We narrowed to 8,512 results for: gnal
-
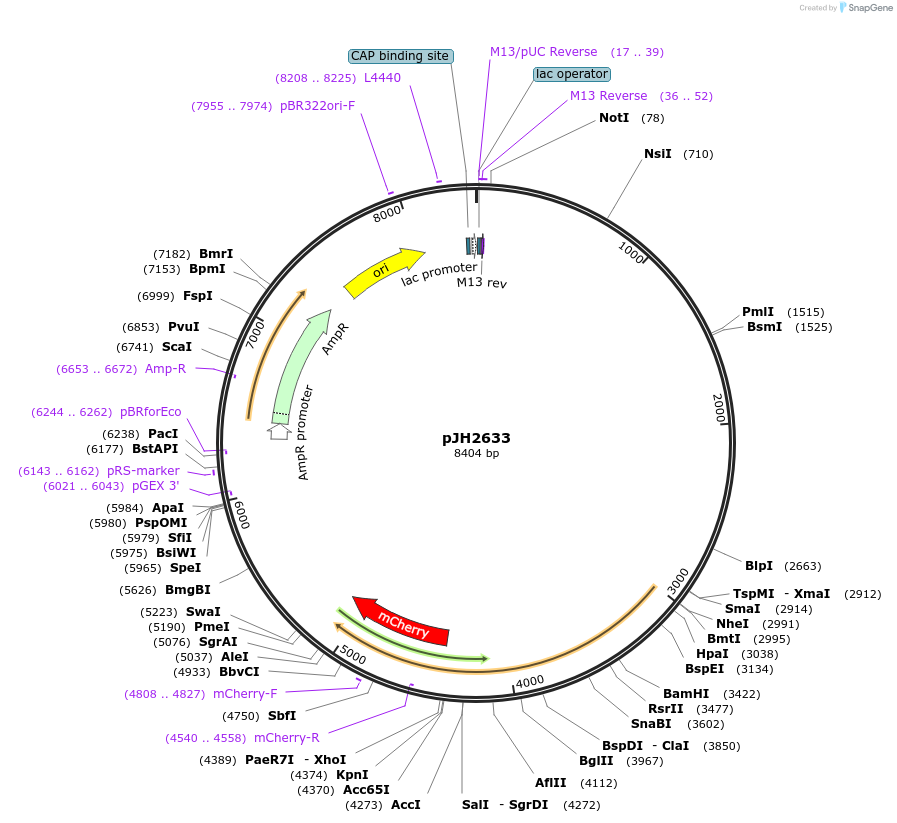
Plasmid#192107PurposePsra-11-TeTx::wCherry unc-54 3' UTR C.elegans AVB and other neurons expression of TeTx RFPDepositorInsertTetanus Toxin Light Chain
TagswCherryExpressionWormPromoterPsra-11Available SinceNov. 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
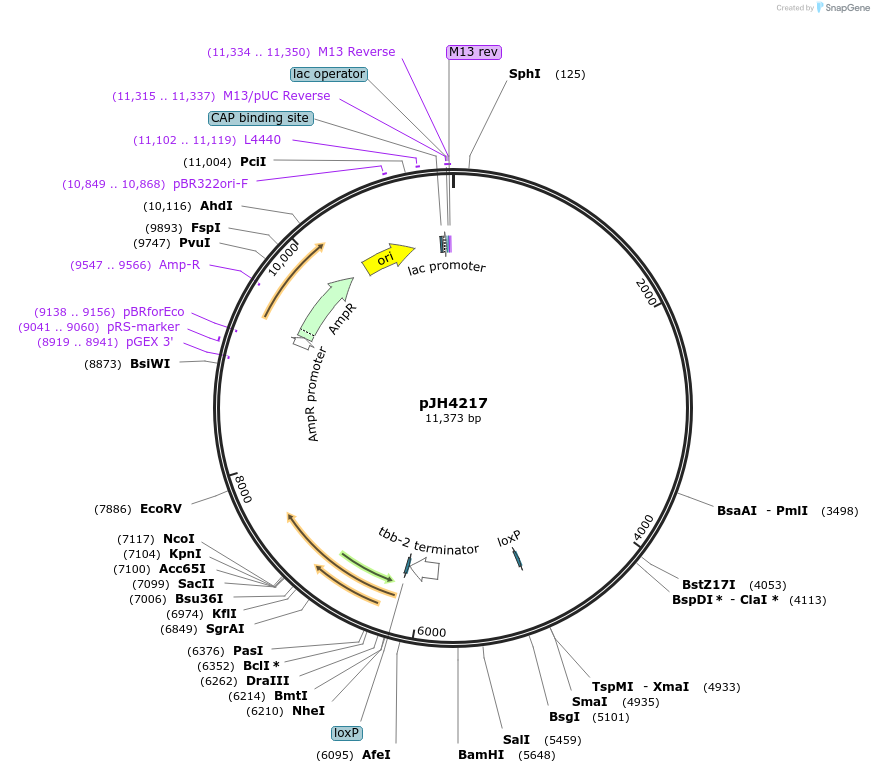
pJH4217
Plasmid#191533PurposePrig-3-LoxP::EBFP::LoxP::GtACR2::wCherry unc-54 3' UTR C.elegans AVA neuron expression of GtACR2 RFPDepositorInsertGtACR2
TagswCherryExpressionWormPromoterPrig-3Available SinceNov. 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
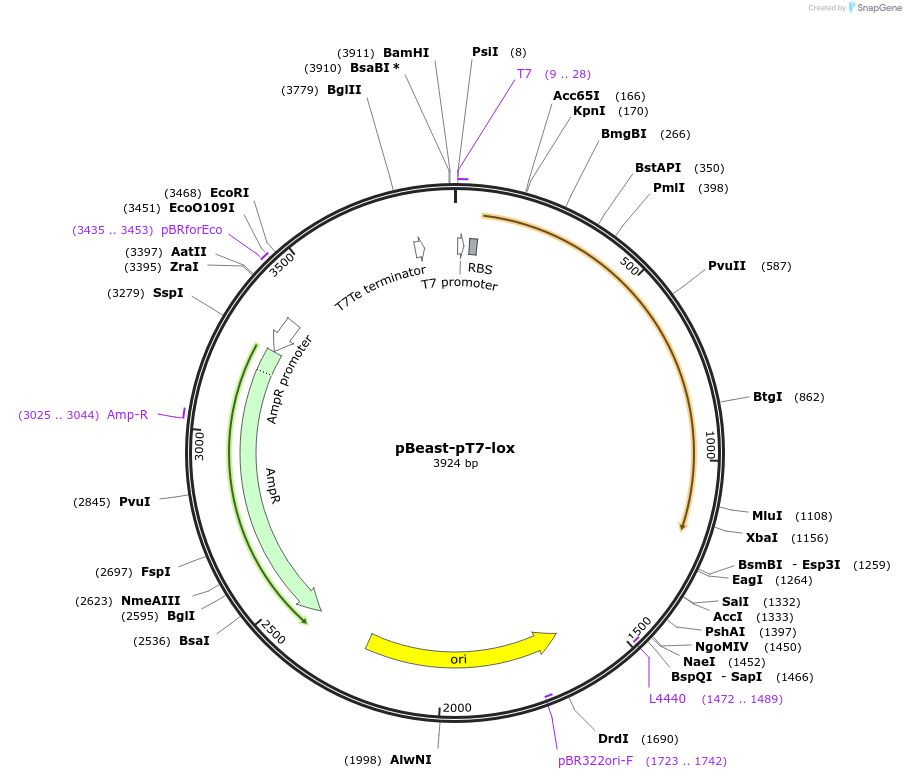
pBeast-pT7-lox
