We narrowed to 9,541 results for: pho;
-
Plasmid#153194Purposeencoded soma targeted NovArchDepositorInsertsNovArch
TagscitrineExpressionMammalianAvailable SinceMay 10, 2021AvailabilityAcademic Institutions and Nonprofits only -
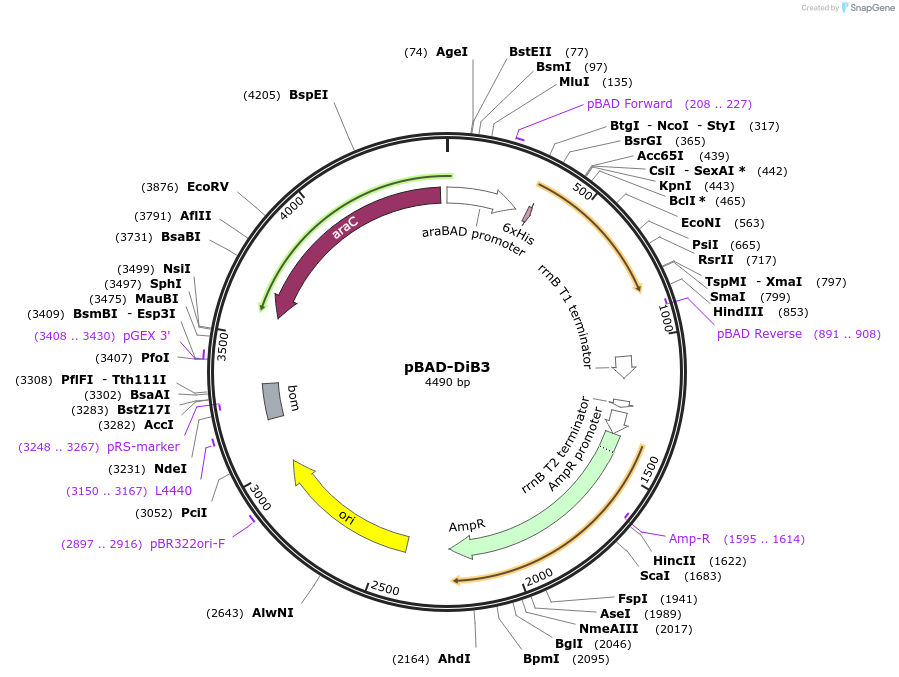
pBAD-DiB3
Plasmid#168802PurposeFluorogen activating protein (FAP) DiB3DepositorInsertFluorogen activating protein DiB3
TagsHis-tagExpressionBacterialPromoteraraBADAvailable SinceMay 5, 2021AvailabilityAcademic Institutions and Nonprofits only -
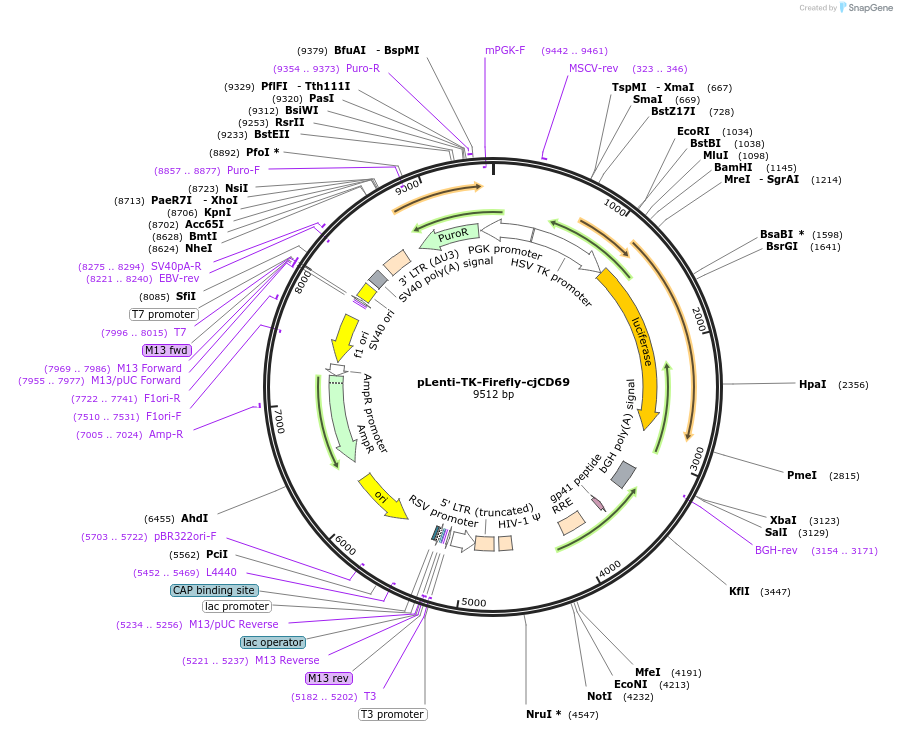
pLenti-TK-Firefly-cjCD69
Plasmid#122279PurposeLentiviral vector for stable expression of Firefly luciferase.DepositorInsertsFirefly luciferase
cjCD69 3'UTR
UseLentiviral and LuciferaseExpressionMammalianAvailable SinceApril 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
