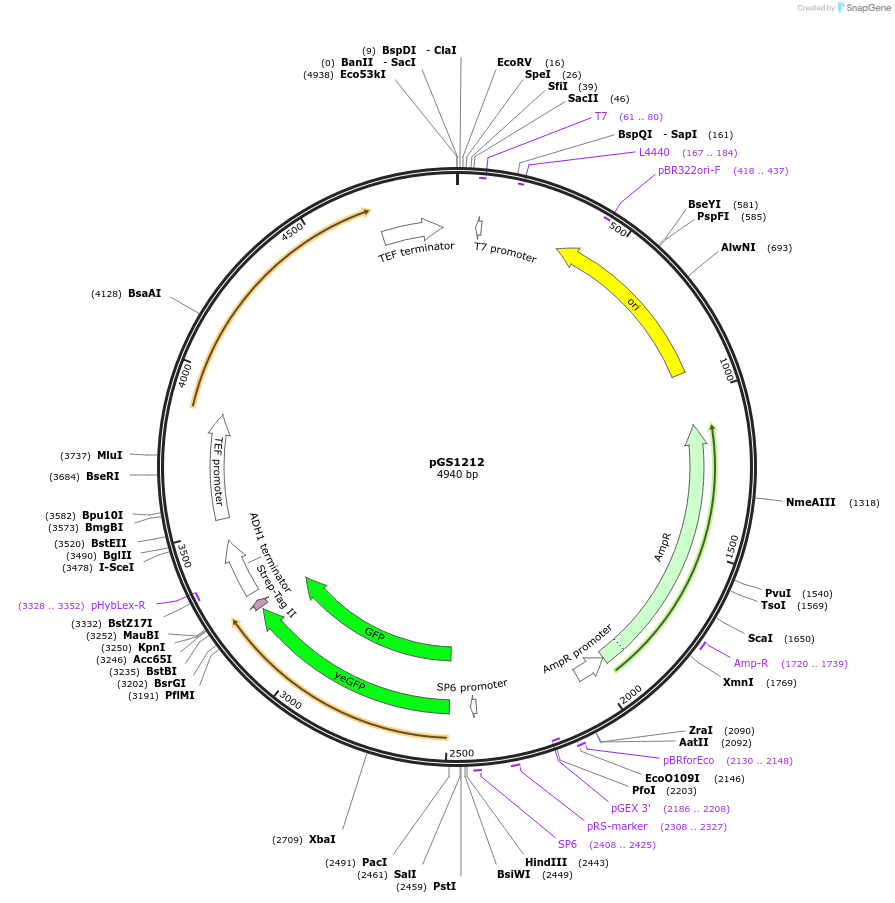
pGS1212
Plasmid#63785PurposeVector to create a cassette for homologous recombination in S. cerevisiae; adds a C-terminal eGFP Strep-tag II followed by URA3 selection markerDepositorInserteGFP-Strep-tag II followed by URA3 selection marker
UseE. coli cloning vectorTagseGFP-Strep-tag IIPromoternoneAvailable SinceApril 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
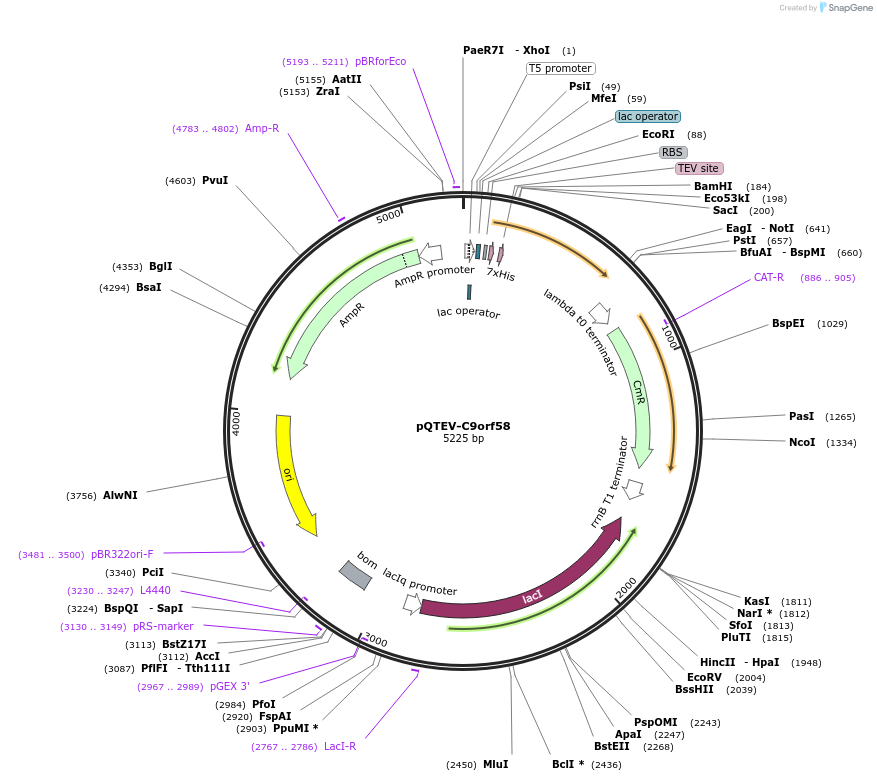
pQTEV-C9orf58
Plasmid#34819DepositorAvailable SinceMarch 22, 2013AvailabilityAcademic Institutions and Nonprofits only -
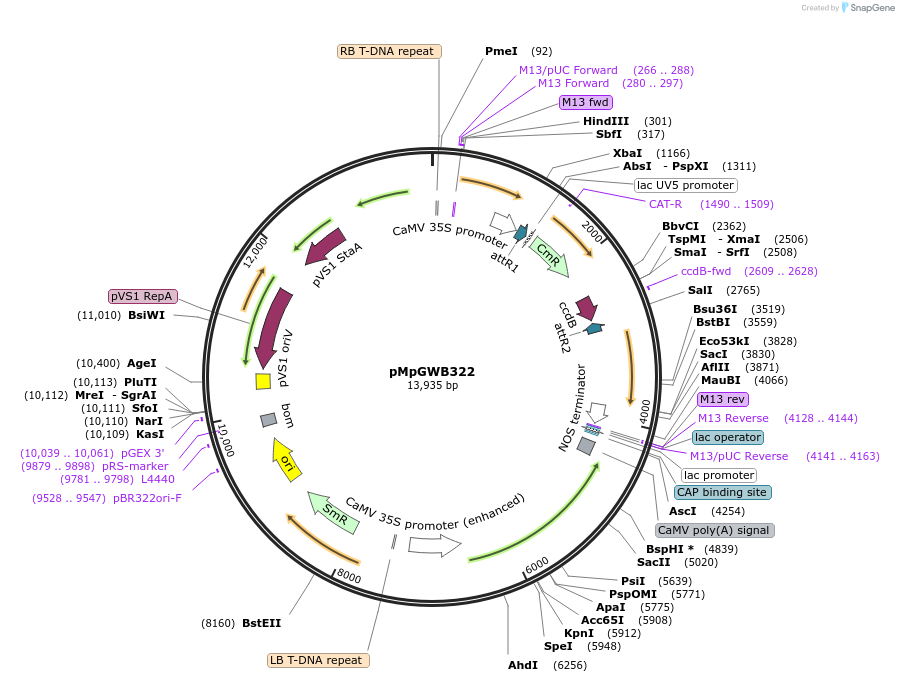
pMpGWB322
