We narrowed to 10,326 results for: Ada;
-
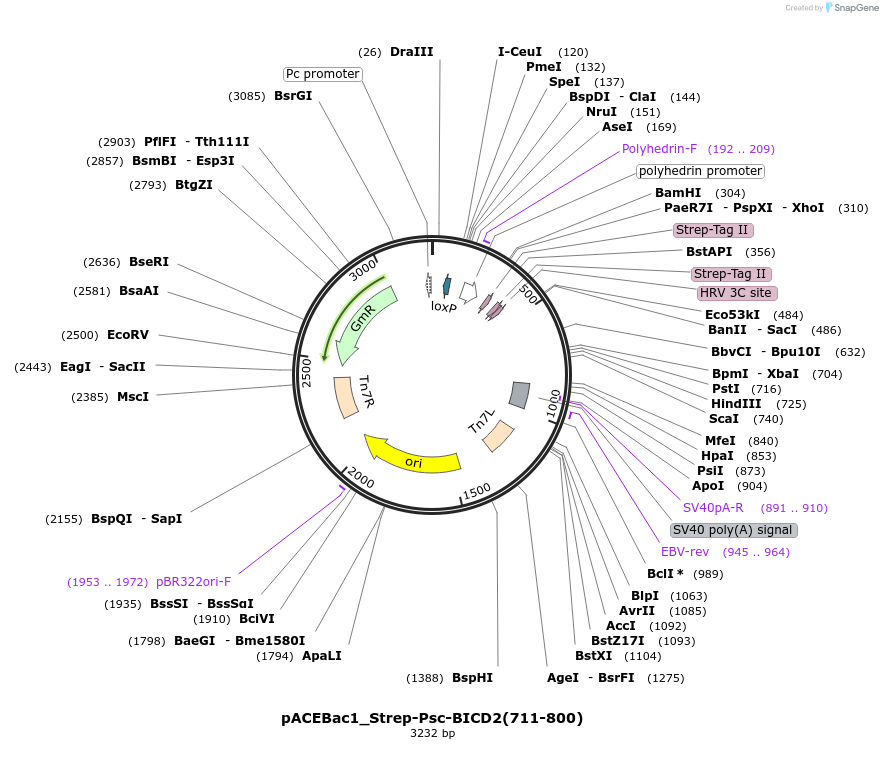
Plasmid#228833PurposeInsect cell expression of CC3 domain of BICD2 (aa 711-800)DepositorAvailable SinceDec. 17, 2024AvailabilityAcademic Institutions and Nonprofits only
-
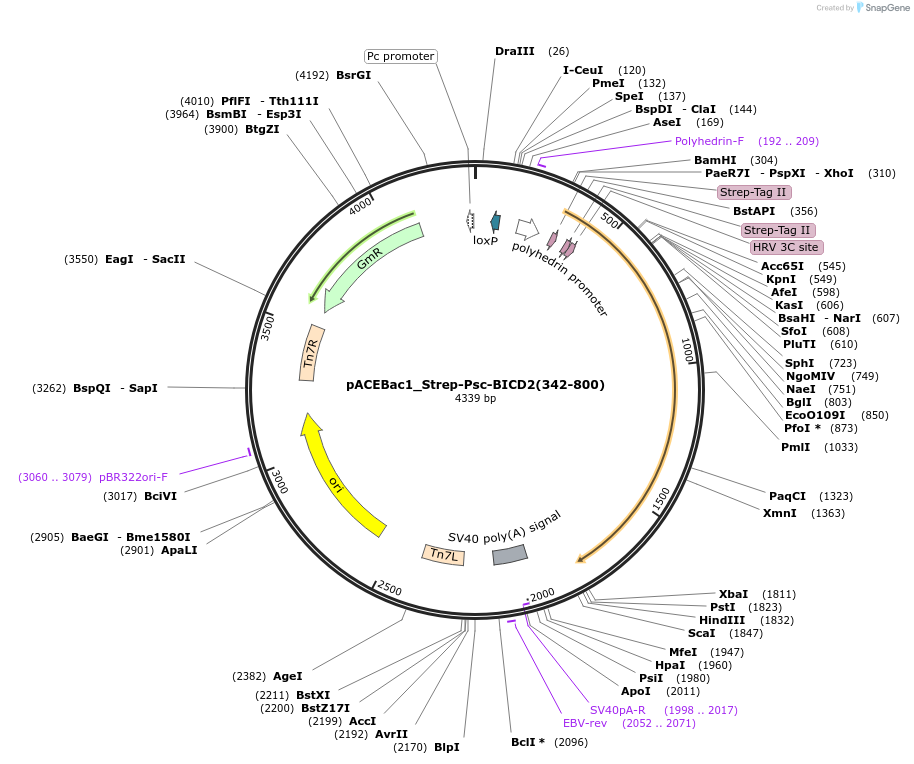
pACEBac1_Strep-Psc-BICD2(342-800)
Plasmid#228832PurposeInsect cell expression of C-terminal portion of BICD2 (aa 342-800)DepositorAvailable SinceDec. 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
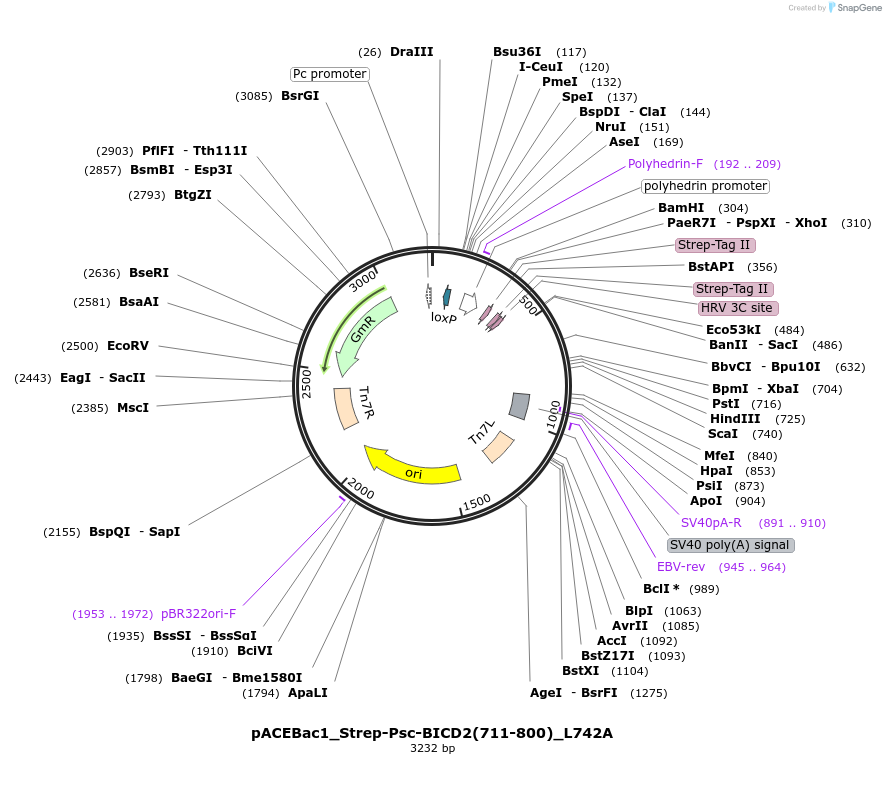
pACEBac1_Strep-Psc-BICD2(711-800)_L742A
Plasmid#228839PurposeInsect cell expression of the mutant C-terminus of BICD2DepositorAvailable SinceDec. 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