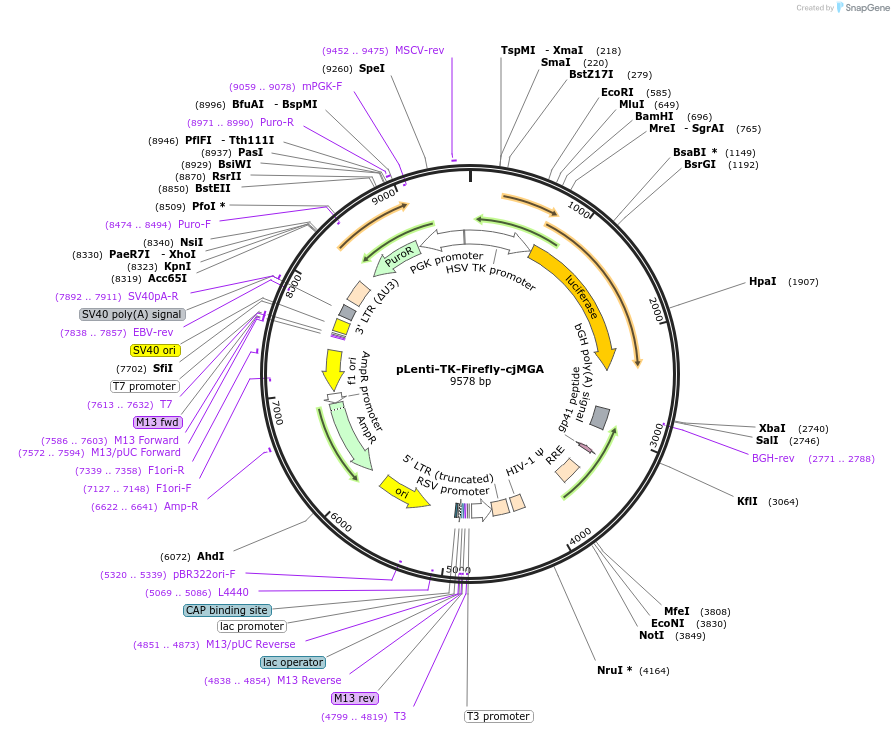
pLenti-TK-Firefly-cjMGA
Plasmid#122280PurposeLentiviral vector for stable expression of Firefly luciferase.DepositorInsertsFirefly luciferase
cjMGA 3'UTR
UseLentiviral and LuciferaseExpressionMammalianAvailable SinceApril 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
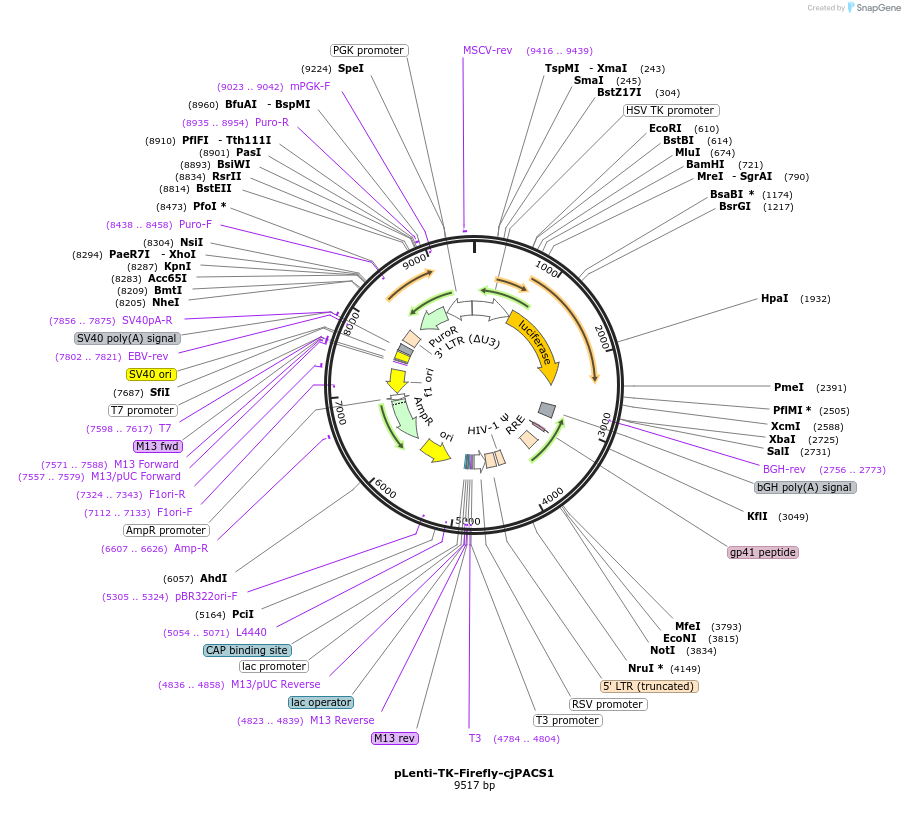
pLenti-TK-Firefly-cjPACS1
Plasmid#122281PurposeLentiviral vector for stable expression of Firefly luciferase.DepositorInsertsFirefly luciferase
cjPACS1 3'UTR
UseLentiviral and LuciferaseExpressionMammalianAvailable SinceApril 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
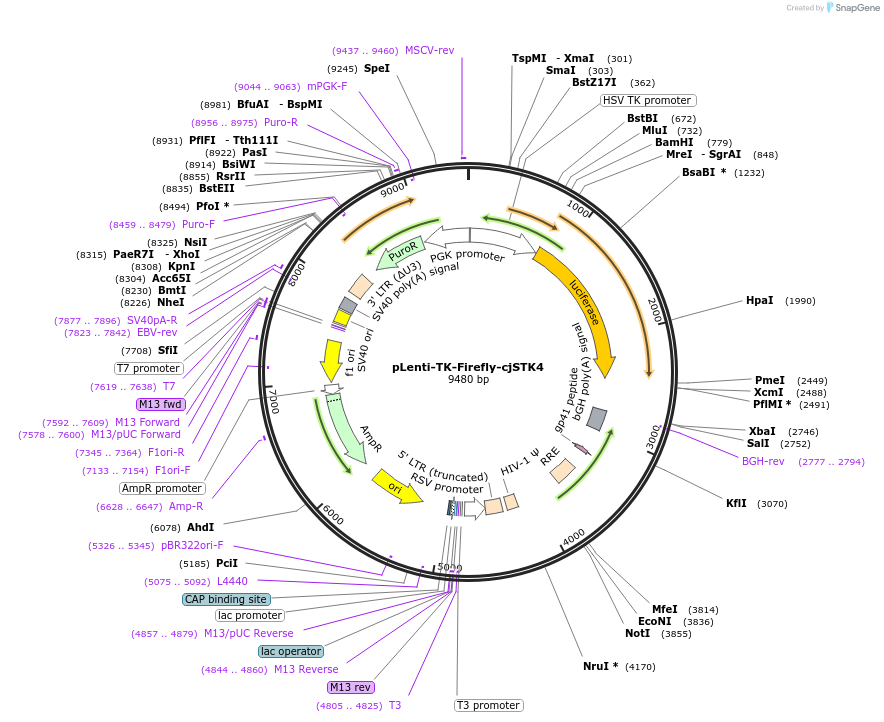
pLenti-TK-Firefly-cjSTK4
Plasmid#122282PurposeLentiviral vector for stable expression of Firefly luciferase.DepositorInsertsFirefly luciferase
cjSTK4 3'UTR