Plasmid#68650PurposeGateway binary vector designed for transgenic research with Marchantia polymorpha as well as other plantsDepositorTypeEmpty backboneTagsModified EAR motif plant-specific repression doma…ExpressionPlantPromoterCauliflower mosaic virus 35S promoterAvailable SinceJune 24, 2016AvailabilityAcademic Institutions and Nonprofits only -
HOXB4-FLAG-WG-SP65
Plasmid#21087DepositorInsertHOXB4 (HOXB4 Human)
UseIn vitro t/tMutationW to G mutation in YPWM Pbx1 interaction motif, p…Available SinceNov. 10, 2009AvailabilityAcademic Institutions and Nonprofits only -
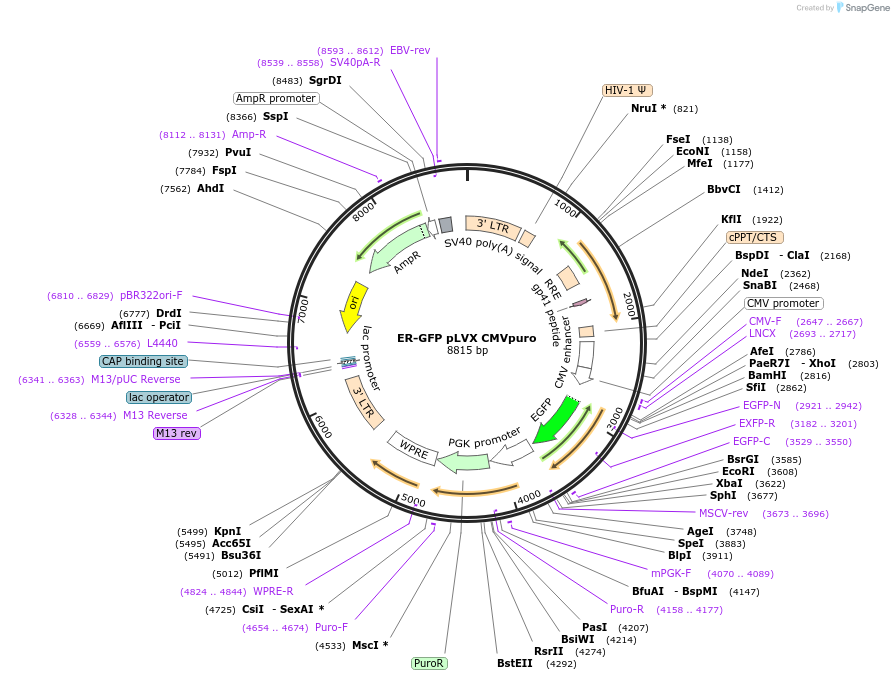
ER-GFP pLVX CMVpuro