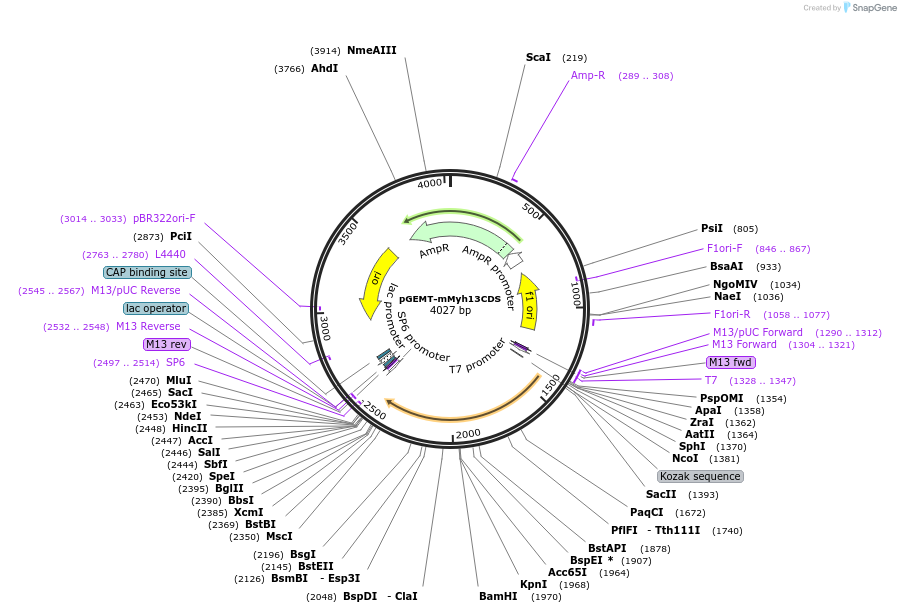
pGEMT-mMyh13CDS
Plasmid#192853PurposePlasmid to produce ISH probe for mMyh13 gene with CDS sequenceDepositorInsertmyosin heavy chain 13 (Myh13 Mouse)
Available SinceDec. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
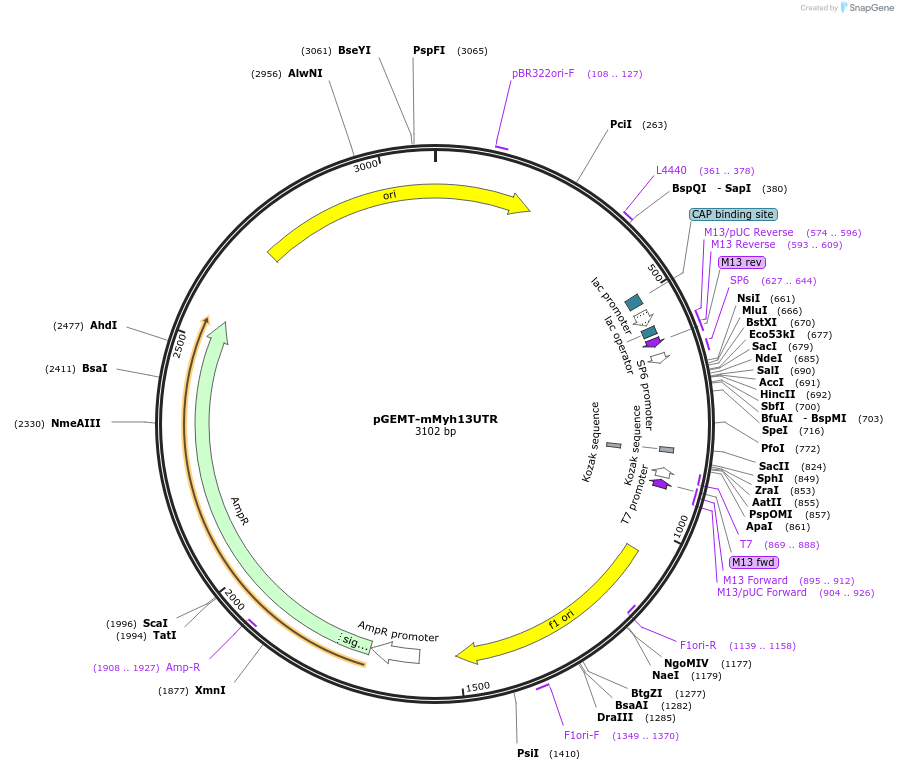
pGEMT-mMyh13UTR
Plasmid#192854PurposePlasmid to produce ISH probe for mMyh13 gene with UTR sequenceDepositorInsertmyosin heavy chain 13 (Myh13 Mouse)
Available SinceDec. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
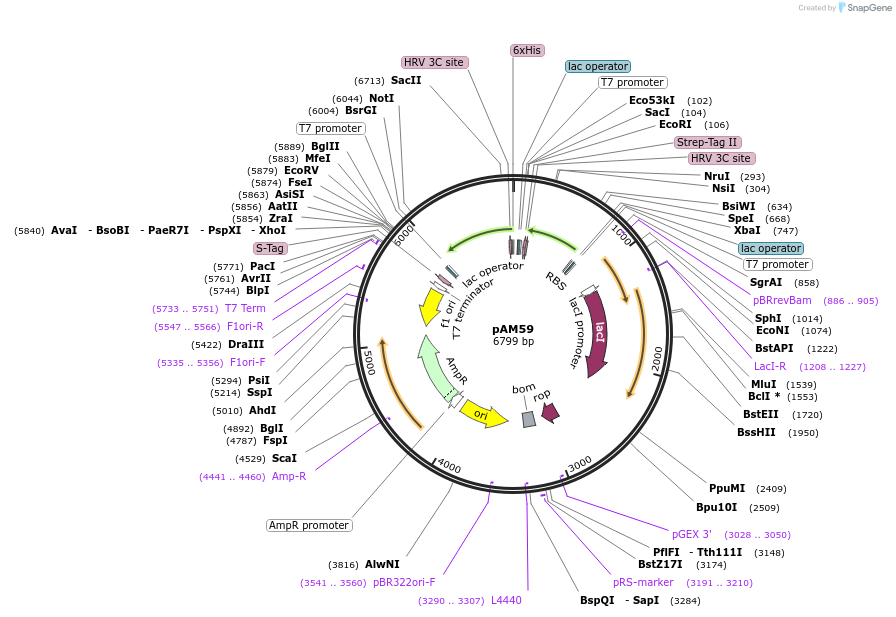
pAM59