UseLentiviral and LuciferaseExpressionMammalianAvailable SinceApril 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
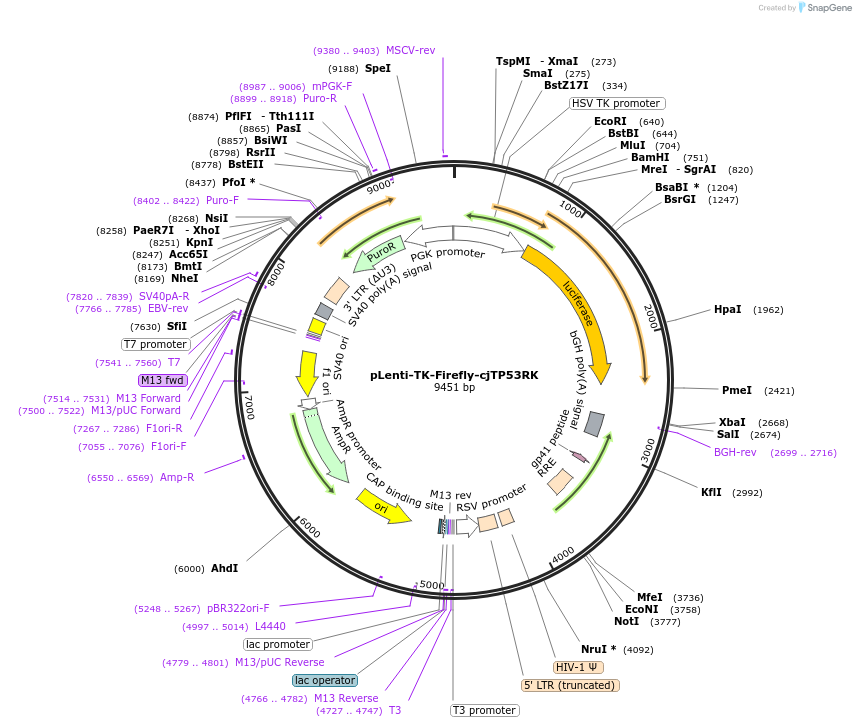
pLenti-TK-Firefly-cjTP53RK
Plasmid#122283PurposeLentiviral vector for stable expression of Firefly luciferase.DepositorInsertsFirefly luciferase
cjTP53RK 3'UTR
UseLentiviral and LuciferaseExpressionMammalianAvailable SinceApril 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
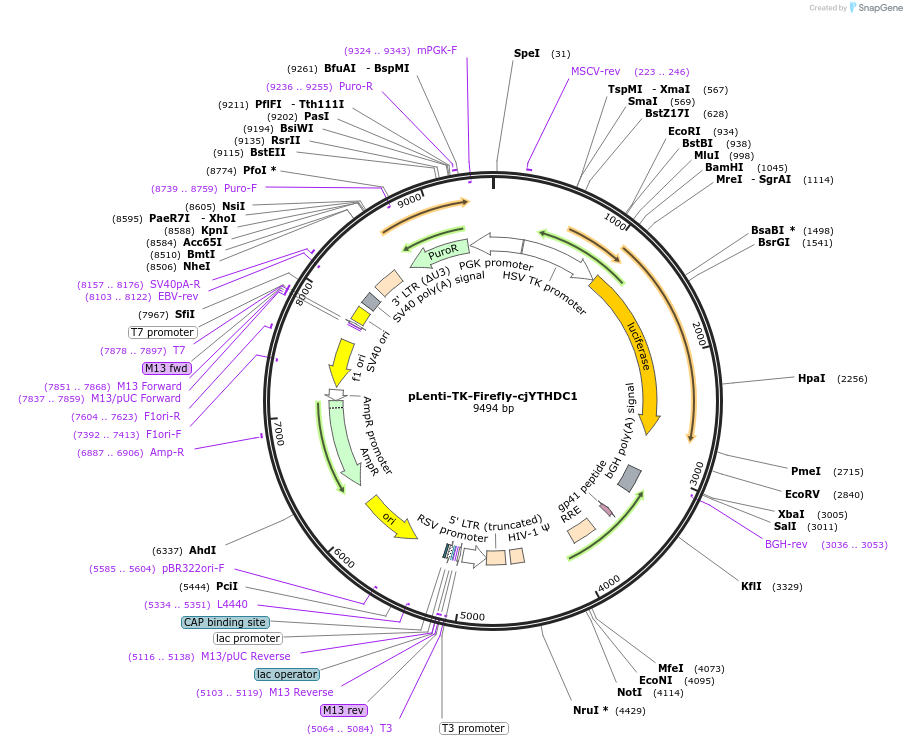
pLenti-TK-Firefly-cjYTHDC1
Plasmid#122284PurposeLentiviral vector for stable expression of Firefly luciferase.DepositorInsertsFirefly luciferase
cjYTHDC1 3'UTR
UseLentiviral and LuciferaseExpressionMammalianAvailable SinceApril 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
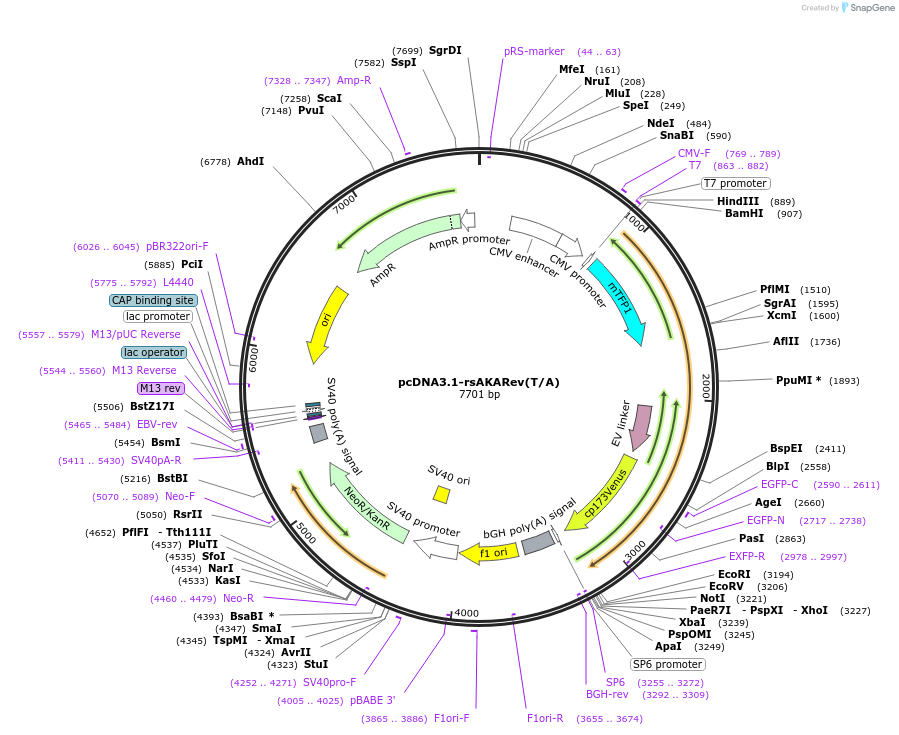
pcDNA3.1-rsAKARev(T/A)