Plasmid#246912PurposeLentiviral-based expression of EGFP with an ER retention signal sequence in mammalian cellsDepositorInsertEGFP
UseLentiviralExpressionMammalianPromoterCMVAvailable SinceNov. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
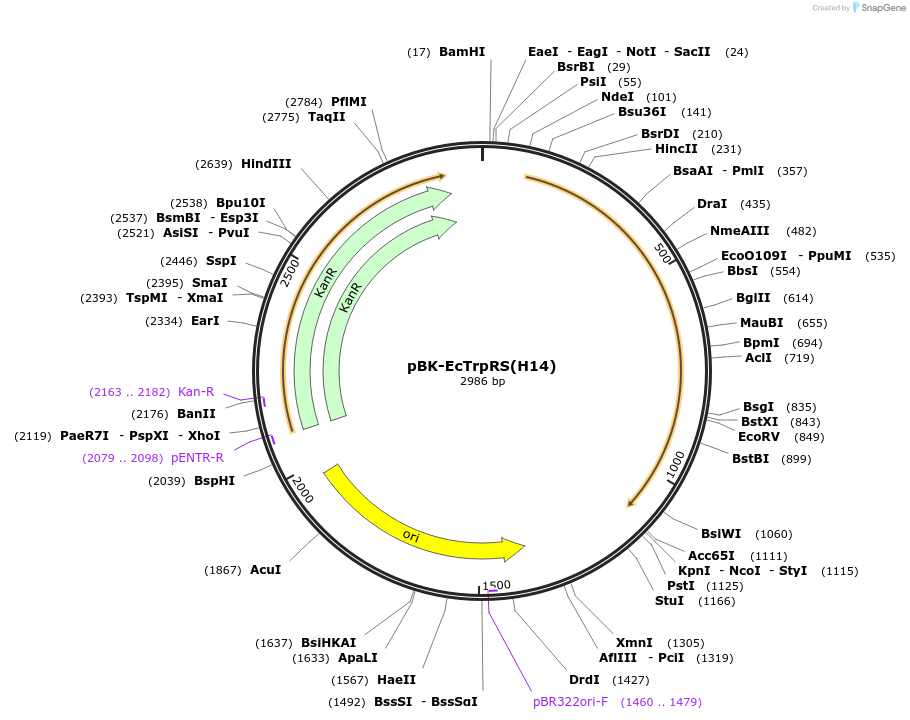
pBK-EcTrpRS(H14)
Plasmid#231129PurposeExpresses mutant E. coli TrpRS from a weak glnS promoter for proteome-wide incorporation of tryptophan analogues.DepositorInsertE. coli tryptophanyl tRNA synthetase
ExpressionBacterialMutationS8A, V144G, V146CAvailable SinceOct. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
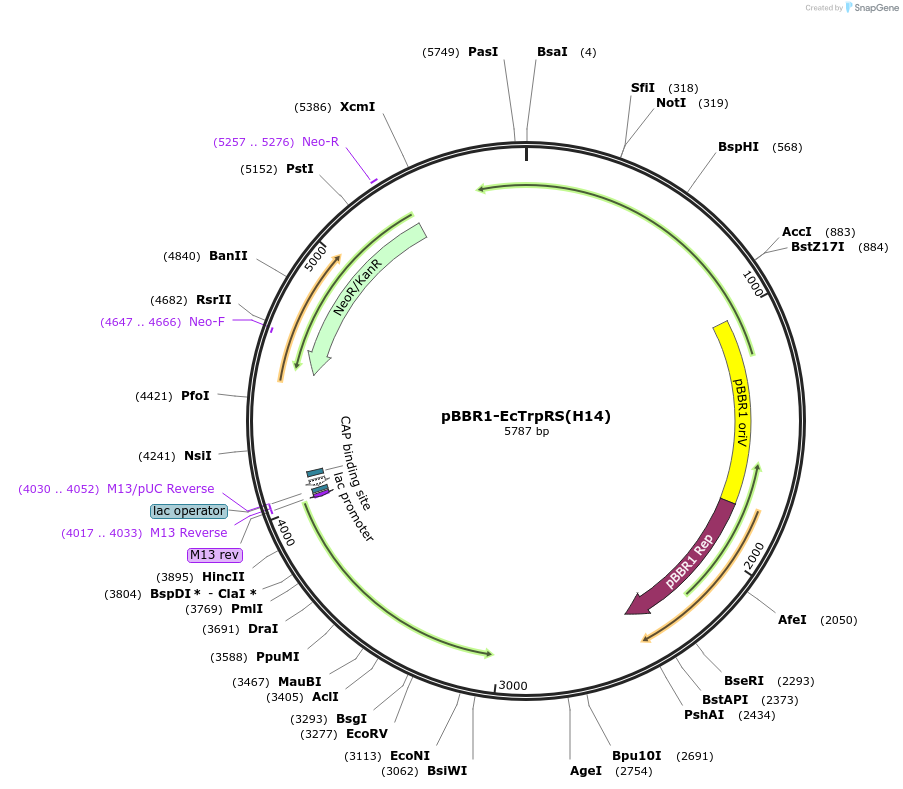
pBBR1-EcTrpRS(H14)