Plasmid#227655PurposepETDuet-1: Rpn8(1-179)-PreScission-Strep, H6-PreScission-Rpn11(2-239, Δ78–81)DepositorTagsH6-PreScission and PreScission-StrepExpressionBacterialMutation1-179 and amino acids 2-239, Δ78–81PromoterT7Available SinceDec. 4, 2024AvailabilityAcademic Institutions and Nonprofits only -
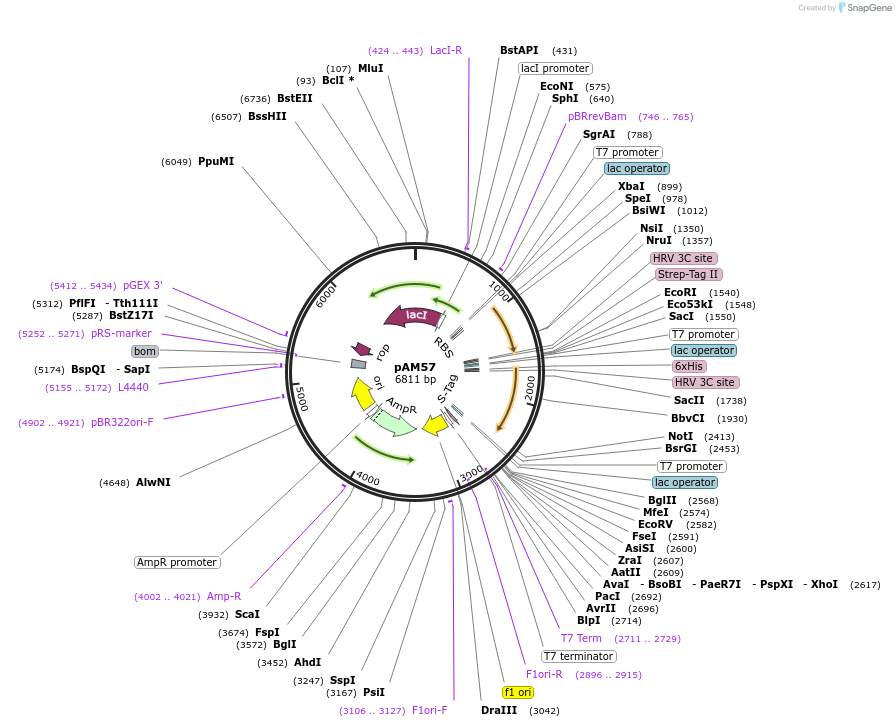
pAM57
Plasmid#227654PurposepETDuet-1: Rpn8(1-179)-PreScission-Strep, H6-PreScission-Rpn11(2-239, V80A)DepositorTagsH6-PreScission and PreScission-StrepExpressionBacterialMutation1-179 and amino acids 2-239, V80APromoterT7Available SinceDec. 4, 2024AvailabilityAcademic Institutions and Nonprofits only -
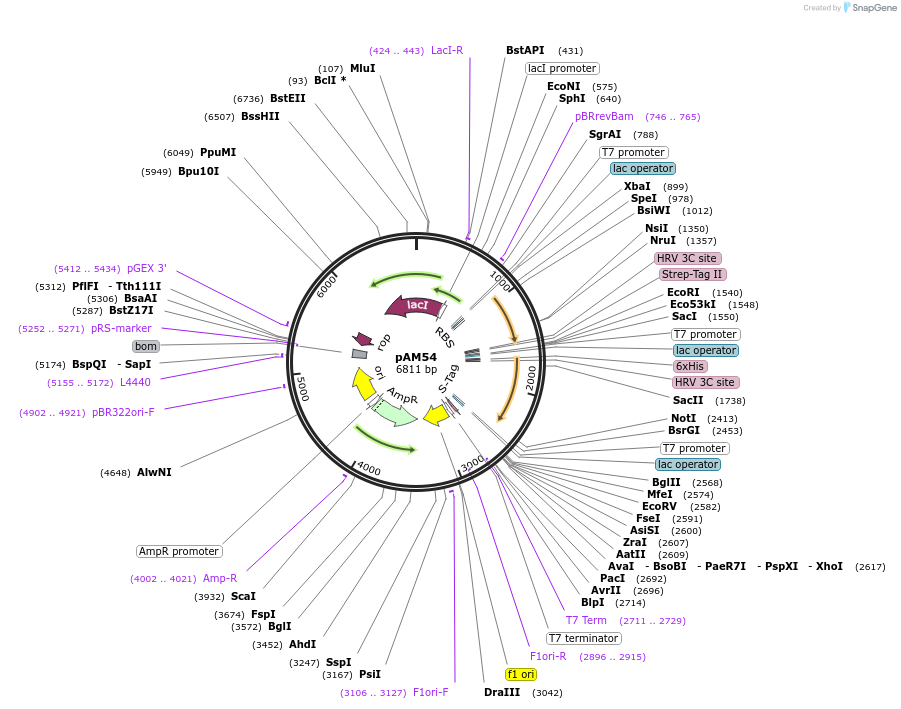
pAM54