Plasmid#166232PurposeExpresses reversibly switchable A-kinase activity reporter with EV-linker inactive T/A mutant (rsAKARev(T/A)) in mammalian cellsDepositorInsertReversibly switchable A-kinase activity reporter with EV-linker inactive T/A mutant
ExpressionMammalianMutationchanged Threonine 506 in rsAKARev to AlaninePromoterCMVAvailable SinceApril 5, 2021AvailabilityAcademic Institutions and Nonprofits only -
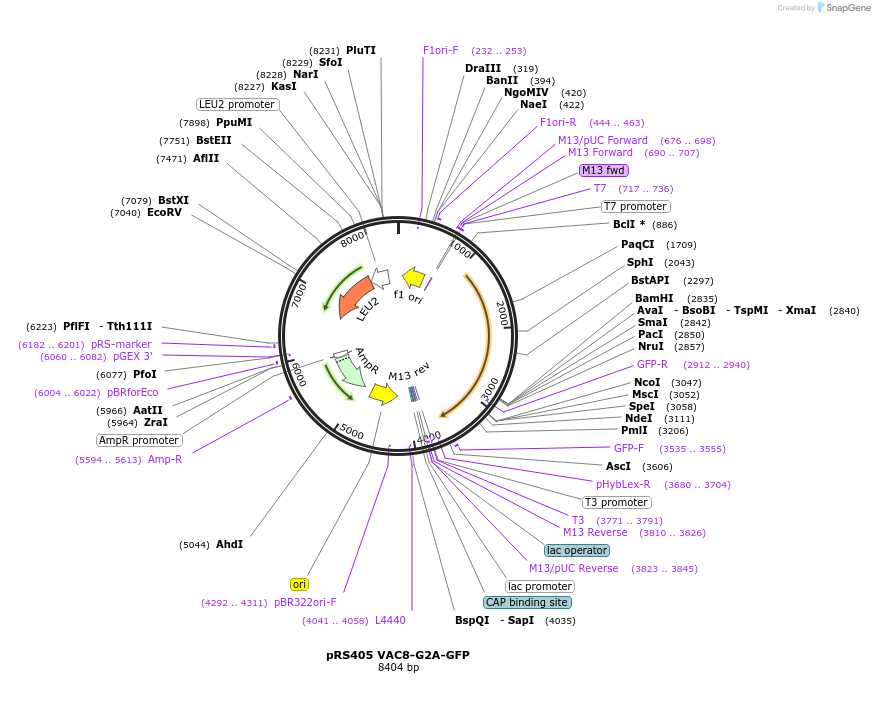
pRS405 VAC8-G2A-GFP
Plasmid#166837PurposeExpresses a myristoylation mutant Vac8-GFP in yeasts, mutation is Gly2AlaDepositorAvailable SinceApril 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
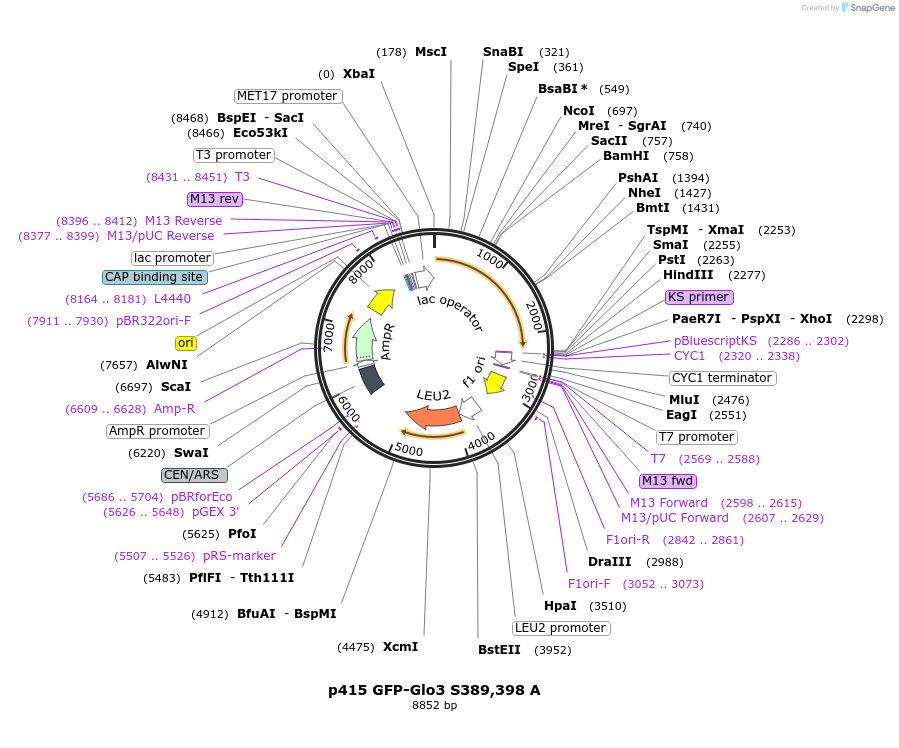
p415 GFP-Glo3 S389,398 A
Plasmid#112660PurposeExpression S.cerevisiaeDepositorInsertGFP- Glo3 S389,398A
