We narrowed to 12,662 results for: Cre expression vector
-
Plasmid#160030PurposeIntegrative vector for K. marxianus, targeting the ARO3 locus. Uracil selection. Low integration efficiency in an NHEJ-deficient background.DepositorTypeEmpty backboneUseKluyveromyces marxianusExpressionYeastPromotern/aAvailable SinceDec. 4, 2020AvailabilityAcademic Institutions and Nonprofits only
-
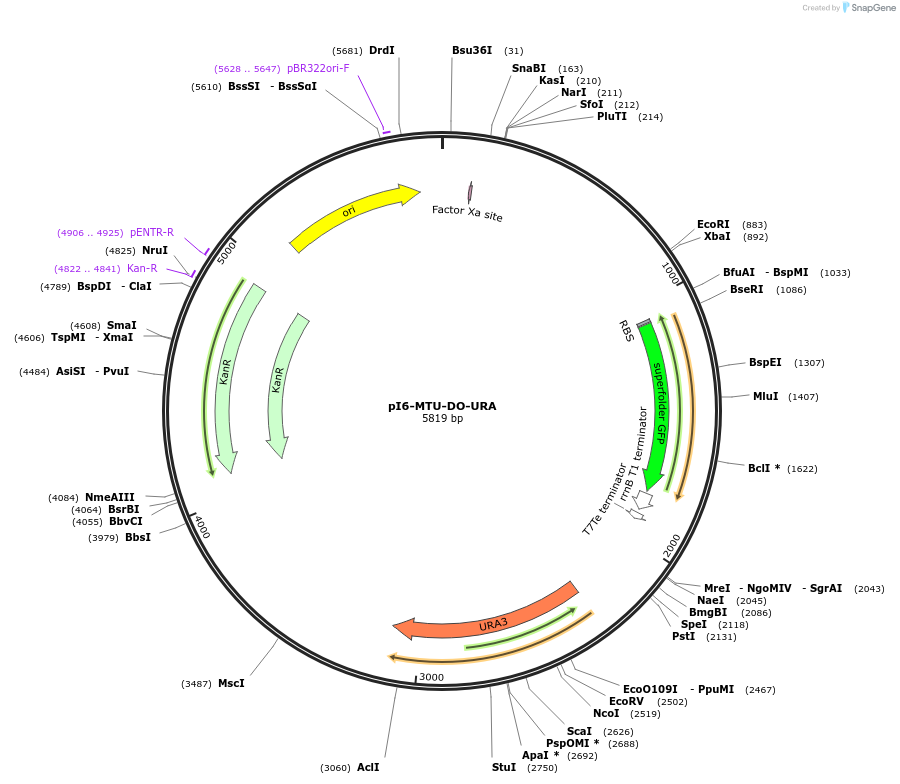
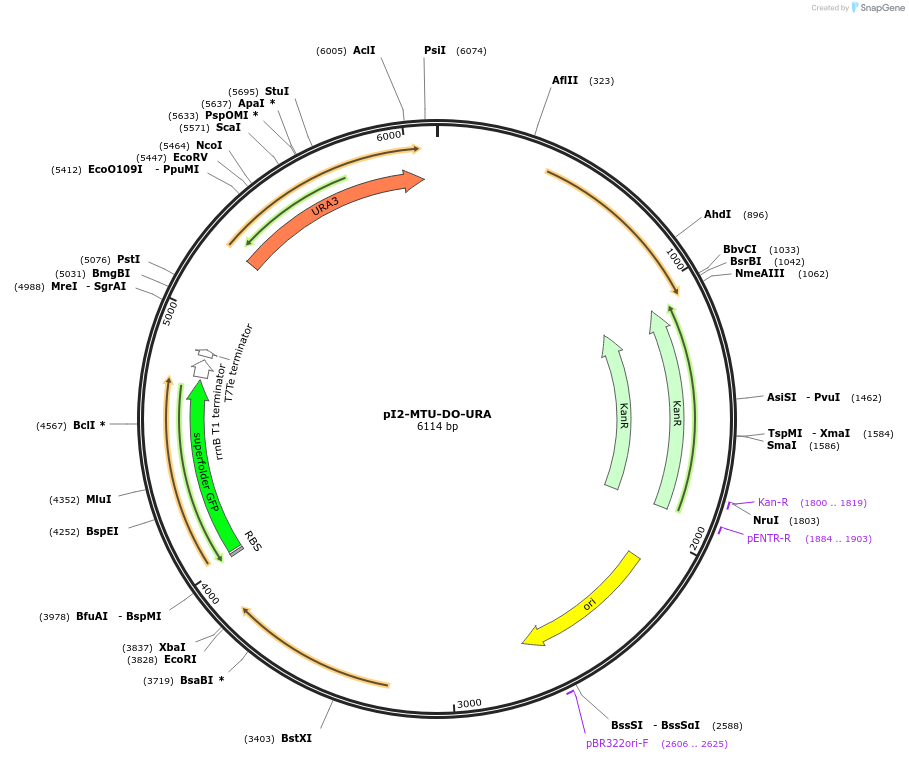
pI2-MTU-DO-URA
Plasmid#160015PurposeIntegrative vector for K. marxianus, targeting integration site I2 described in (Rajkumar et al.,2019). Uracil selection.DepositorTypeEmpty backboneUseKluyveromyces marxianusExpressionYeastPromotern/aAvailable SinceMarch 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
pI2-MTU-DO-URA
Plasmid#160015PurposeIntegrative vector for K. marxianus, targeting integration site I2 described in (Rajkumar et al.,2019). Uracil selection.DepositorTypeEmpty backboneUseKluyveromyces marxianusExpressionYeastPromotern/aAvailable SinceMarch 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