Plasmid#227653PurposepETDuet-1: Rpn8(1-179)-PreScission-Strep, H6-PreScission-Rpn11(2-239, L132A)DepositorTagsH6-PreScission and PreScission-StrepExpressionBacterialMutation1-179 and amino acids 2-239, L132APromoterT7Available SinceDec. 4, 2024AvailabilityAcademic Institutions and Nonprofits only -
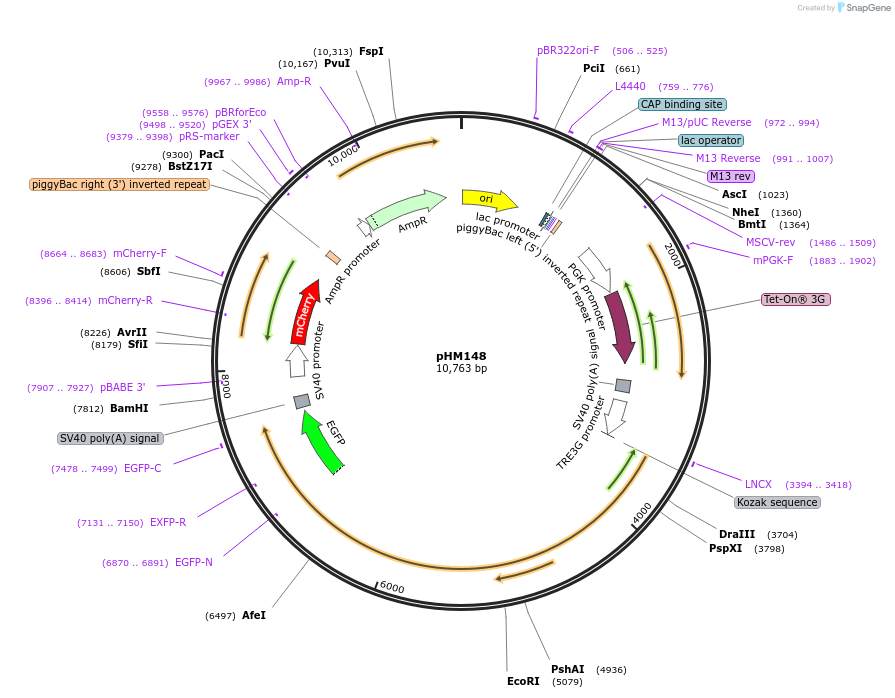
pHM148
Plasmid#225532PurposeEncodes constitutive rtTA, tetOn-Pcdh19-GFP, and constitutive mCherryDepositorAvailable SinceNov. 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
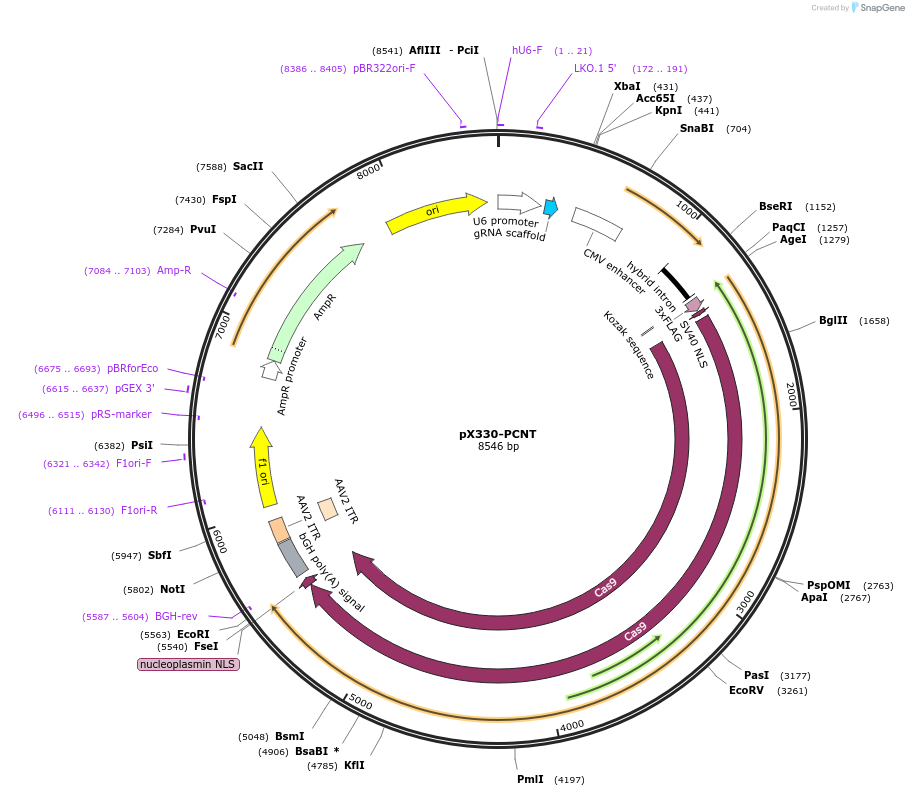
pX330-PCNT
Plasmid#227284PurposeExpresses SpCas9 and a sgRNA targeting the N-terminus of PCNT for knock-in.DepositorAvailable SinceNov. 13, 2024AvailabilityAcademic Institutions and Nonprofits only -