Plasmid#231131PurposeExpresses mutant E. coli TrpRS from a derepressed lac promoter for proteome-wide incorporation of tryptophan analogues in E. coli/K. pneumoniae.DepositorInsertE. coli tryptophanyl tRNA synthetase
UseKlebsiella pneumoniae expressionExpressionBacterialMutationS8A, V144G, V146CAvailable SinceOct. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
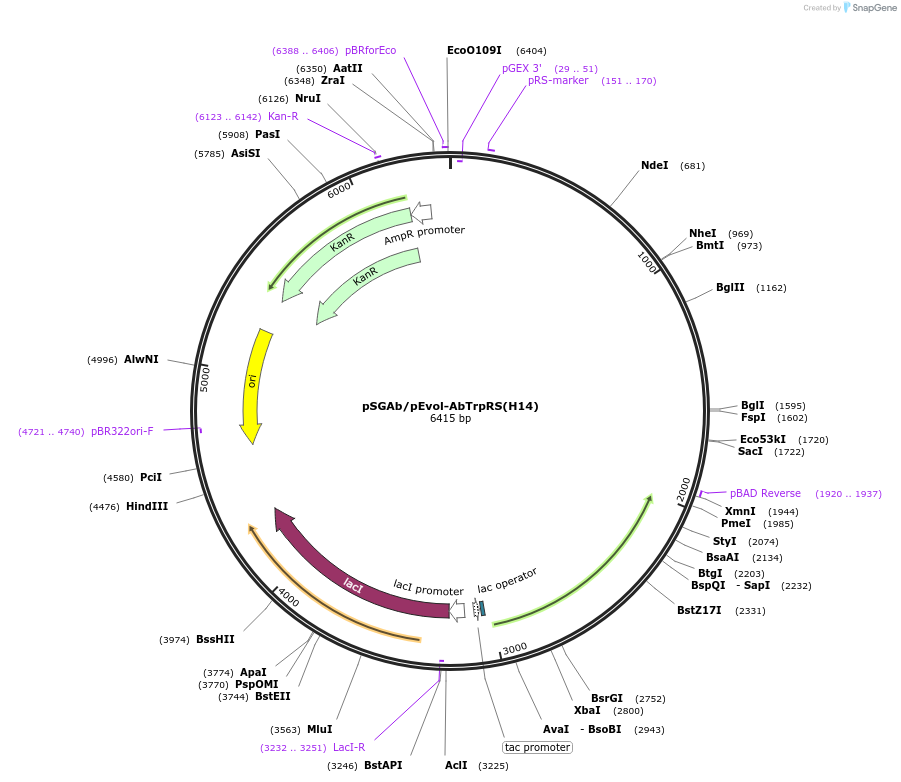
pSGAb/pEvol-AbTrpRS(H14)
Plasmid#231133PurposeExpresses mutant A. baumannii TrpRS from an inducible lac promoter for proteome-wide incorporation of tryptophan analogues.DepositorInsertA. baumannii tryptophanyl tRNA synthetase