Plasmid#190059PurposeExpress the bacterial lactate oxidase enzyme under the control of the strong constitutif phage promoter T7DepositorInsertlox
UseSynthetic Biology; Cell-free protein synthesisAvailable SinceNov. 22, 2022AvailabilityAcademic Institutions and Nonprofits only -
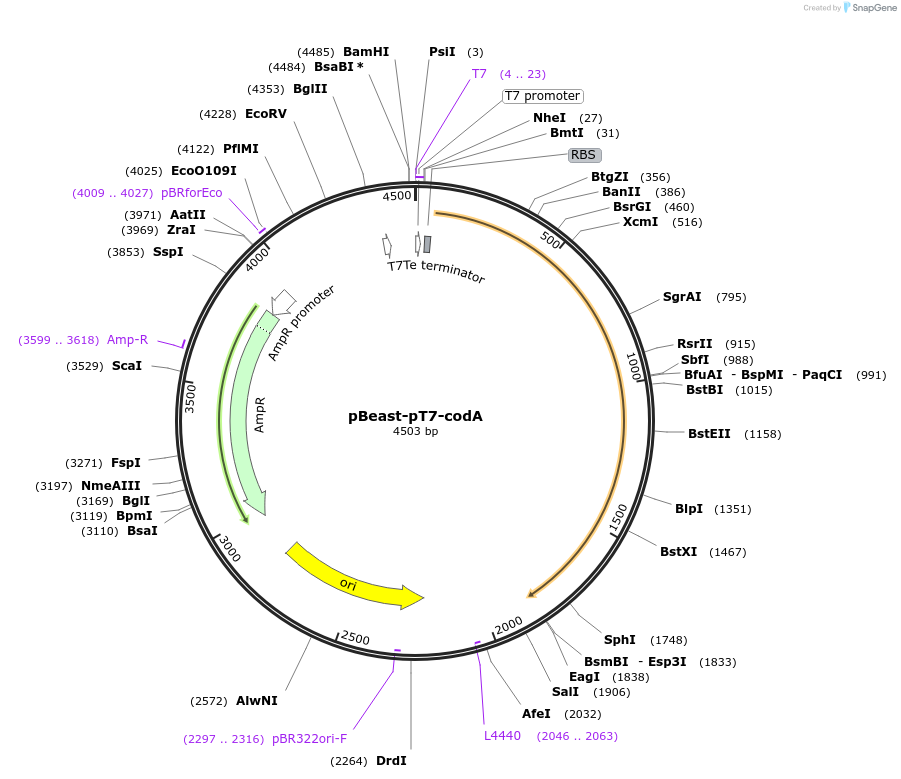
pBeast-pT7-codA
Plasmid#190058PurposeExpress the bacterial choline oxidase enzyme under the control of the strong constitutif phage promoter T7DepositorInsertcodA
UseSynthetic Biology; Cell-free protein synthesisAvailable SinceNov. 22, 2022AvailabilityAcademic Institutions and Nonprofits only -
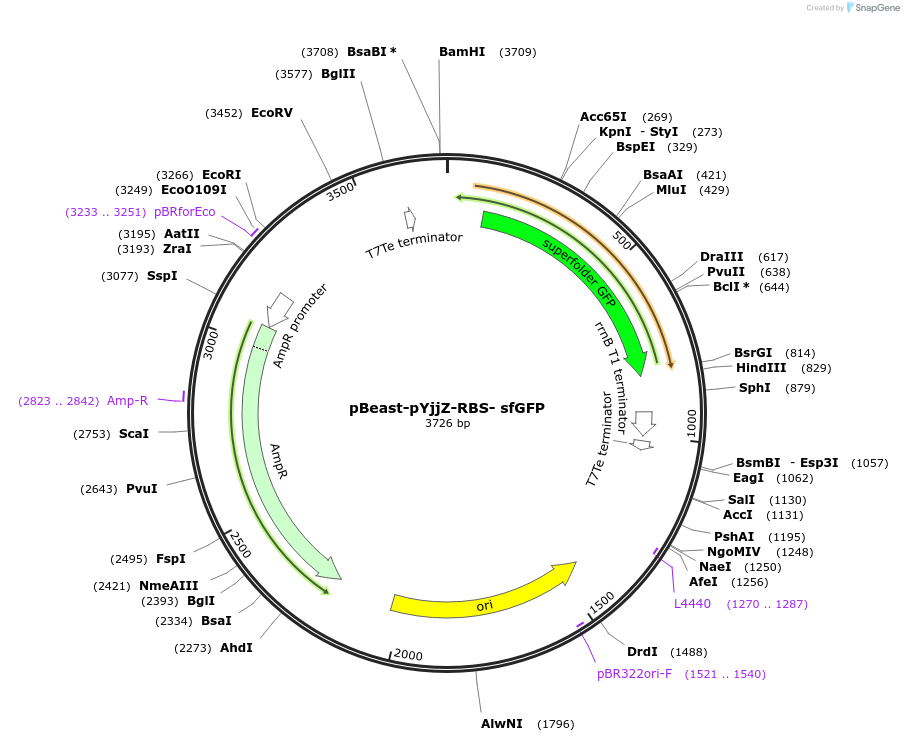
pBeast-pYjjZ-RBS- sfGFP