TagssfGFPExpressionYeastMutationmutations (S389A, S398A) in Glo3 motif, non-phosp…PromoterMet25Available SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
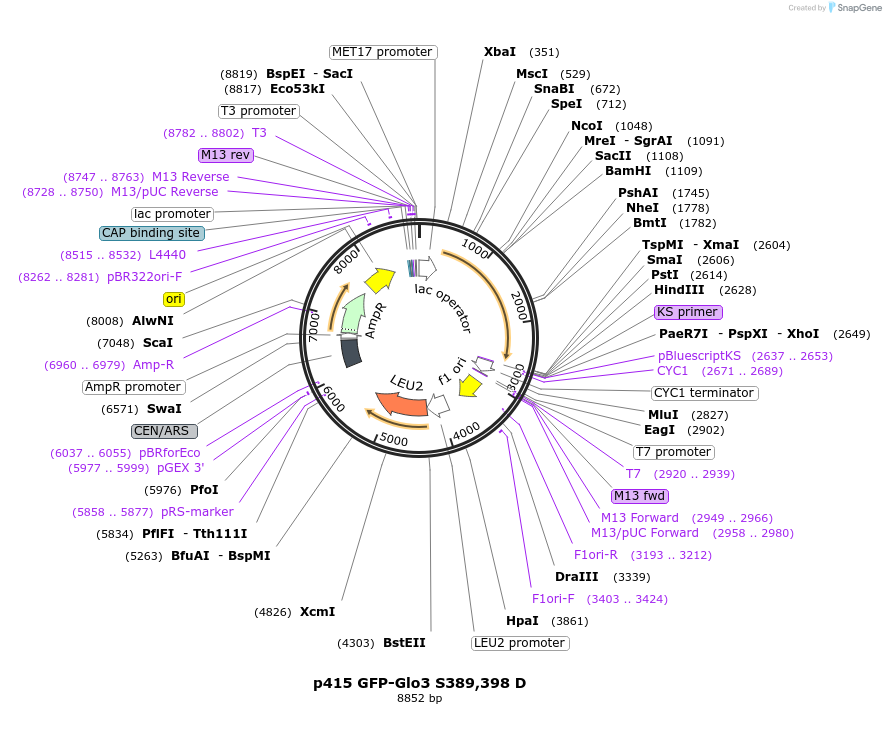
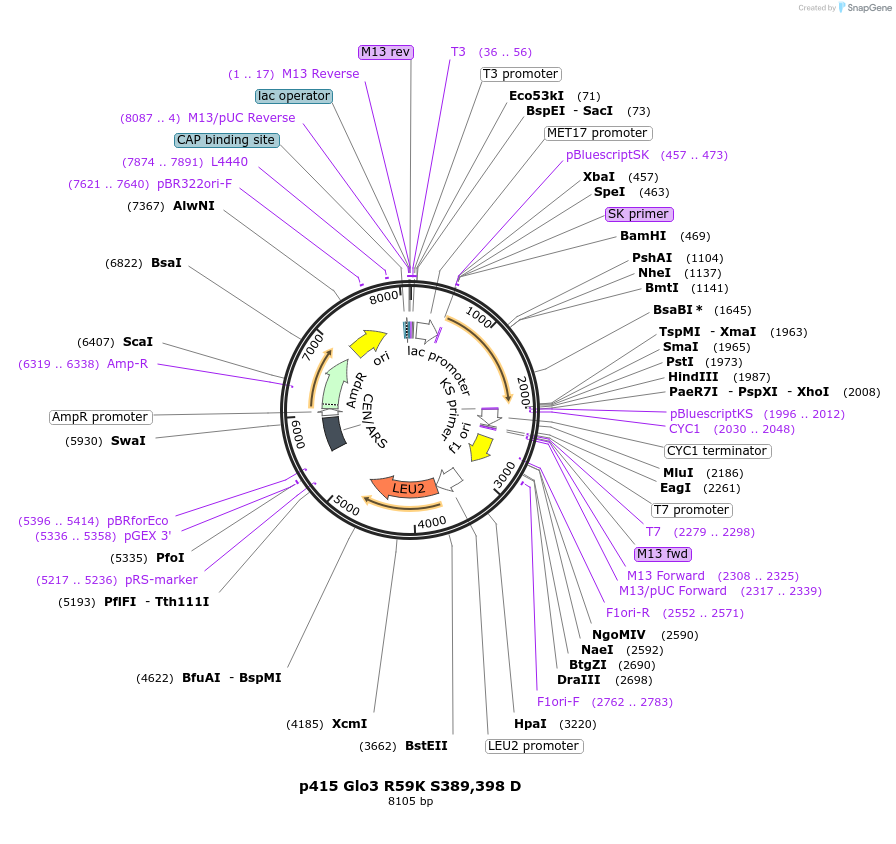
p415 GFP-Glo3 S389,398 D
Plasmid#112661PurposeExpression S.cerevisiaeDepositorInsertGFP-Glo3 S389,398D
TagssfGFPExpressionYeastMutationmutations (S389D, S398D) in Glo3 motif, phosphomi…PromoterMet25Available SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
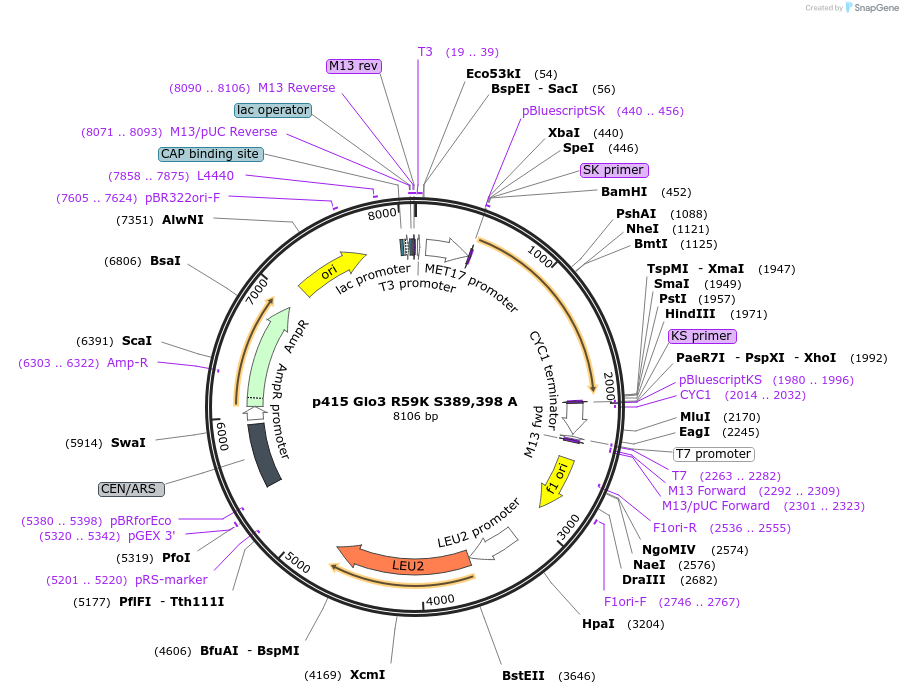
p415 Glo3 R59K S389,398 A
Plasmid#112656PurposeExpression S.cerevisiaeDepositorInsertGlo3 R59K S389,398A
ExpressionYeastMutationR59K, mutations (S389A, S398A) in Glo3 motif, non…PromoterMet25Available SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
p415 Glo3 R59K S389,398 D
Plasmid#112657PurposeExpression S.cerevisiaeDepositorInsertGlo3 R59K S389,398D