UseAcinetobacter baumannii expressionExpressionBacterialMutationT12A, V151G, V153CAvailable SinceOct. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
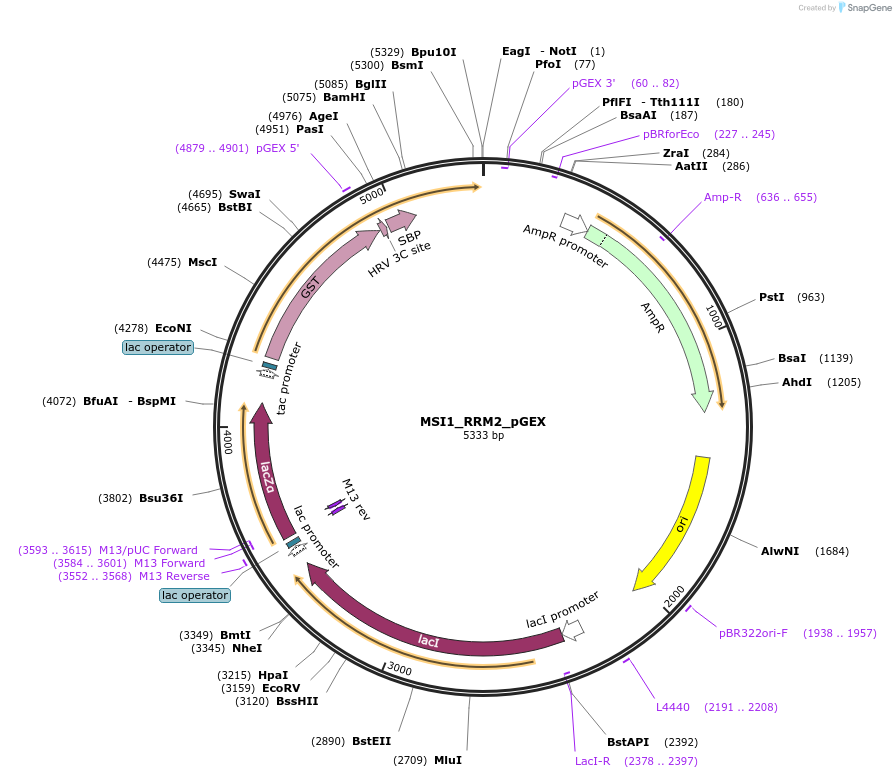
MSI1_RRM2_pGEX
Plasmid#228799PurposeRecombinant expression of mouse MSI1 RRM2 with dual-affinity tagDepositorAvailable SinceAug. 11, 2025AvailabilityAcademic Institutions and Nonprofits only -
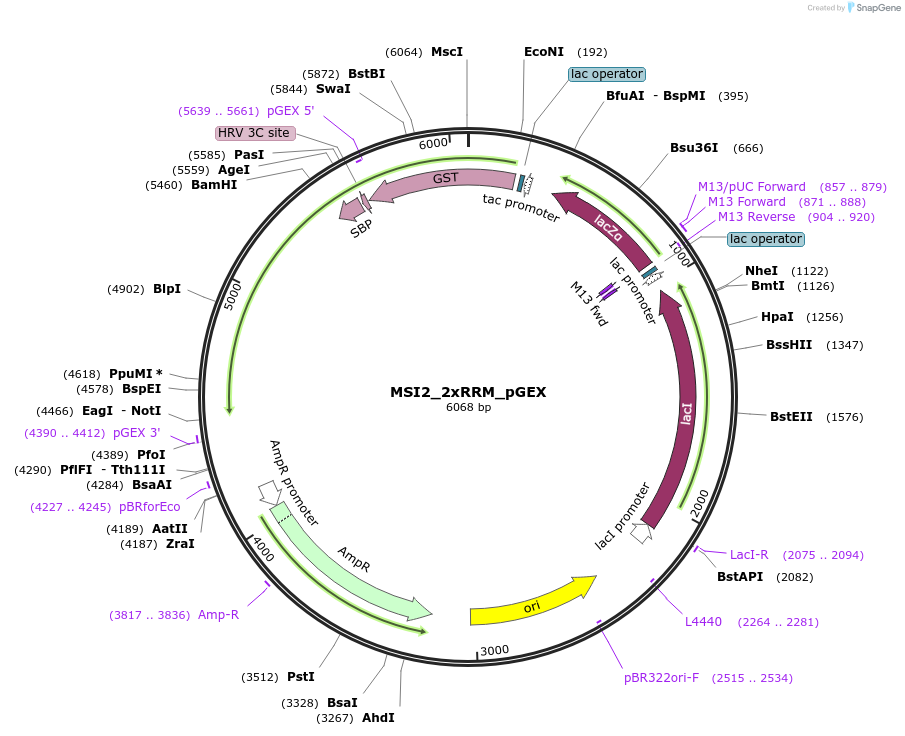
MSI2_2xRRM_pGEX
Plasmid#228800PurposeRecombinant expression of MSI2 with dual-affinity tagDepositorAvailable SinceAug. 11, 2025AvailabilityAcademic Institutions and Nonprofits only -
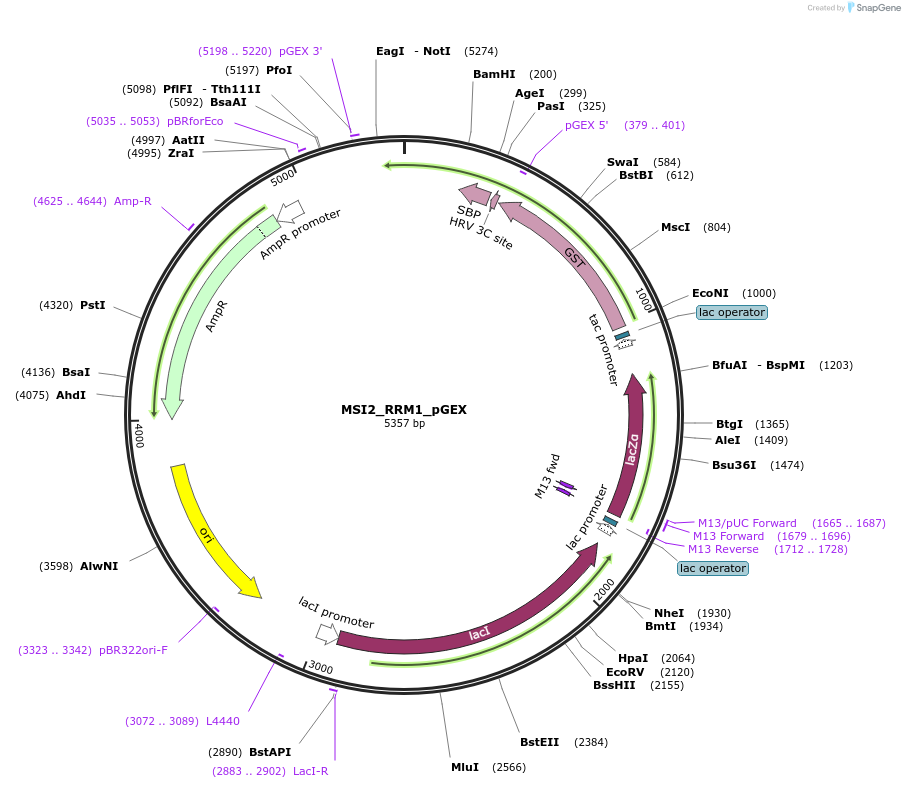
MSI2_RRM1_pGEX