Plasmid#190054PurposeGFP containing reporter plasmid expressing it under the control of the reportedely OxyR/H2O2 inducible pYjjZ promoterDepositorInsertsfGFP
UseSynthetic Biology; Cell-free protein synthesisAvailable SinceNov. 22, 2022AvailabilityAcademic Institutions and Nonprofits only -
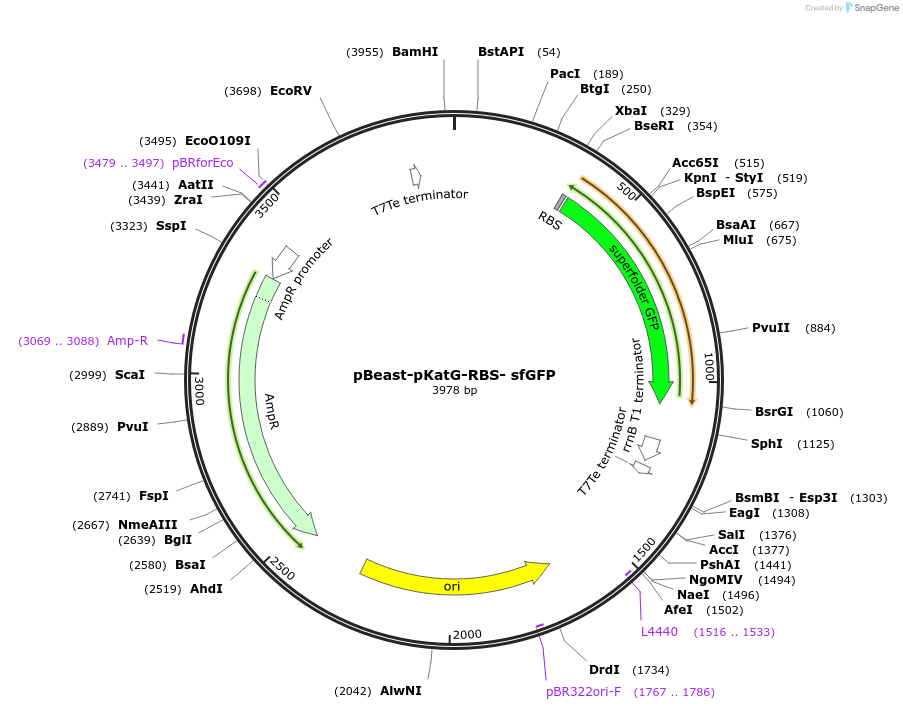
pBeast-pKatG-RBS- sfGFP
Plasmid#190053PurposeGFP containing reporter plasmid expressing it under the control of the OxyR/H2O2 inducible pKatG promoterDepositorInsertsfGFP
UseSynthetic Biology; Cell-free protein synthesisAvailable SinceNov. 22, 2022AvailabilityAcademic Institutions and Nonprofits only -
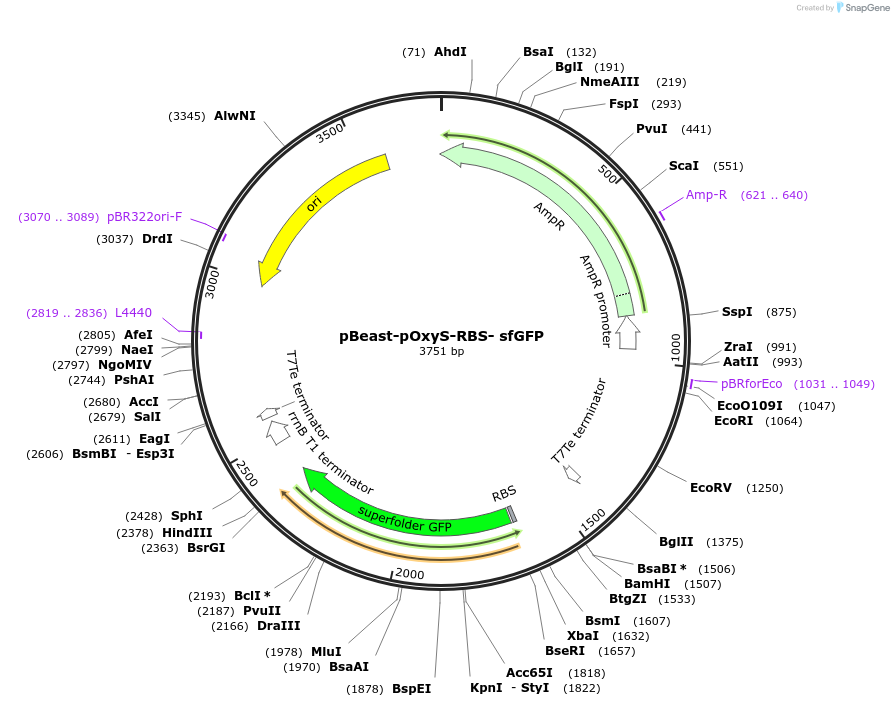
pBeast-pOxyS-RBS- sfGFP