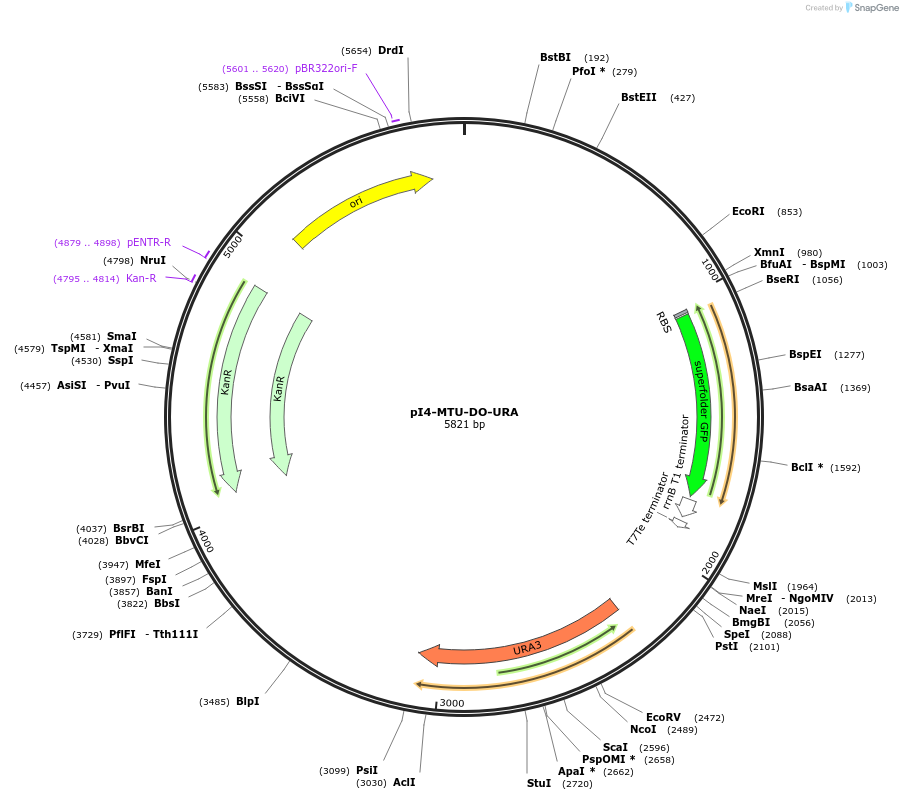
pI4-MTU-DO-URA
Plasmid#160025PurposeIntegrative vector for K. marxianus, targeting integration site I4 described in (Rajkumar et al.,2019). Uracil selection.DepositorTypeEmpty backboneUseKluyveromyces marxianusExpressionYeastPromotern/aAvailable SinceNov. 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
pI4-MTU-DO-URA
Plasmid#160025PurposeIntegrative vector for K. marxianus, targeting integration site I4 described in (Rajkumar et al.,2019). Uracil selection.DepositorTypeEmpty backboneUseKluyveromyces marxianusExpressionYeastPromotern/aAvailable SinceNov. 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
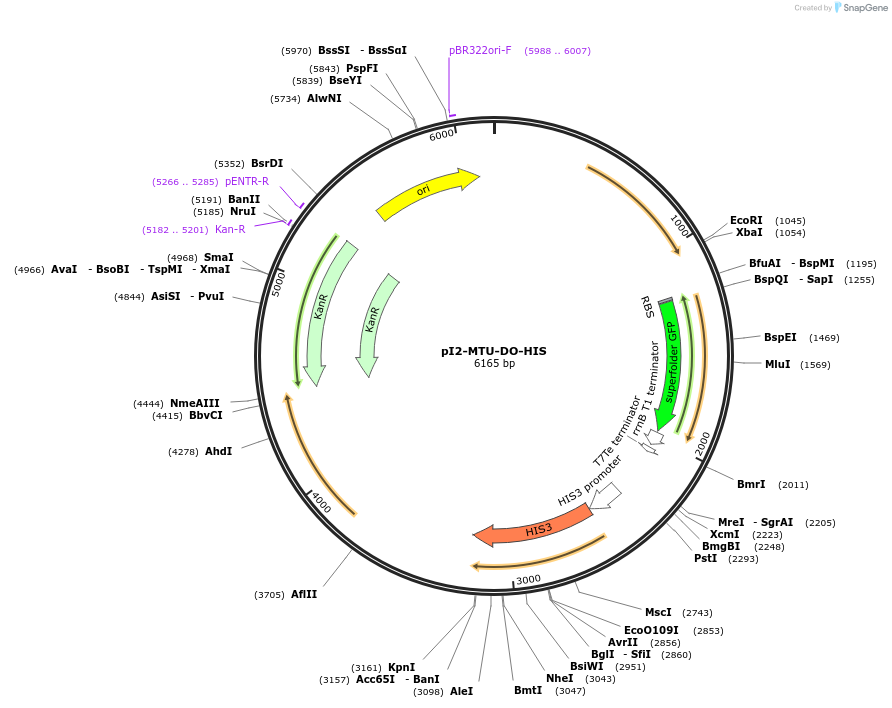
pI2-MTU-DO-HIS