Plasmid#228801PurposeRecombinant expression of MSI2 RRM1 with dual-affinity tagDepositorAvailable SinceAug. 11, 2025AvailabilityAcademic Institutions and Nonprofits only -
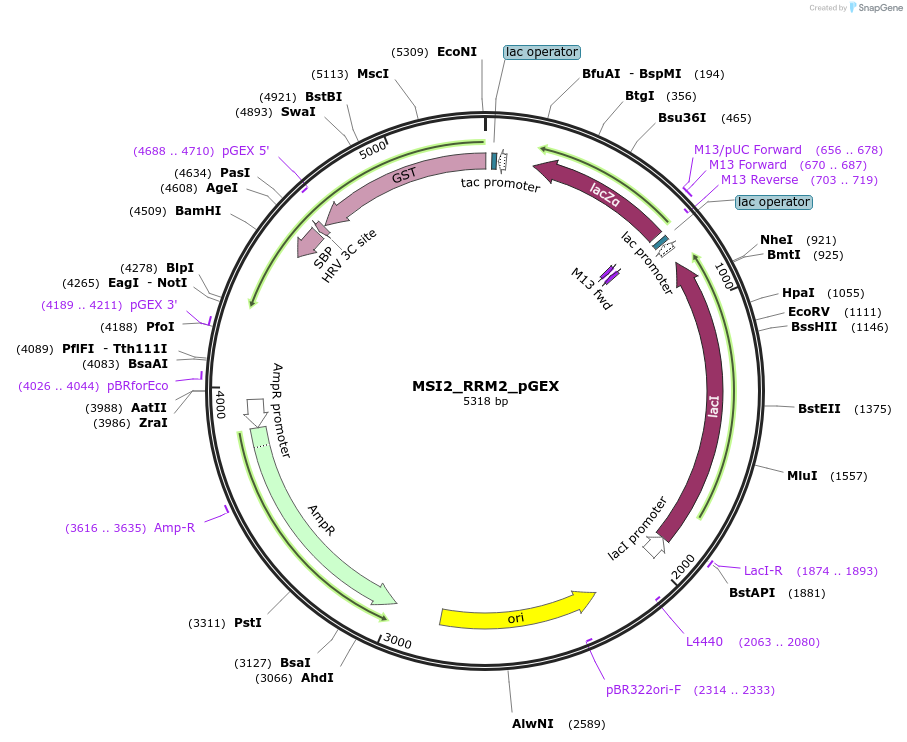
MSI2_RRM2_pGEX
Plasmid#228802PurposeRecombinant expression of MSI2 RRM2 with dual-affinity tagDepositorAvailable SinceAug. 11, 2025AvailabilityAcademic Institutions and Nonprofits only -
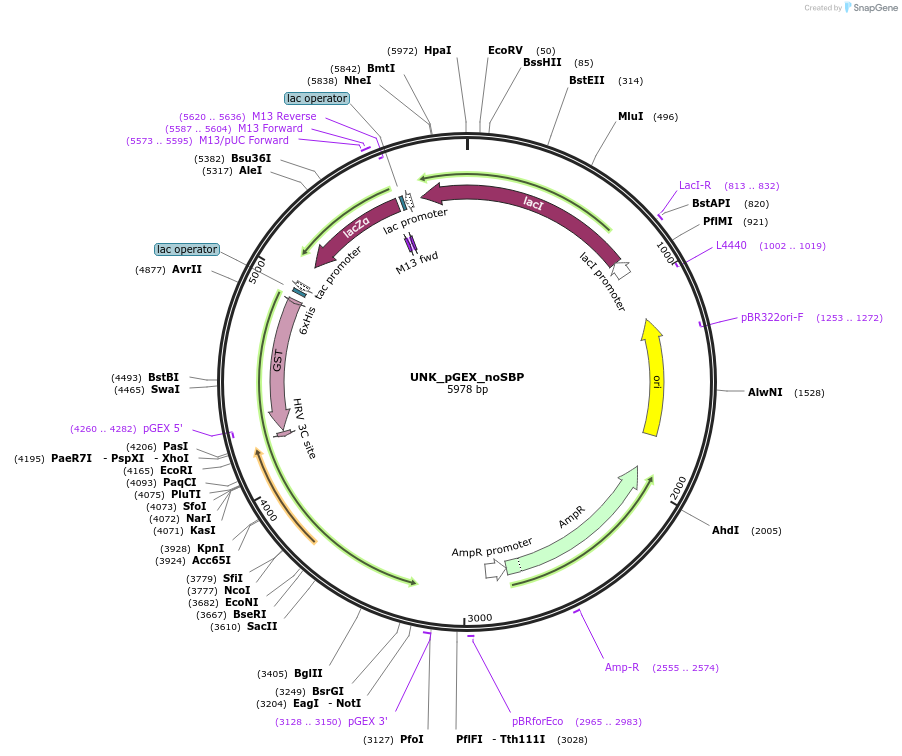
UNK_pGEX_noSBP
Plasmid#228803PurposeRecombinant expression of UNK with mono-affinity tagDepositorAvailable SinceAug. 11, 2025AvailabilityAcademic Institutions and Nonprofits only -
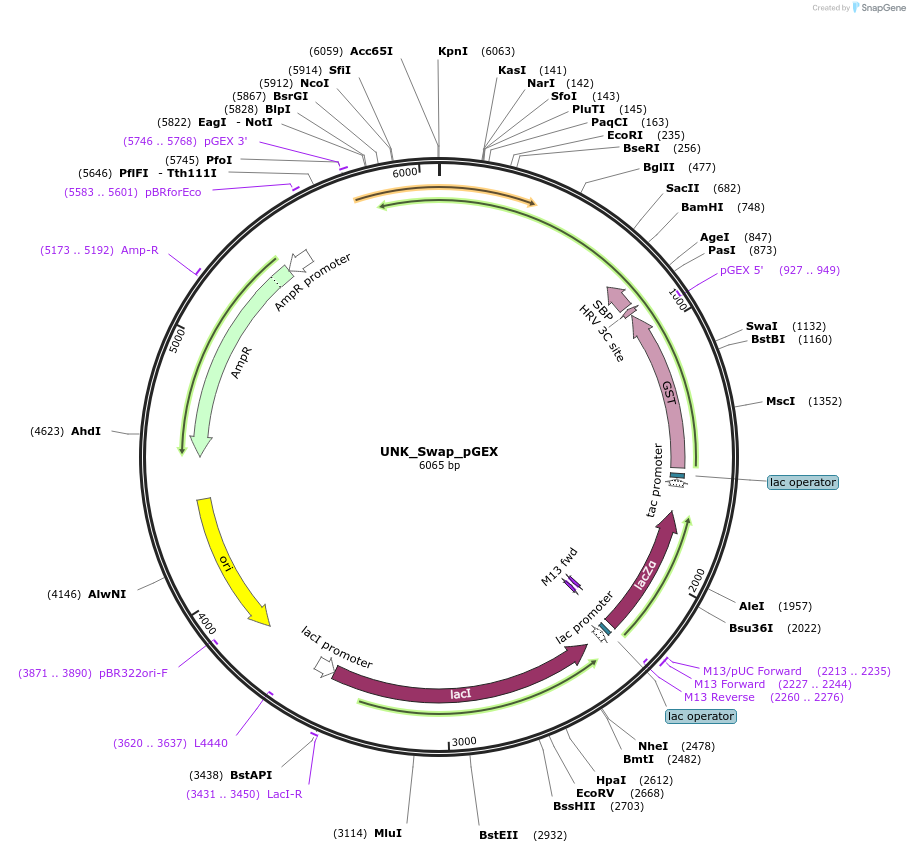
UNK_Swap_pGEX