Plasmid#190052PurposeGFP containing reporter plasmid expressing it under the control of the OxyR/H2O2 inducible pOxyS promoterDepositorInsertsfGFP
UseSynthetic Biology; Cell-free protein synthesisAvailable SinceNov. 22, 2022AvailabilityAcademic Institutions and Nonprofits only -
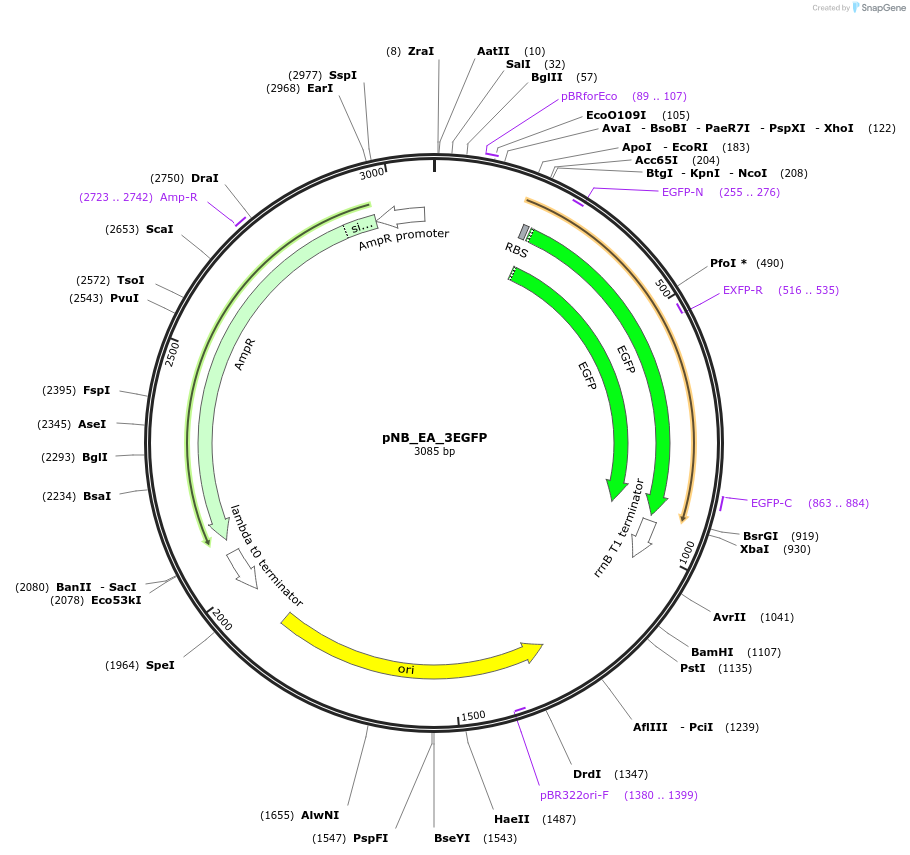
pNB_EA_3EGFP
Plasmid#185967PurposeExpresses EGFP from PR promoter in E.coli Dh5alpha cellDepositorInsertPr-Egfp cassette in forward direction in Network Brick
UseSynthetic BiologyPromoterPRAvailable SinceAug. 29, 2022AvailabilityAcademic Institutions and Nonprofits only -
pAc-His-zfCsf1b
Plasmid#168104PurposeExpress soluble His-tagged zebrafish Csf1b in insect cellsDepositorAvailable SinceAug. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