Plasmid#160016PurposeIntegrative vector for K. marxianus, targeting integration site I2 described in (Rajkumar et al.,2019). Histidine selection.DepositorTypeEmpty backboneUseKluyveromyces marxianusExpressionYeastPromotern/aAvailable SinceNov. 20, 2020AvailabilityAcademic Institutions and Nonprofits only -
pI2-MTU-DO-HIS
Plasmid#160016PurposeIntegrative vector for K. marxianus, targeting integration site I2 described in (Rajkumar et al.,2019). Histidine selection.DepositorTypeEmpty backboneUseKluyveromyces marxianusExpressionYeastPromotern/aAvailable SinceNov. 20, 2020AvailabilityAcademic Institutions and Nonprofits only -
CyclinB-GFP
Plasmid#54465PurposeCyclinB-GFP in pIVT vector for in vitro transcriptionDepositorInsertCyclinB
TagsGFPExpressionMammalianMutationK67E and I150T mutations compared to GenBank refe…Available SinceJan. 13, 2016AvailabilityAcademic Institutions and Nonprofits only -
CyclinB-GFP
Plasmid#54465PurposeCyclinB-GFP in pIVT vector for in vitro transcriptionDepositorInsertCyclinB
TagsGFPExpressionMammalianMutationK67E and I150T mutations compared to GenBank refe…Available SinceJan. 13, 2016AvailabilityAcademic Institutions and Nonprofits only -
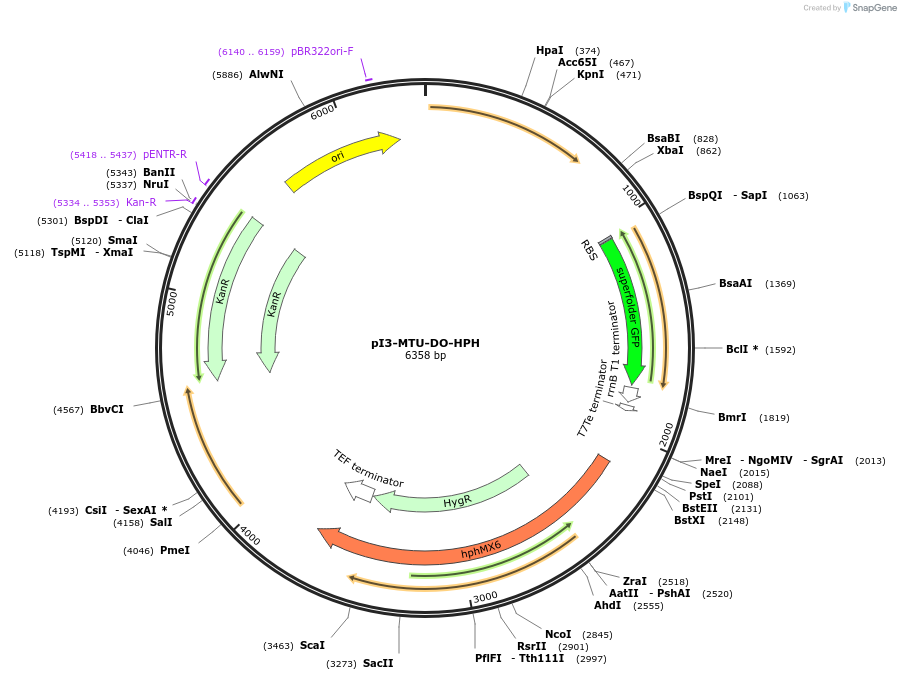
pI3-MTU-DO-HPH
Plasmid#160207PurposeIntegrative vector for K. marxianus, targeting integration site I3 described in (Rajkumar et al.,2019). Hygromycin selection.DepositorTypeEmpty backboneUseKluyveromyces marxianusExpressionYeastPromotern/aAvailable SinceNov. 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
pI3-MTU-DO-HPH
Plasmid#160207PurposeIntegrative vector for K. marxianus, targeting integration site I3 described in (Rajkumar et al.,2019). Hygromycin selection.DepositorTypeEmpty backboneUseKluyveromyces marxianusExpressionYeastPromotern/aAvailable SinceNov. 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
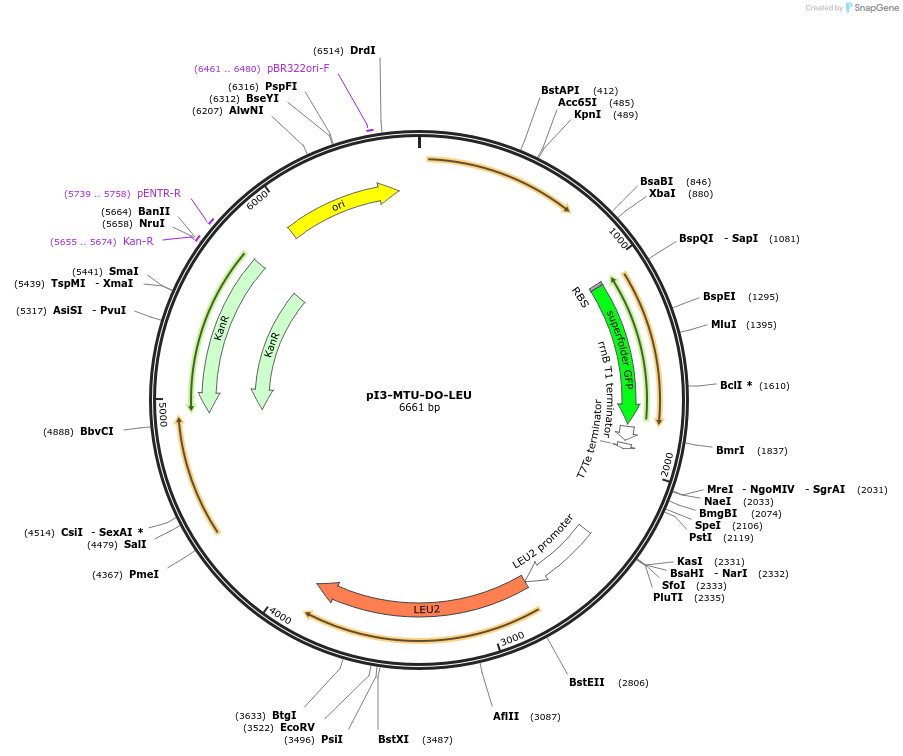
pI3-MTU-DO-LEU