ExpressionYeastMutationR59K, mutations (S389D, S398D) in Glo3 motif, pho…PromoterMet25Available SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
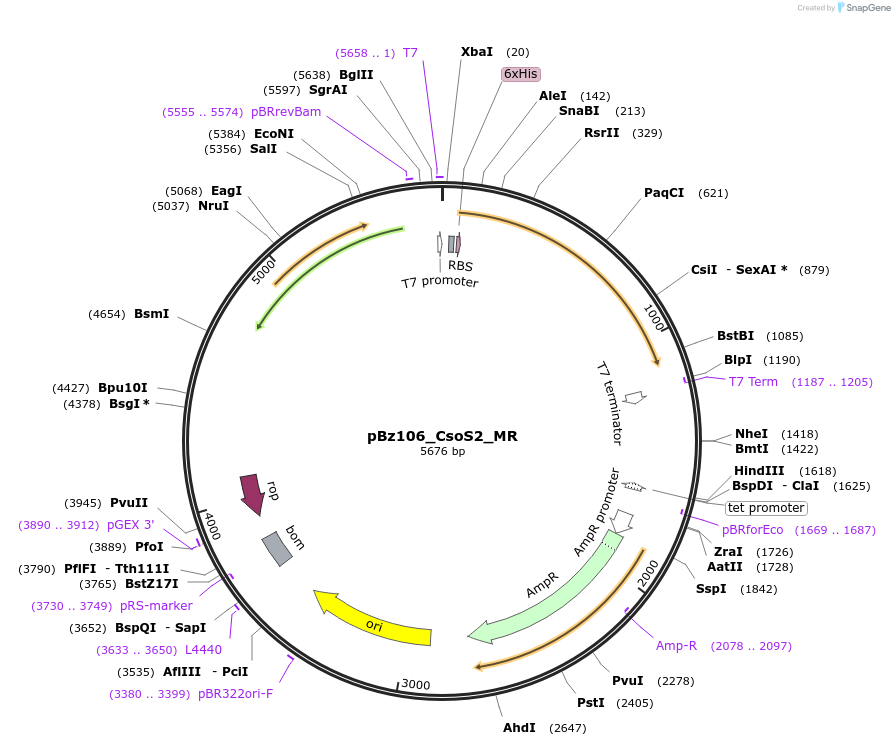
pBz106_CsoS2_MR
Plasmid#140843PurposeHis-tagged CsoS2 Middle Region (MR), pET-14b backboneDepositorInsertCsoS2 Middle Region (MR)
TagsHexahistidine tagExpressionBacterialMutationonly amino acids 257-603PromoterT7Available SinceJune 22, 2020AvailabilityAcademic Institutions and Nonprofits only -
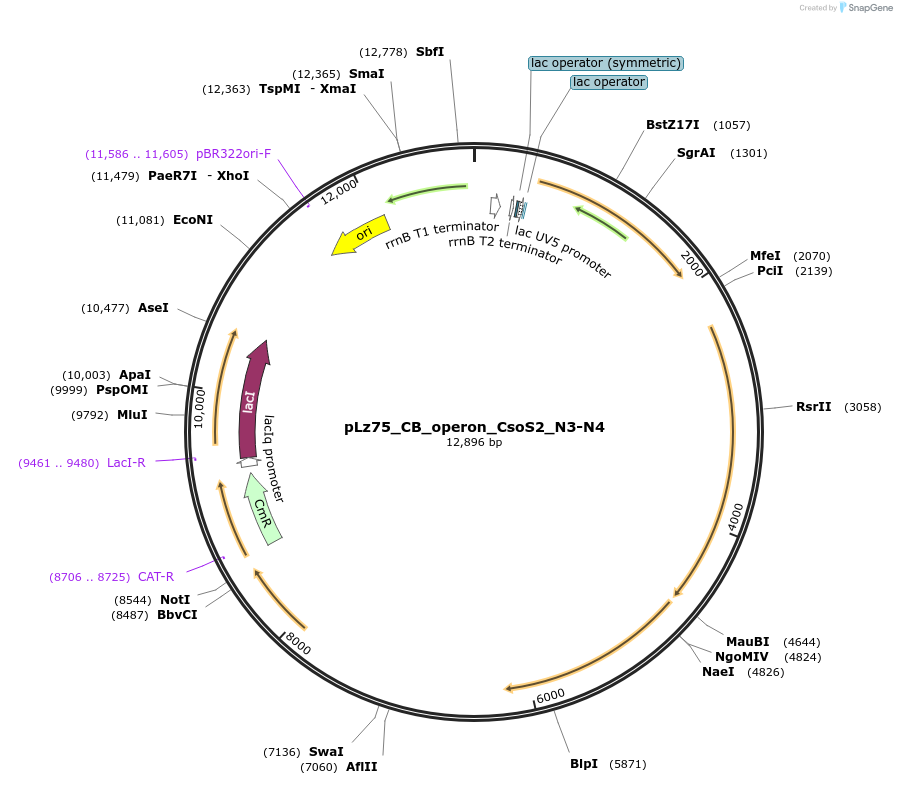
pLz75_CB_operon_CsoS2_N3-N4
Plasmid#140857PurposeSame as pLz37 but with CsoS2 truncated such that NTD repeats N1 and N2 are removedDepositorInsertCbbL, CbbS, CsoS2 (delN1-N2), CsoSCA, CsoS4A, CsoS4B, CsoS1C, CsoS1A, CsoS1B, CsoS1D