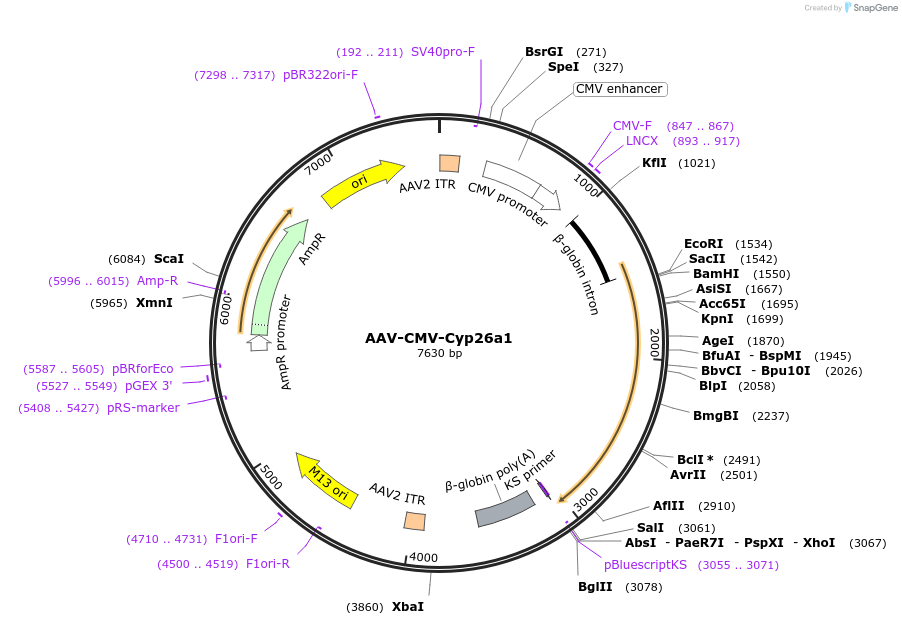
AAV-CMV-Cyp26a1
Plasmid#185563PurposeAAV construct to ubiquitously express mouse Cyp26a1DepositorAvailable SinceJune 24, 2022AvailabilityAcademic Institutions and Nonprofits only -
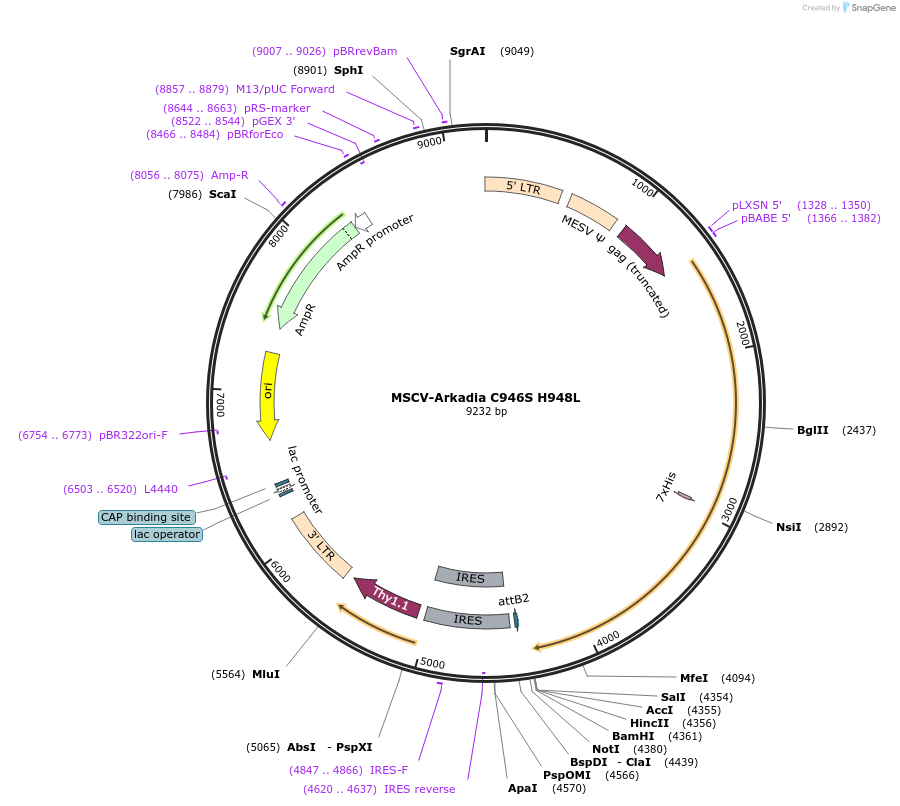
MSCV-Arkadia C946S H948L
Plasmid#180373Purposeexpressing mouse Arkadia (isoform 3) C946S H948L E3 ligase dead mutant proteinsDepositorInsertArkadia (Rnf111 Mouse)
UseRetroviralExpressionMammalianMutationC946S H948LPromoterLTR promoterAvailable SinceJune 14, 2022AvailabilityAcademic Institutions and Nonprofits only -
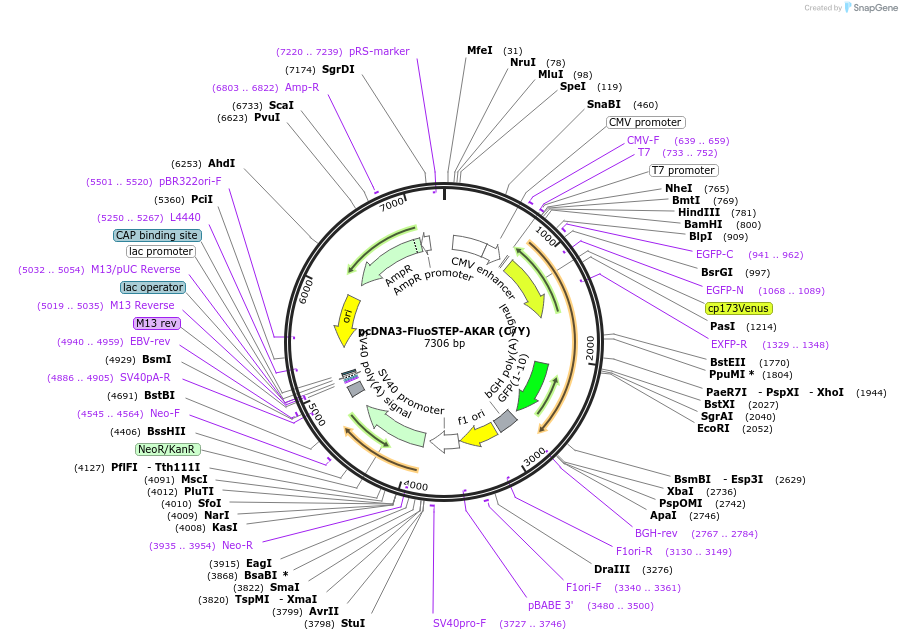
pcDNA3-FluoSTEP-AKAR (C/Y)