Plasmid#160176PurposeIntegrative vector for K. marxianus, targeting integration site I3 described in (Rajkumar et al.,2019). Leucine selection.DepositorTypeEmpty backboneUseKluyveromyces marxianusExpressionYeastPromotern/aAvailable SinceNov. 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
pI3-MTU-DO-LEU
Plasmid#160176PurposeIntegrative vector for K. marxianus, targeting integration site I3 described in (Rajkumar et al.,2019). Leucine selection.DepositorTypeEmpty backboneUseKluyveromyces marxianusExpressionYeastPromotern/aAvailable SinceNov. 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
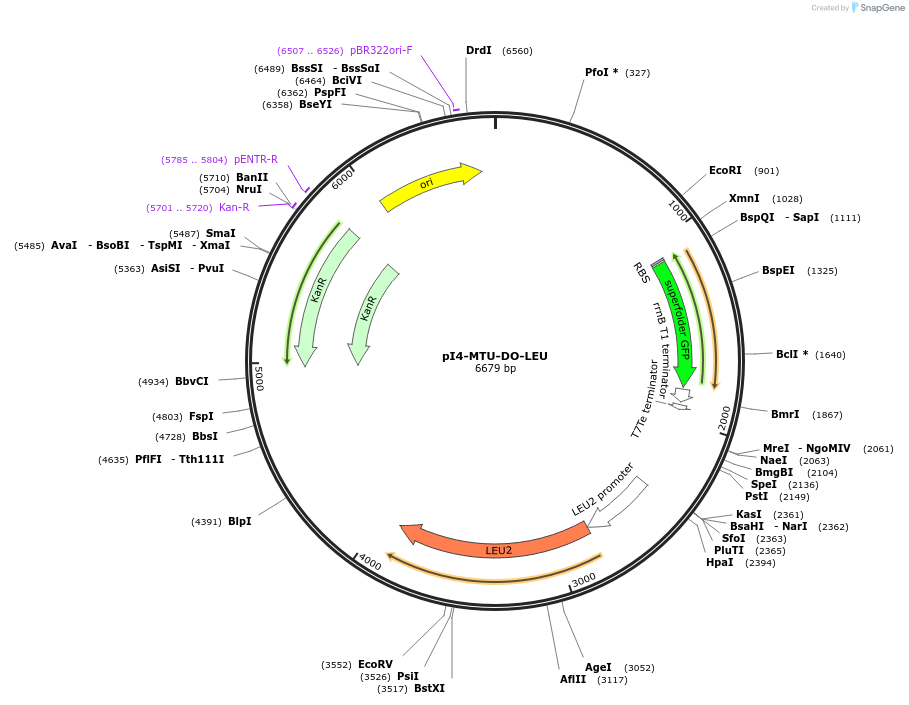
pI4-MTU-DO-LEU
Plasmid#160029PurposeIntegrative vector for K. marxianus, targeting integration site I4 described in (Rajkumar et al.,2019). Leucine selection.DepositorTypeEmpty backboneUseKluyveromyces marxianusExpressionYeastPromotern/aAvailable SinceNov. 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
pI4-MTU-DO-LEU