Plasmid#228804PurposeRecombinant expression of UNK Swap with dual-affinity tagDepositorAvailable SinceAug. 11, 2025AvailabilityAcademic Institutions and Nonprofits only -
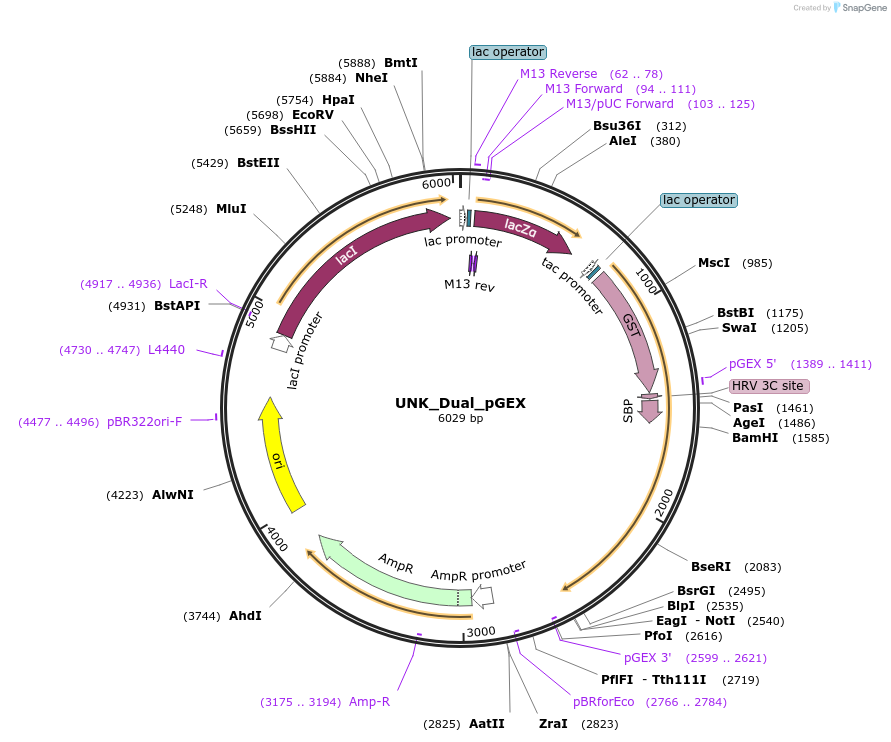
UNK_Dual_pGEX
Plasmid#228805PurposeRecombinant expression of UNK Dual with dual-affinity tagDepositorAvailable SinceAug. 11, 2025AvailabilityAcademic Institutions and Nonprofits only -
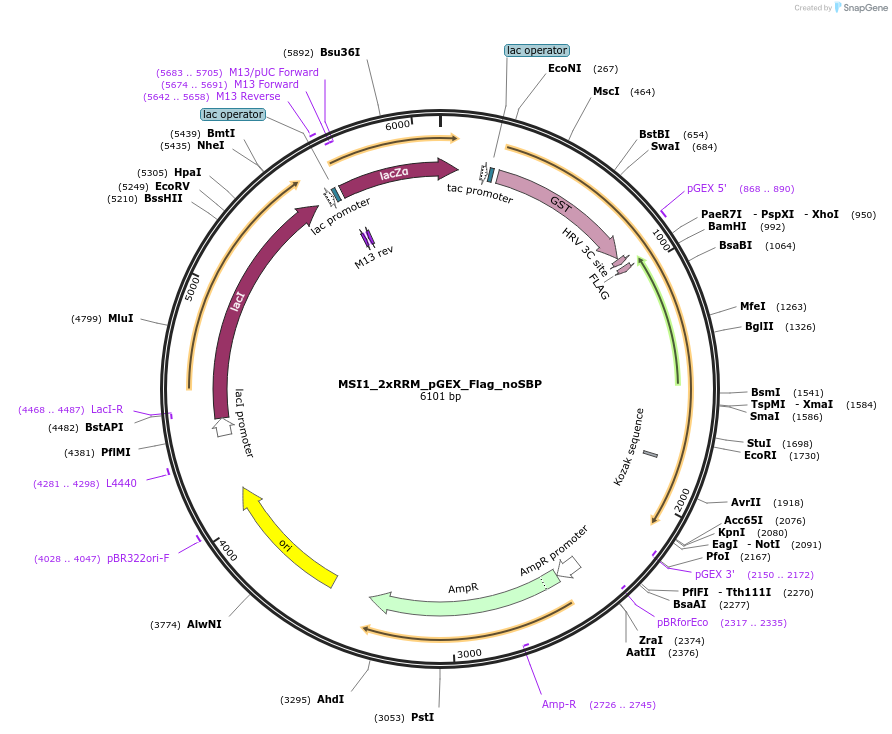
MSI1_2xRRM_pGEX_Flag_noSBP
Plasmid#228796PurposeRecombinant expression of mouse MSI1 with dual-affinity tagDepositorAvailable SinceAug. 11, 2025AvailabilityAcademic Institutions and Nonprofits only -
MSI1_SWAP_2xRRM_pGEX