Plasmid#181862PurposeFluoSTEP-AKAR using split CFP and cpVenus(E172) as the FRET donor and acceptor. Must pair with GFP11-tagged POI to reconstitute donor fluorescence.DepositorInsertFluoSTEP-AKAR (C-Y)
TagsCFP(1-10) and cpVenus(E127)ExpressionMammalianPromoterCMVAvailable SinceMay 26, 2022AvailabilityAcademic Institutions and Nonprofits only -
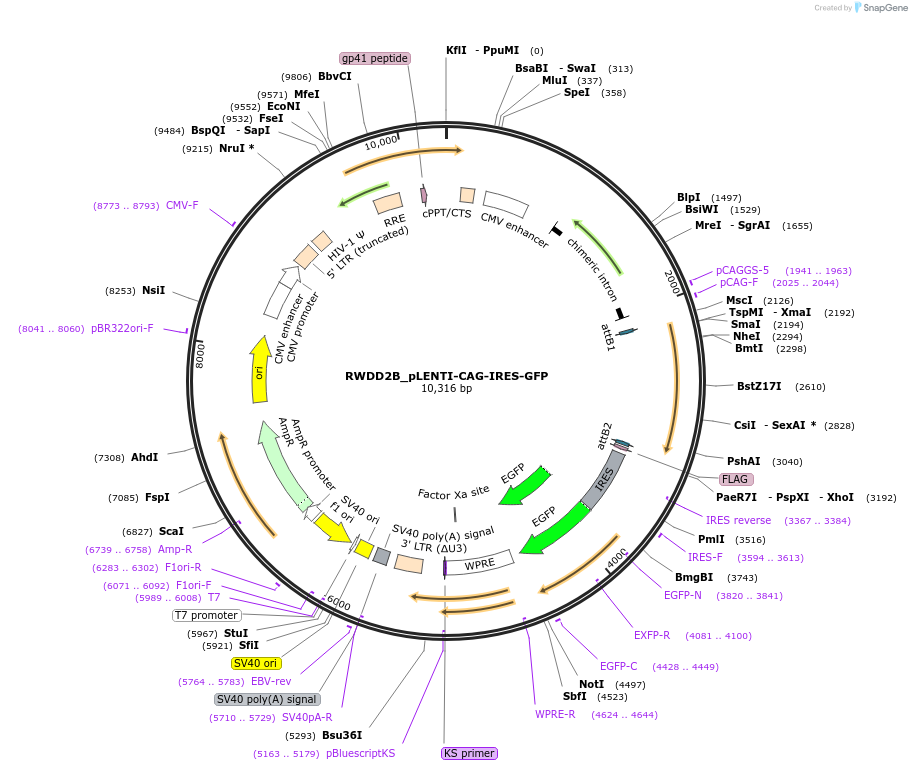
RWDD2B_pLENTI-CAG-IRES-GFP
Plasmid#177001PurposeMammalian lentiviral expression vector encoding RWDD2BDepositorAvailable SinceApril 19, 2022AvailabilityAcademic Institutions and Nonprofits only -
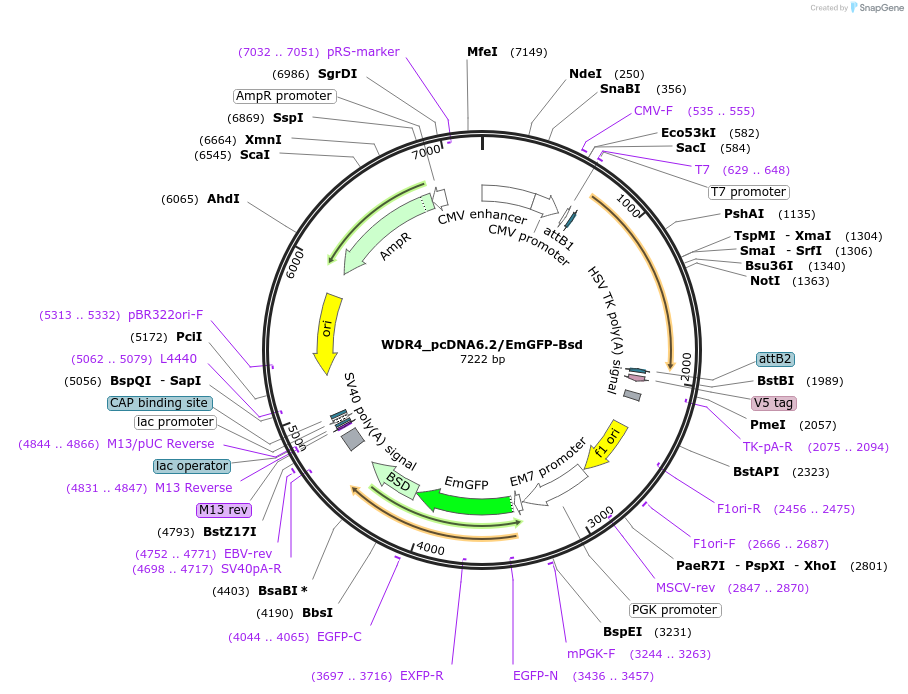
WDR4_pcDNA6.2/EmGFP-Bsd
Plasmid#176984PurposeMammalian expression vector encoding WDR4 and EmGFP-BsdDepositorAvailable SinceApril 19, 2022AvailabilityAcademic Institutions and Nonprofits only -