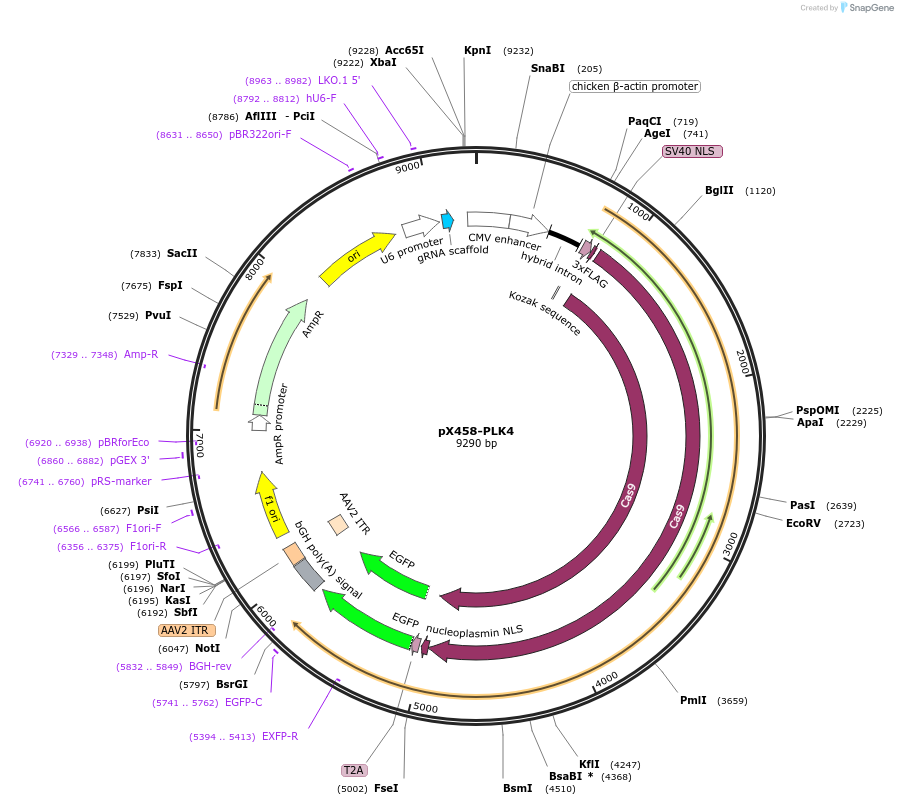
pX458-PLK4
Plasmid#227310PurposeExpresses SpCas9 and a sgRNA targeting the C-terminus of PLK4 for knock-in.DepositorAvailable SinceNov. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
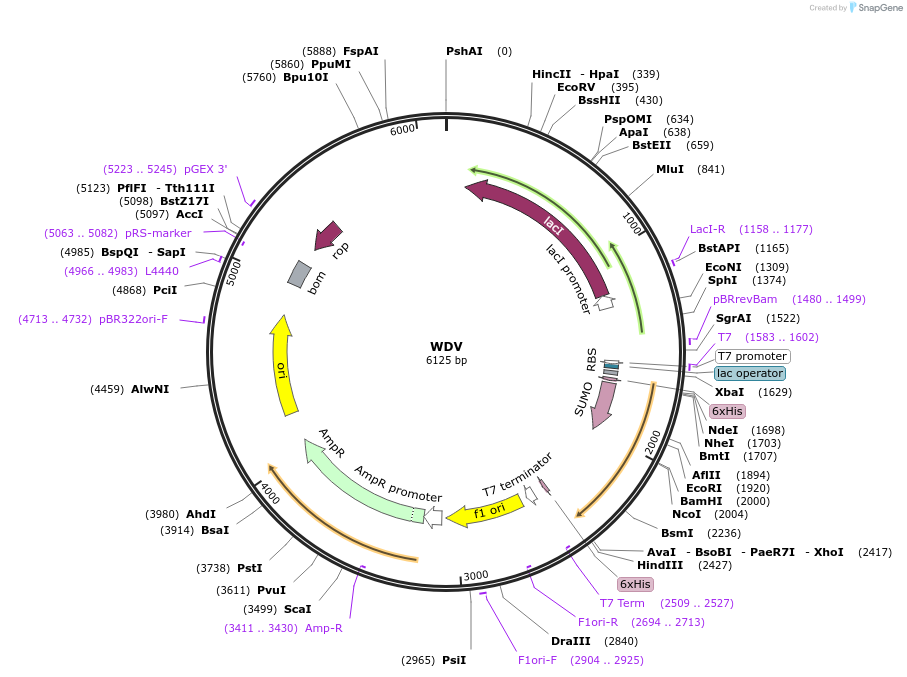
WDV
Plasmid#226983PurposeExpresses the HUH endonuclease WDV, used as a no ligand control for DNA tension sensor experimentsDepositorAvailable SinceOct. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
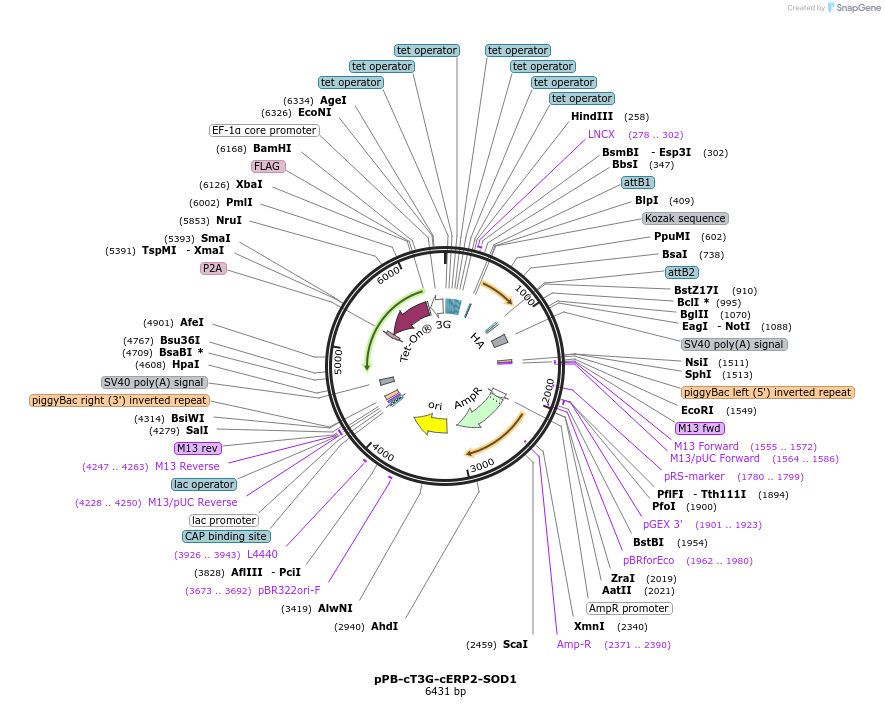
pPB-cT3G-cERP2-SOD1
Plasmid#222568PurposePiggyBac transposon plasmid for doxycycline inducible expression of SOD1DepositorInsertSOD1 (SOD1 Human)