Plasmid#160029PurposeIntegrative vector for K. marxianus, targeting integration site I4 described in (Rajkumar et al.,2019). Leucine selection.DepositorTypeEmpty backboneUseKluyveromyces marxianusExpressionYeastPromotern/aAvailable SinceNov. 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
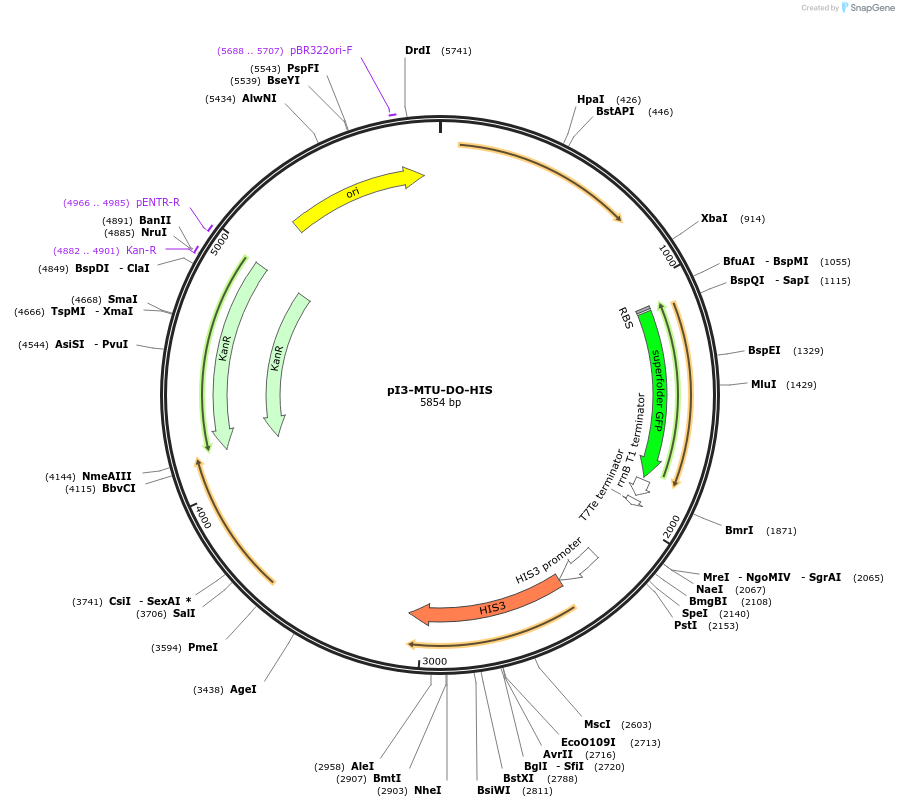
pI3-MTU-DO-HIS
Plasmid#160021PurposeIntegrative vector for K. marxianus, targeting integration site I3 described in (Rajkumar et al.,2019). Histidine selection.DepositorTypeEmpty backboneUseKluyveromyces marxianusExpressionYeastPromotern/aAvailable SinceNov. 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
pI3-MTU-DO-HIS
Plasmid#160021PurposeIntegrative vector for K. marxianus, targeting integration site I3 described in (Rajkumar et al.,2019). Histidine selection.DepositorTypeEmpty backboneUseKluyveromyces marxianusExpressionYeastPromotern/aAvailable SinceNov. 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
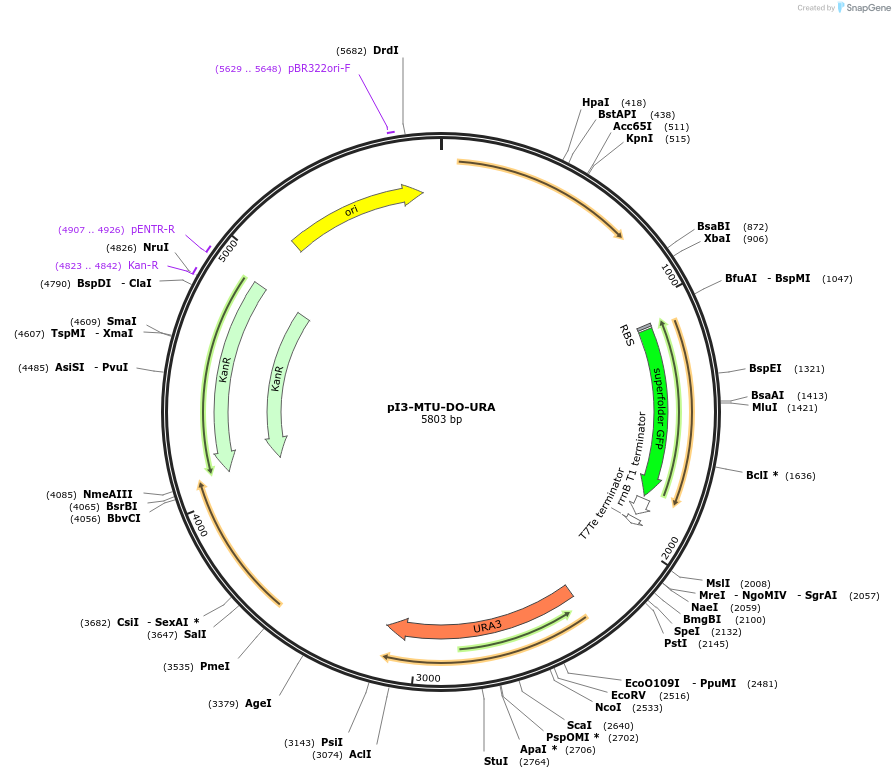
pI3-MTU-DO-URA