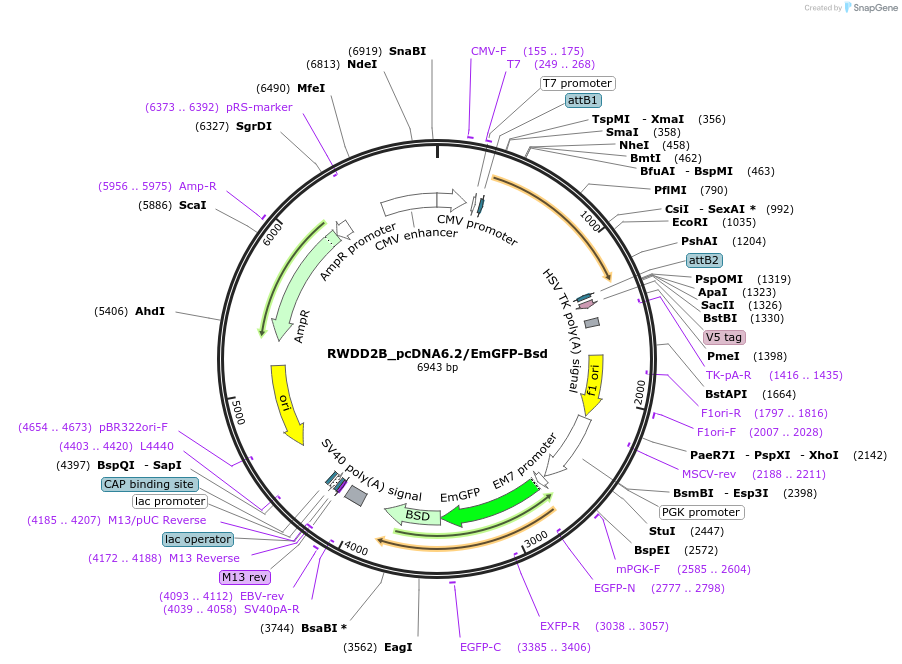
RWDD2B_pcDNA6.2/EmGFP-Bsd
Plasmid#176956PurposeMammalian expression vector encoding RWDD2B and EmGFP-BsdDepositorAvailable SinceApril 19, 2022AvailabilityAcademic Institutions and Nonprofits only -
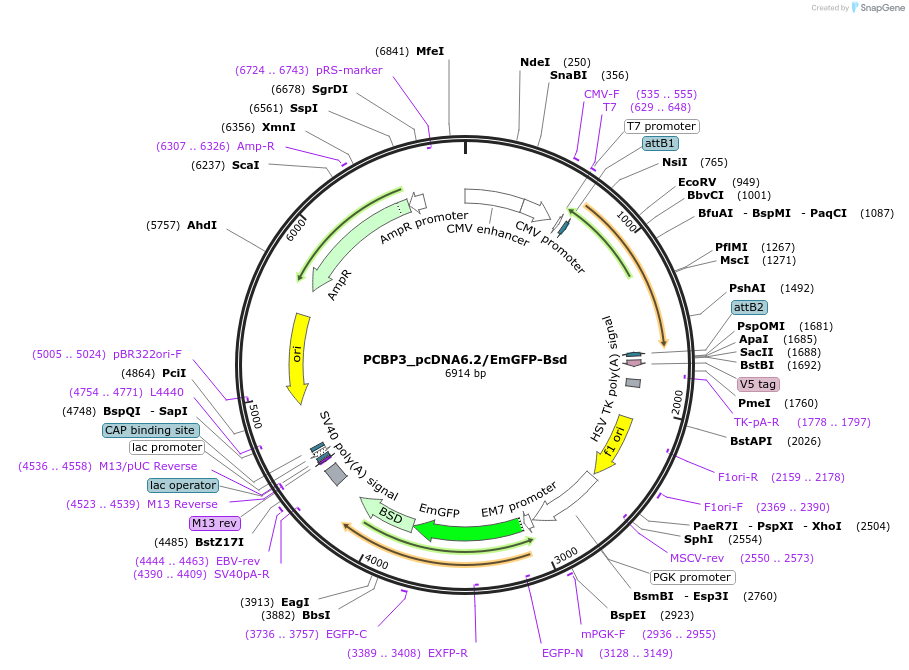
PCBP3_pcDNA6.2/EmGFP-Bsd
Plasmid#176958PurposeMammalian expression vector encoding PCBP3 and EmGFP-BsdDepositorAvailable SinceApril 19, 2022AvailabilityAcademic Institutions and Nonprofits only -
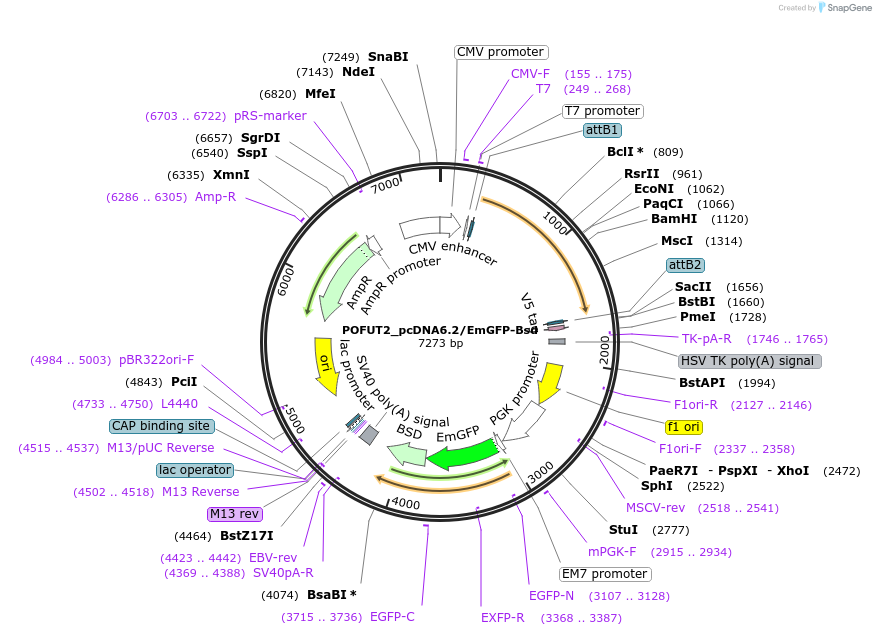
POFUT2_pcDNA6.2/EmGFP-Bsd
Plasmid#176960PurposeMammalian expression vector encoding POFUT2 and EmGFP-BsdDepositorAvailable SinceApril 19, 2022AvailabilityAcademic Institutions and Nonprofits only -