Plasmid#228797PurposeRecombinant expression of mouse MSI1 Swap with dual-affinity tagDepositorAvailable SinceAug. 11, 2025AvailabilityAcademic Institutions and Nonprofits only -
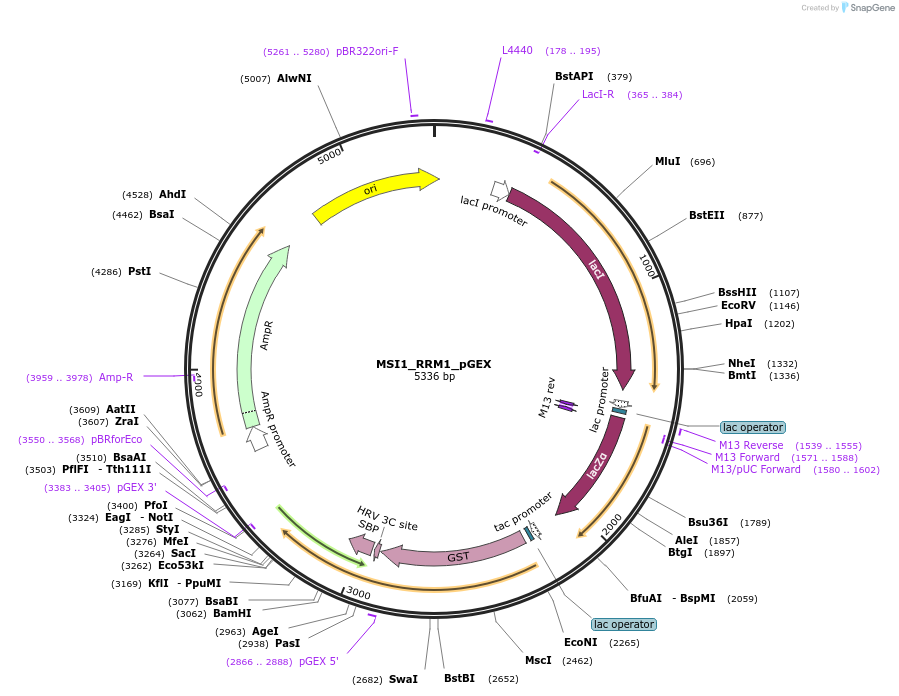
MSI1_RRM1_pGEX
Plasmid#228798PurposeRecombinant expression of mouse MSI1 RRM1 with dual-affinity tagDepositorAvailable SinceAug. 11, 2025AvailabilityAcademic Institutions and Nonprofits only -
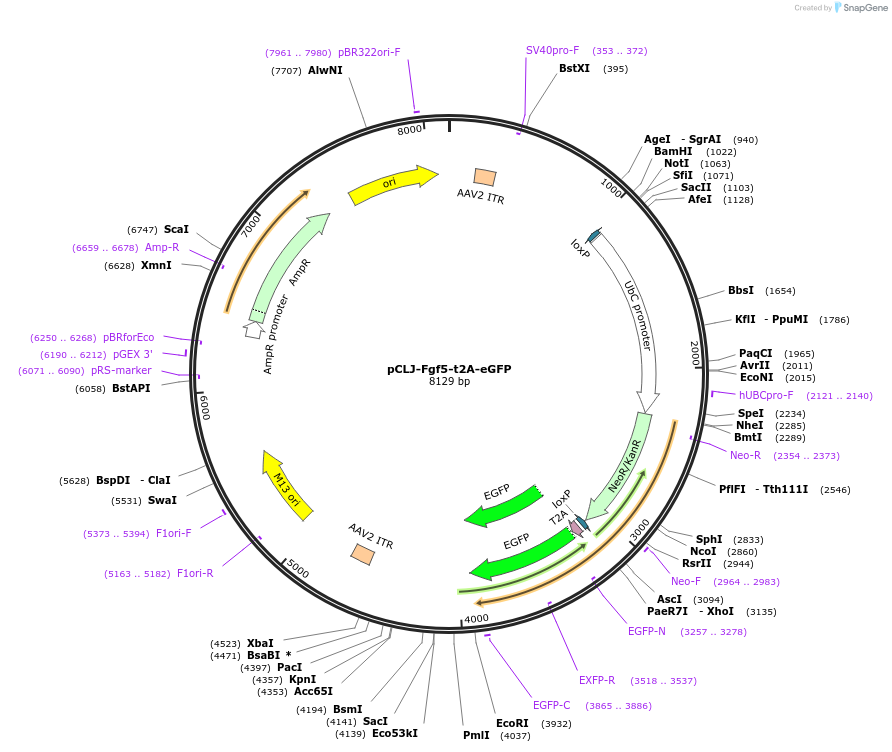
pCLJ-Fgf5-t2A-eGFP
Plasmid#227504PurposeInserts t2a-eGFP at the 3' end of the Fgf5 gene alsong with a removable selection cassetteDepositorInsertEGFP
UseAAV and Cre/LoxAvailable SinceMay 5, 2025AvailabilityAcademic Institutions and Nonprofits only