ExpressionMammalianAvailable SinceAug. 16, 2024AvailabilityAcademic Institutions and Nonprofits only -
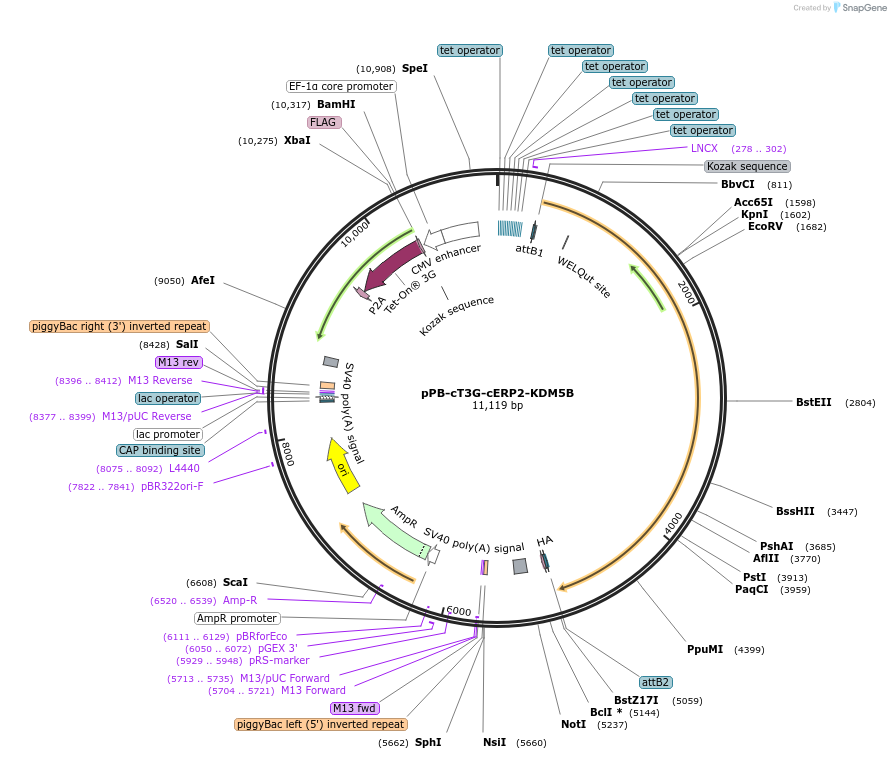
pPB-cT3G-cERP2-KDM5B
Plasmid#222555PurposePiggyBac transposon plasmid for doxycycline inducible expression of KDM5BDepositorInsertKDM5B (KDM5B Human)
ExpressionMammalianAvailable SinceAug. 13, 2024AvailabilityAcademic Institutions and Nonprofits only -
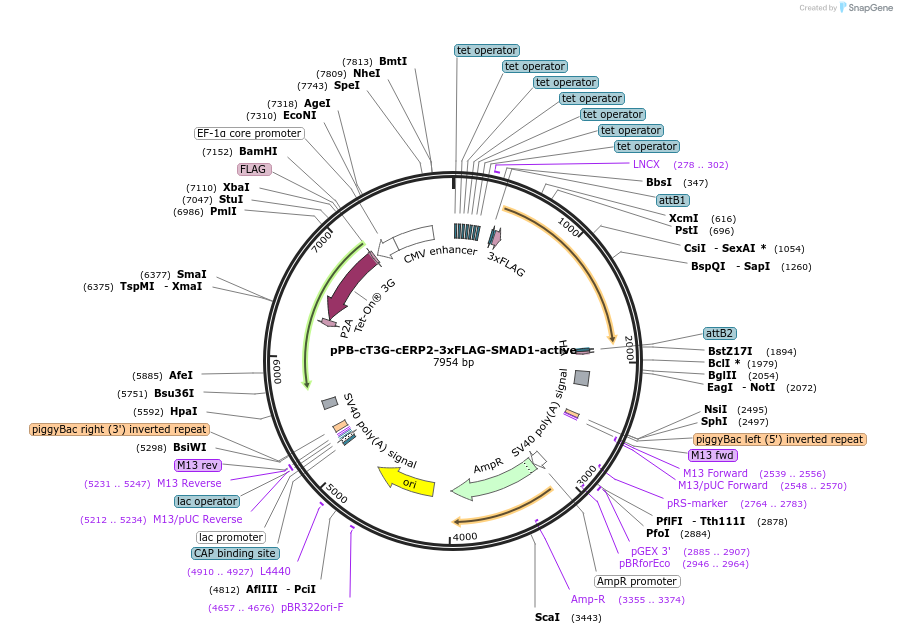
pPB-cT3G-cERP2-3xFLAG-SMAD1-active
Plasmid#222579PurposePiggyBac transposon plasmid for doxycycline inducible expression of 3xFLAG-SMAD1DepositorAvailable SinceAug. 13, 2024AvailabilityAcademic Institutions and Nonprofits only -
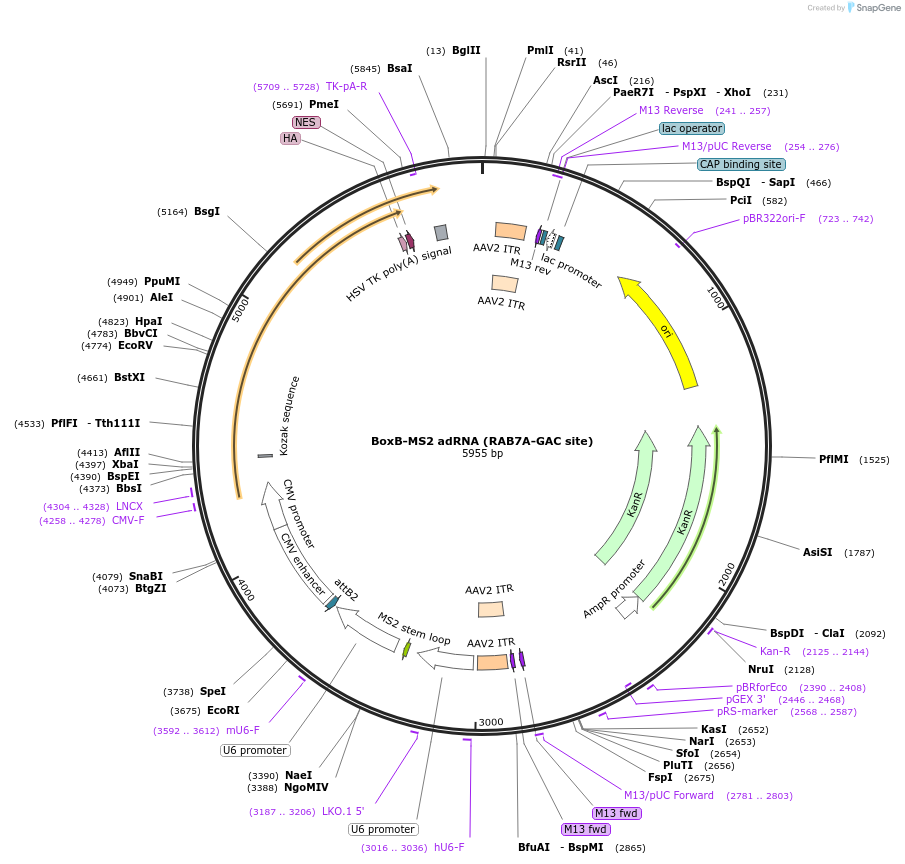