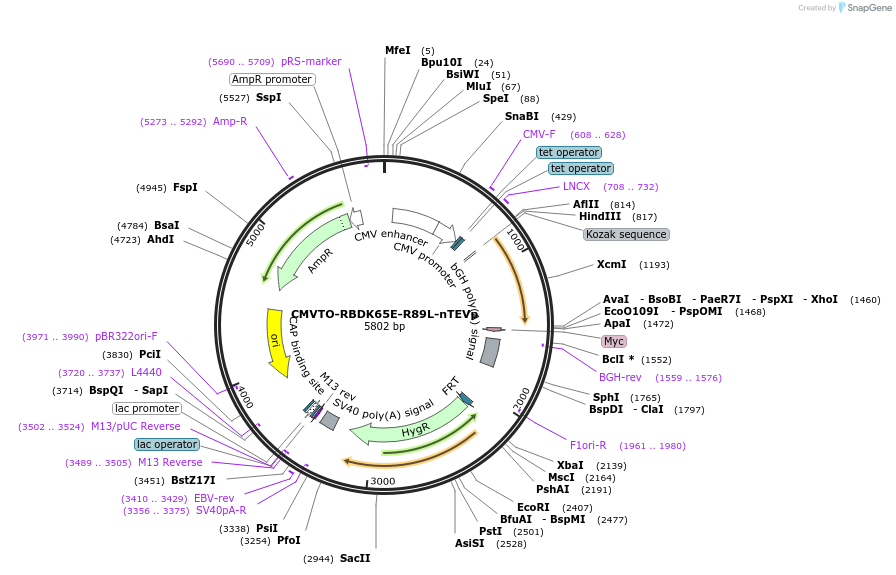
CMVTO-RBDK65E-R89L-nTEVp
Plasmid#179670PurposeMutant Ras Binding Domain split-nTEVPDepositorAvailable SinceMarch 23, 2022AvailabilityAcademic Institutions and Nonprofits only -
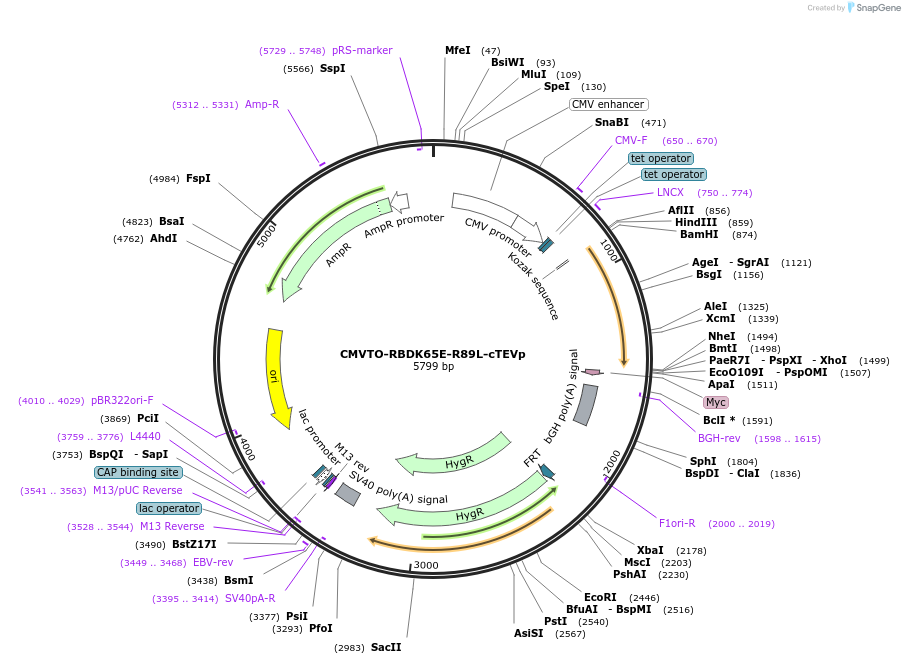
CMVTO-RBDK65E-R89L-cTEVp
Plasmid#179671PurposeMutant Ras Binding Domain split-cTEVPDepositorAvailable SinceMarch 23, 2022AvailabilityAcademic Institutions and Nonprofits only -
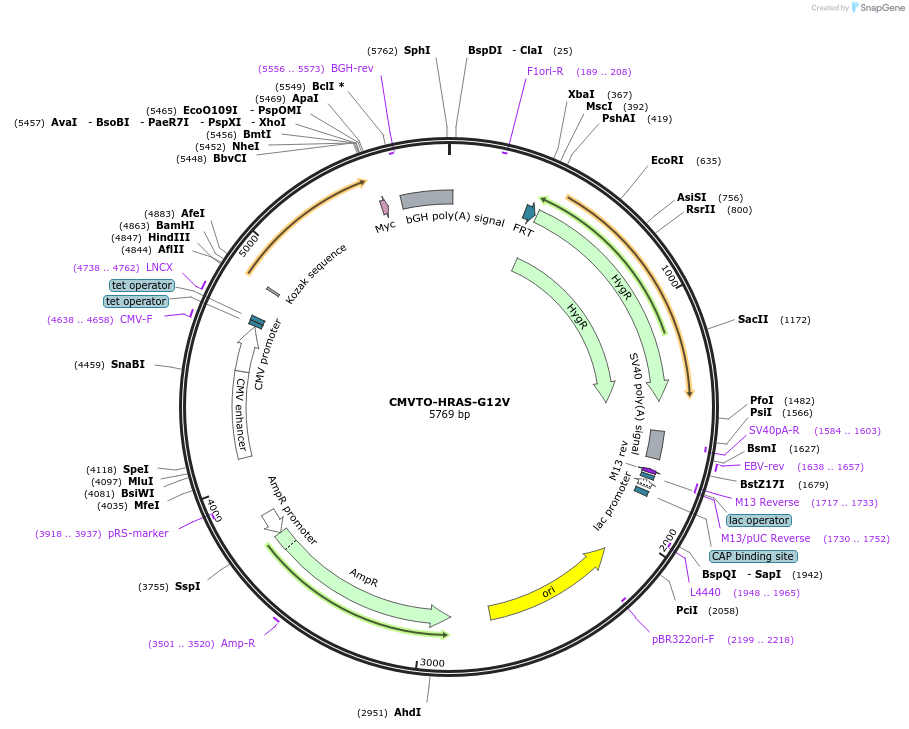
CMVTO-HRAS-G12V
Plasmid#179652PurposeHRAS active mutant (G12V mutation)DepositorAvailable SinceMarch 23, 2022AvailabilityAcademic Institutions and Nonprofits only