ExpressionBacterialMutationdeleted amino acids 1-109 of CsoS2Available SinceJune 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
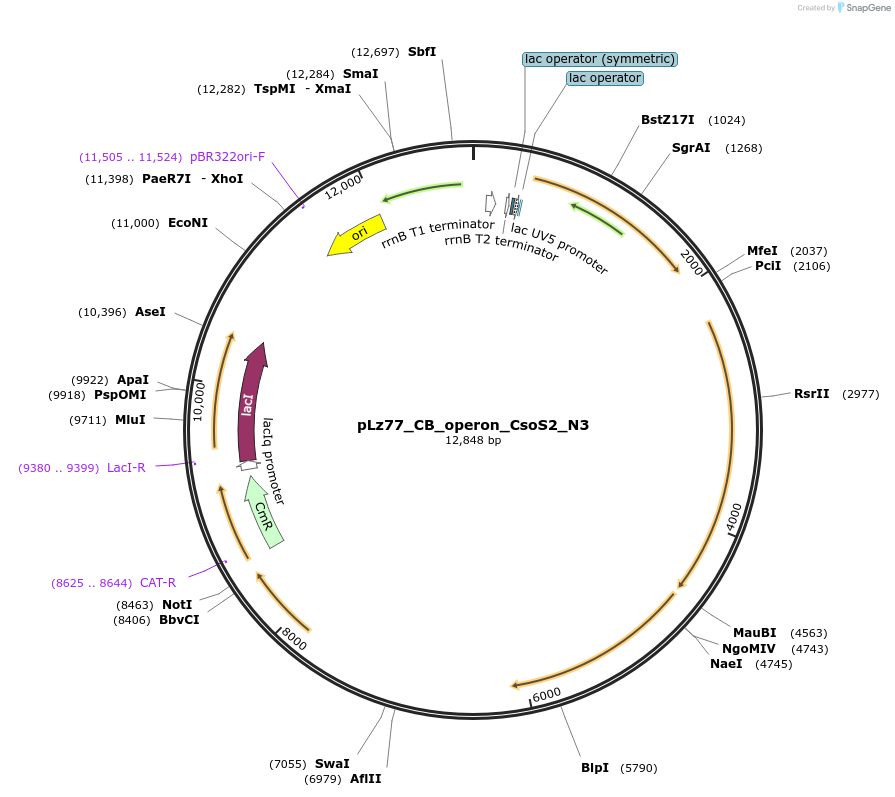
pLz77_CB_operon_CsoS2_N3
Plasmid#140859PurposeSame as pLz37 but with CsoS2 discontinuously truncated such that repeats N1, N2, and N4 are removedDepositorInsertCbbL, CbbS, CsoS2 (delN1-N2, N4), CsoSCA, CsoS4A, CsoS4B, CsoS1C, CsoS1A, CsoS1B, CsoS1D
ExpressionBacterialMutationdeleted amino acids 1-108 and 220-235 of CsoS2Available SinceJune 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
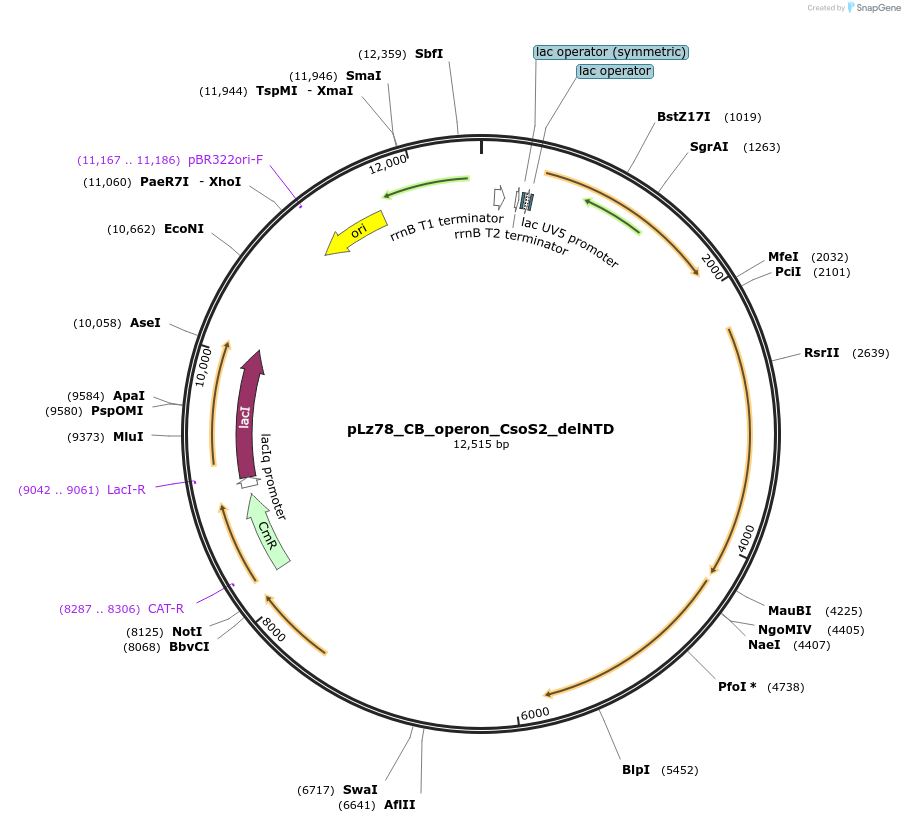
pLz78_CB_operon_CsoS2_delNTD
Plasmid#140860PurposeSame as pLz37 but with CsoS2 truncated to remove the entire NTD (i.e. N1, N2, N3, and N4 are removed)DepositorInsertCbbL, CbbS, CsoS2 (delNTD), CsoSCA, CsoS4A, CsoS4B, CsoS1C, CsoS1A, CsoS1B, CsoS1D