BoxB-MS2 adRNA (RAB7A-GAC site)
Plasmid#170149PurposeAAV vector carrying a BoxB-MS2 adRNA targeting the RAB7A transcriptDepositorInsertBoxB-RAB7A(GAC)-MS2
UseAAVExpressionMammalianPromoterHuman U6Available SinceAug. 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
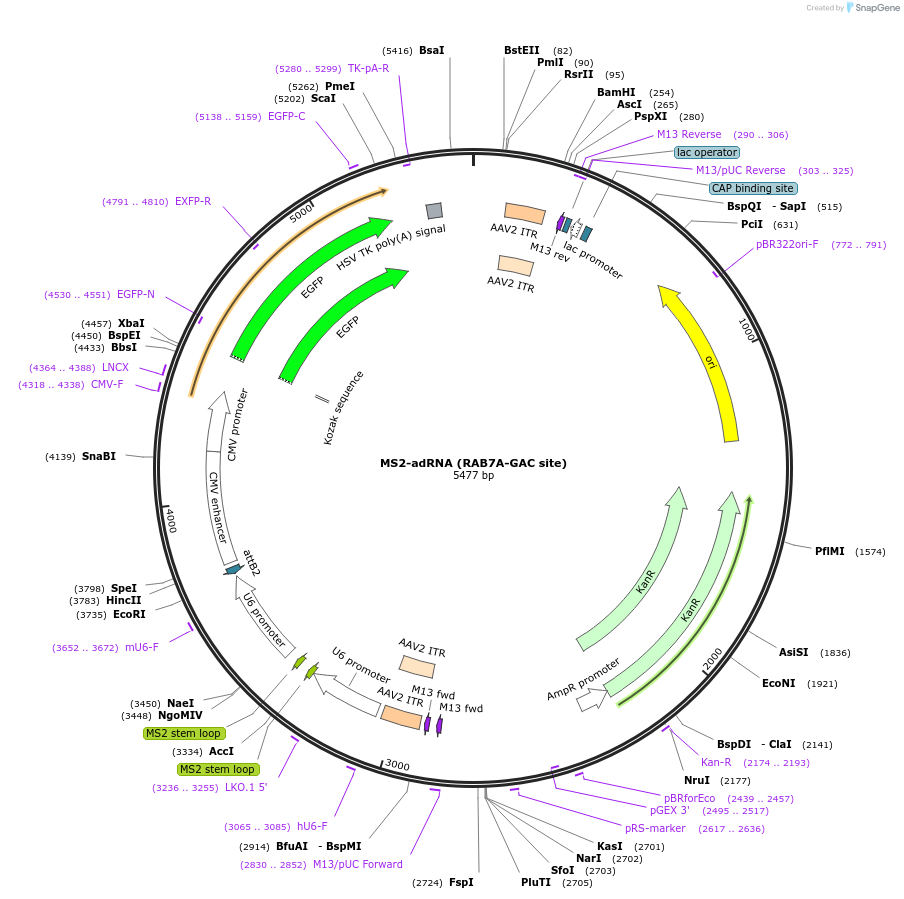
MS2-adRNA (RAB7A-GAC site)
Plasmid#170140PurposeAAV vector carrying a MS2 adRNA targeting the RAB7A transcriptDepositorInsertMS2-RAB7A(GAC)-MS2
UseAAVExpressionMammalianPromoterHuman U6Available SinceAug. 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
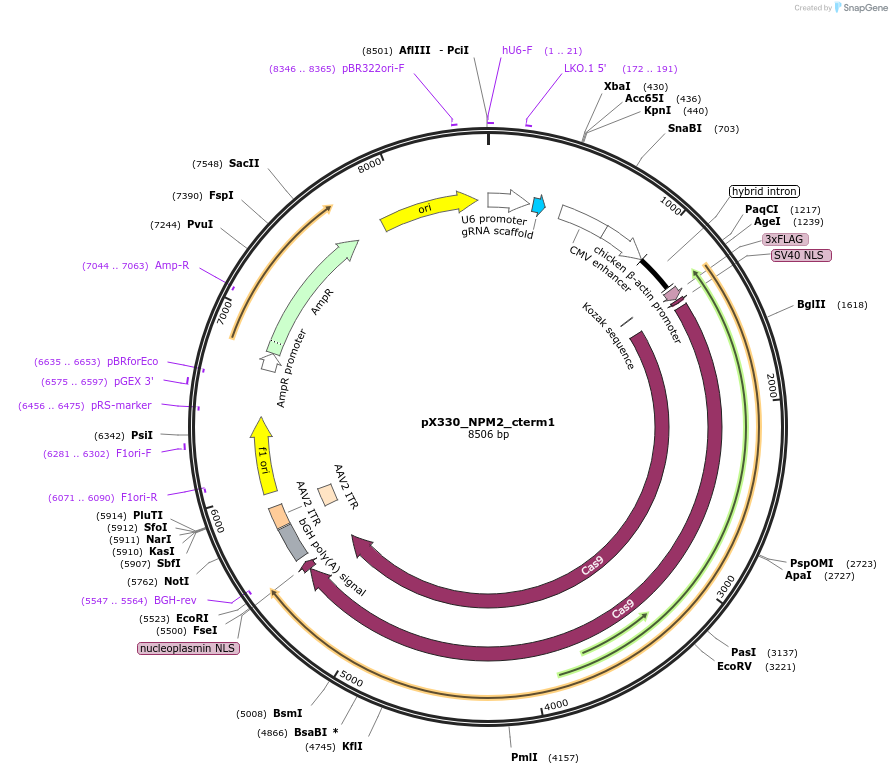
pX330_NPM2_cterm1