Plasmid#160020PurposeIntegrative vector for K. marxianus, targeting integration site I3 described in (Rajkumar et al.,2019). Uracil selection.DepositorTypeEmpty backboneUseKluyveromyces marxianusExpressionYeastPromotern/aAvailable SinceNov. 20, 2020AvailabilityAcademic Institutions and Nonprofits only -
pI3-MTU-DO-URA
Plasmid#160020PurposeIntegrative vector for K. marxianus, targeting integration site I3 described in (Rajkumar et al.,2019). Uracil selection.DepositorTypeEmpty backboneUseKluyveromyces marxianusExpressionYeastPromotern/aAvailable SinceNov. 20, 2020AvailabilityAcademic Institutions and Nonprofits only -
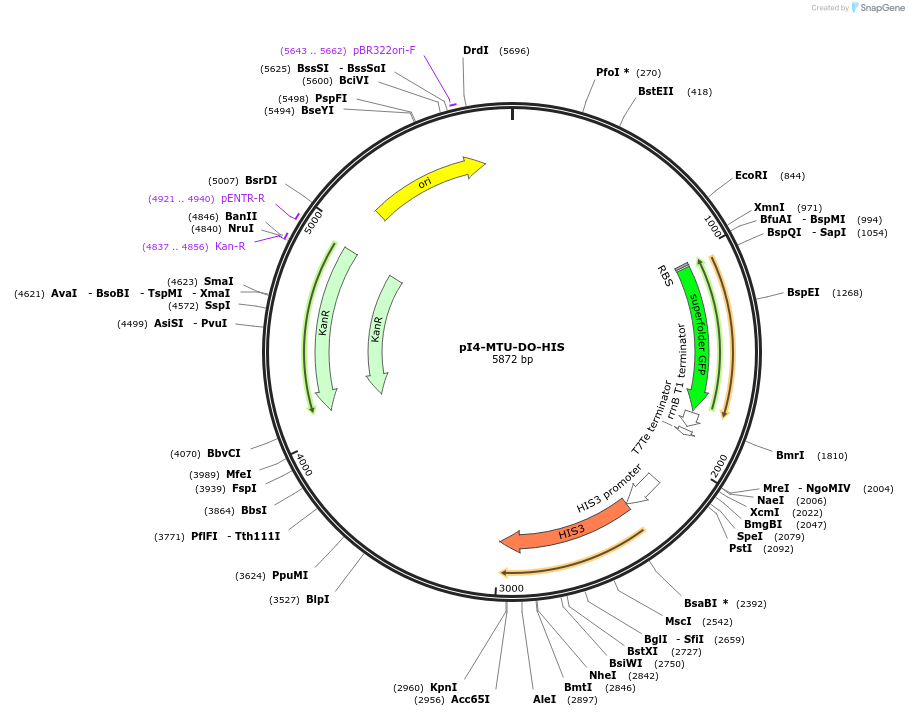
pI4-MTU-DO-HIS
Plasmid#160026PurposeIntegrative vector for K. marxianus, targeting integration site I4 described in (Rajkumar et al.,2019). Histidine selection.DepositorTypeEmpty backboneUseKluyveromyces marxianusExpressionYeastPromotern/aAvailable SinceMarch 8, 2021AvailabilityAcademic Institutions and Nonprofits only