ExpressionBacterialMutationdeleted amino acids 1-235 of CsoS2Available SinceJune 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
pLz76_CB_operon_CsoS2_N4
Plasmid#140858PurposeSame as pLz37 but with CsoS2 truncated such that NTD repeats N1, N2, and N3 are removedDepositorInsertCbbL, CbbS, CsoS2 (delN1-N3), CsoSCA, CsoS4A, CsoS4B, CsoS1C, CsoS1A, CsoS1B, CsoS1D
ExpressionBacterialMutationdeleted amino acids 1-189 of CsoS2Available SinceJune 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
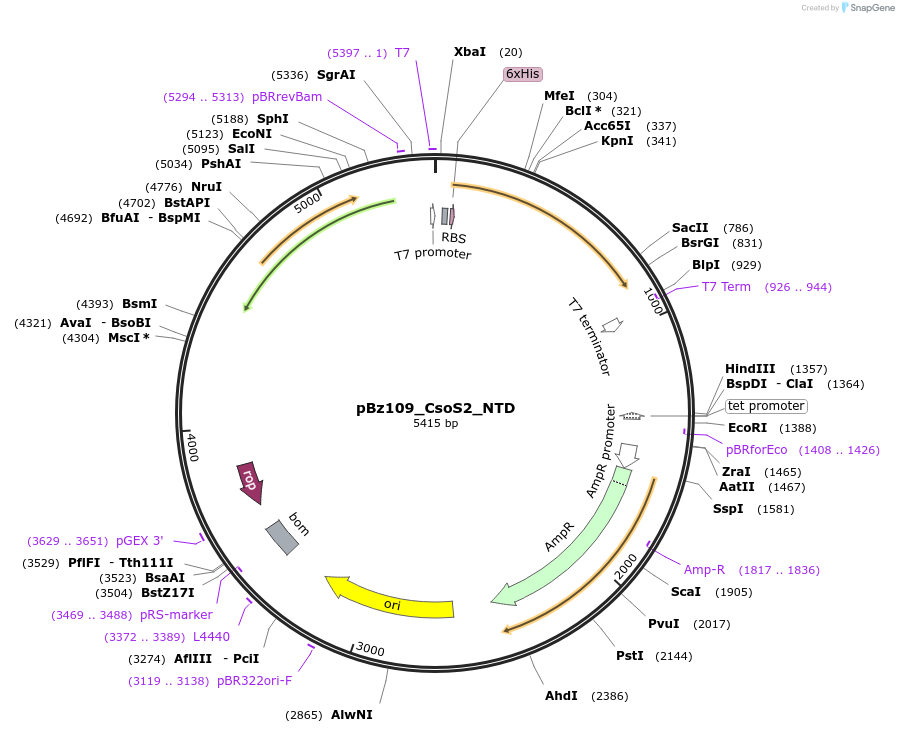
pBz109_CsoS2_NTD
Plasmid#140842PurposeHis-tagged CsoS2 N-terminal domain (NTD), pET-14b backboneDepositorInsertCsoS2 N-terminal domain (NTD)
TagsHexahistidine tagExpressionBacterialMutationdeleted amino acids 262-869PromoterT7Available SinceJune 2, 2020AvailabilityAcademic Institutions and Nonprofits only