Plasmid#222911PurposeCas9/sgRNA plasmid for targeting NPM2DepositorInsertCas9, NPM2 sgRNA 1 (NPM2 Synthetic, Human)
UseCRISPRAvailable SinceJuly 22, 2024AvailabilityAcademic Institutions and Nonprofits only -
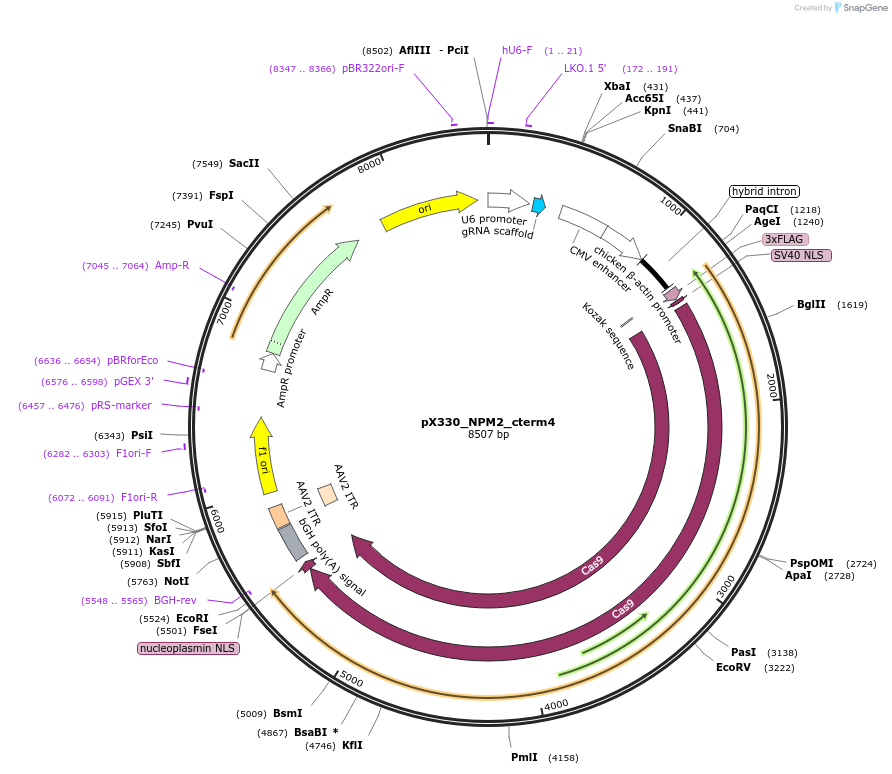
pX330_NPM2_cterm4
Plasmid#222914PurposeCas9/sgRNA plasmid for targeting NPM2DepositorInsertCas9, NPM2 sgRNA 4 (NPM2 Synthetic, Human)
UseCRISPRAvailable SinceJuly 22, 2024AvailabilityAcademic Institutions and Nonprofits only -
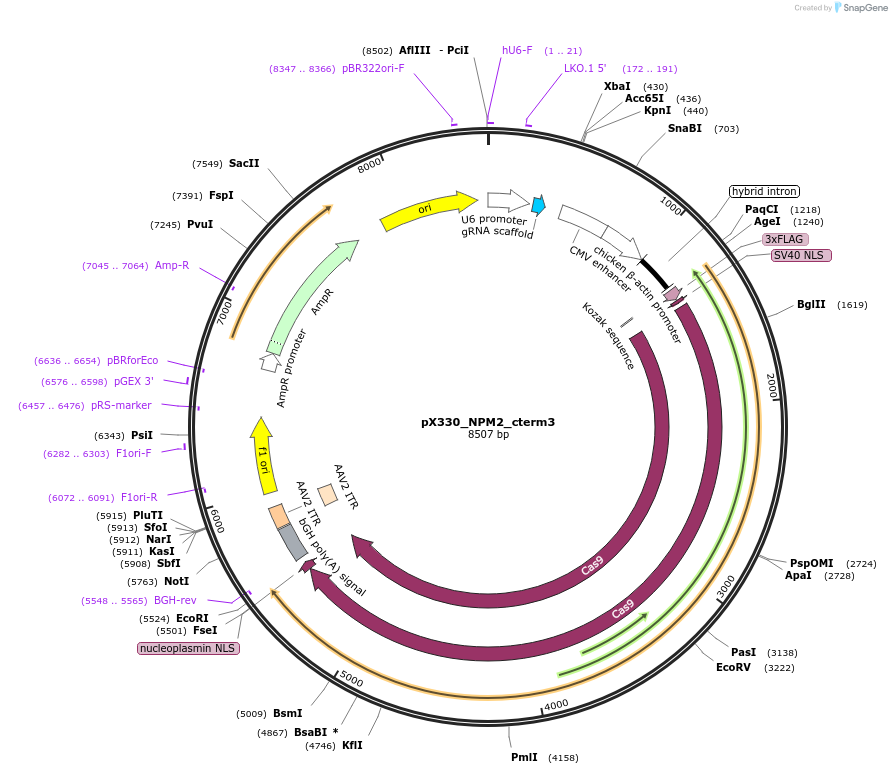
pX330_NPM2_cterm3
Plasmid#222913PurposeCas9/sgRNA plasmid for targeting NPM2DepositorInsertCas9, NPM2 sgRNA 3 (NPM2 Synthetic, Human)
UseCRISPRAvailable SinceJuly 22, 2024AvailabilityAcademic Institutions and Nonprofits only