We narrowed to 13,611 results for: Mpl;
-
Plasmid#136727PurposeTemplate to synthesize riboprobes for in situ hybridization (ISH) for Olfr1239DepositorInsertOlfr1239
UseIn situ probeMutationISH Probe insertAvailable SinceOct. 14, 2020AvailabilityAcademic Institutions and Nonprofits only -
pISH-Olfr1353
Plasmid#136730PurposeTemplate to synthesize riboprobes for in situ hybridization (ISH) for Olfr1353DepositorInsertOlfr1353
UseIn situ probeMutationISH Probe insertAvailable SinceOct. 14, 2020AvailabilityAcademic Institutions and Nonprofits only -
pISH-Olfr1402
Plasmid#136732PurposeTemplate to synthesize riboprobes for in situ hybridization (ISH) for Olfr1402DepositorInsertOlfr466
UseIn situ probeMutationISH Probe insertAvailable SinceOct. 14, 2020AvailabilityAcademic Institutions and Nonprofits only -
pISH-Olfr1412
Plasmid#136733PurposeTemplate to synthesize riboprobes for in situ hybridization (ISH) for Olfr1412DepositorInsertOlfr1412
UseIn situ probeMutationISH Probe insertAvailable SinceOct. 14, 2020AvailabilityAcademic Institutions and Nonprofits only -
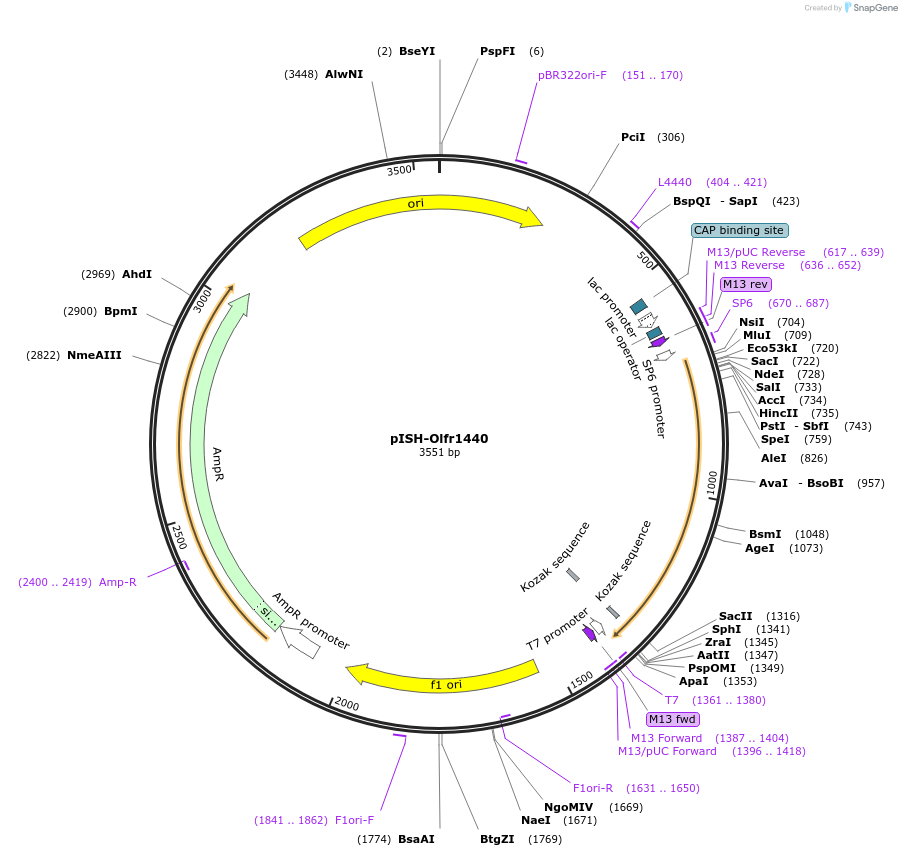
pISH-Olfr1440
Plasmid#136734PurposeTemplate to synthesize riboprobes for in situ hybridization (ISH) for Olfr1440DepositorInsertOlfr1440
UseIn situ probeMutationISH Probe insertAvailable SinceOct. 14, 2020AvailabilityAcademic Institutions and Nonprofits only -
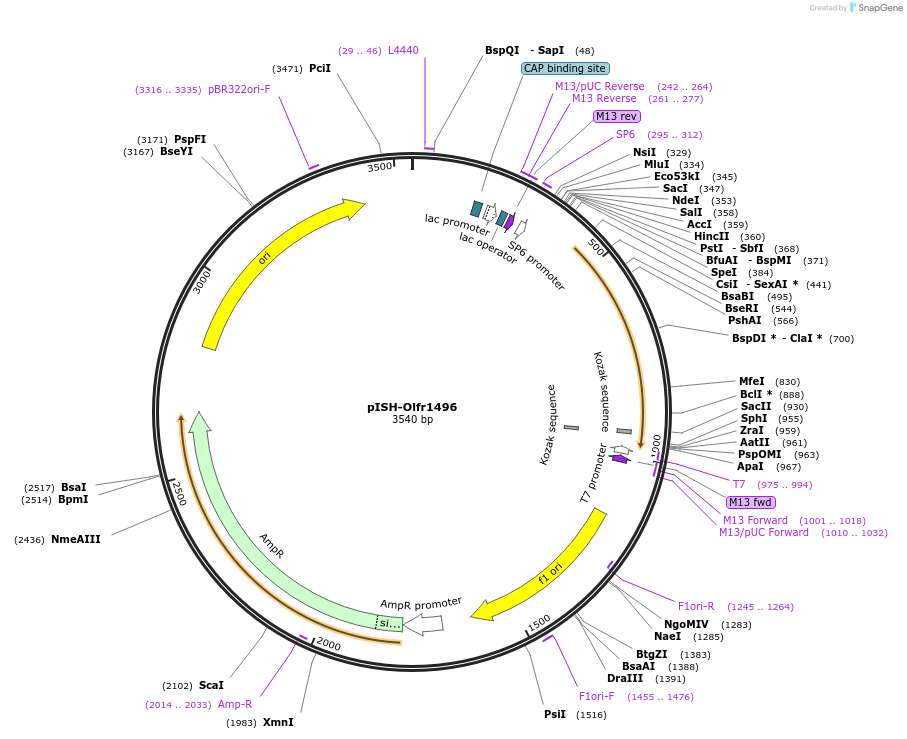
pISH-Olfr1496
Plasmid#136735PurposeTemplate to synthesize riboprobes for in situ hybridization (ISH) for Olfr1496DepositorInsertOlfr1496
UseIn situ probeMutationISH Probe insertAvailable SinceOct. 14, 2020AvailabilityAcademic Institutions and Nonprofits only -
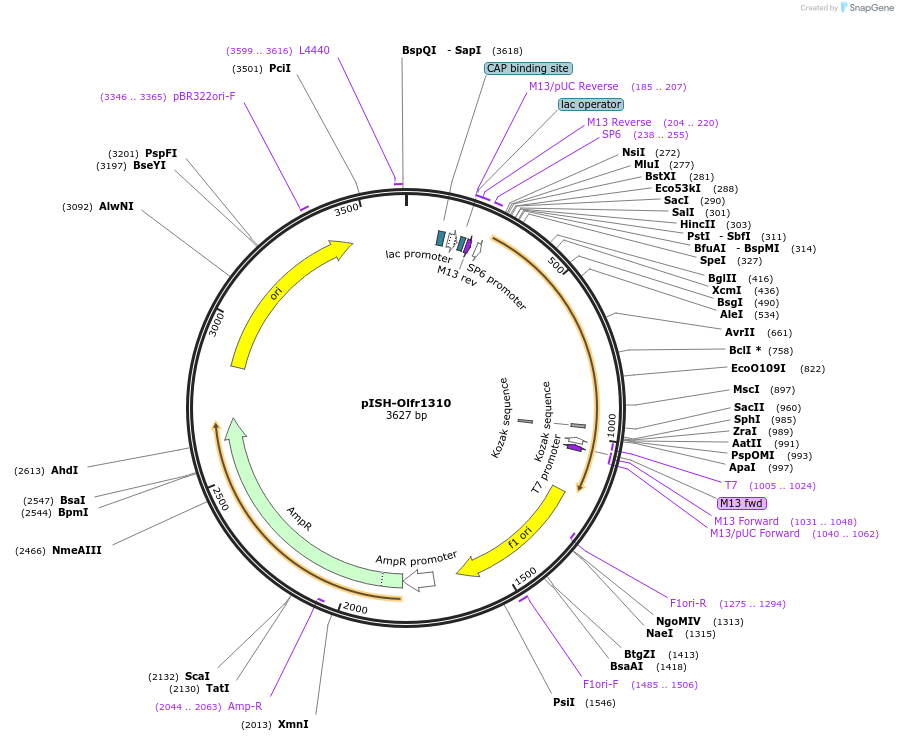
pISH-Olfr1310
Plasmid#136737PurposeTemplate to synthesize riboprobes for in situ hybridization (ISH) for Olfr1310DepositorInsertOlfr1310
UseIn situ probeMutationISH Probe insertAvailable SinceOct. 14, 2020AvailabilityAcademic Institutions and Nonprofits only -
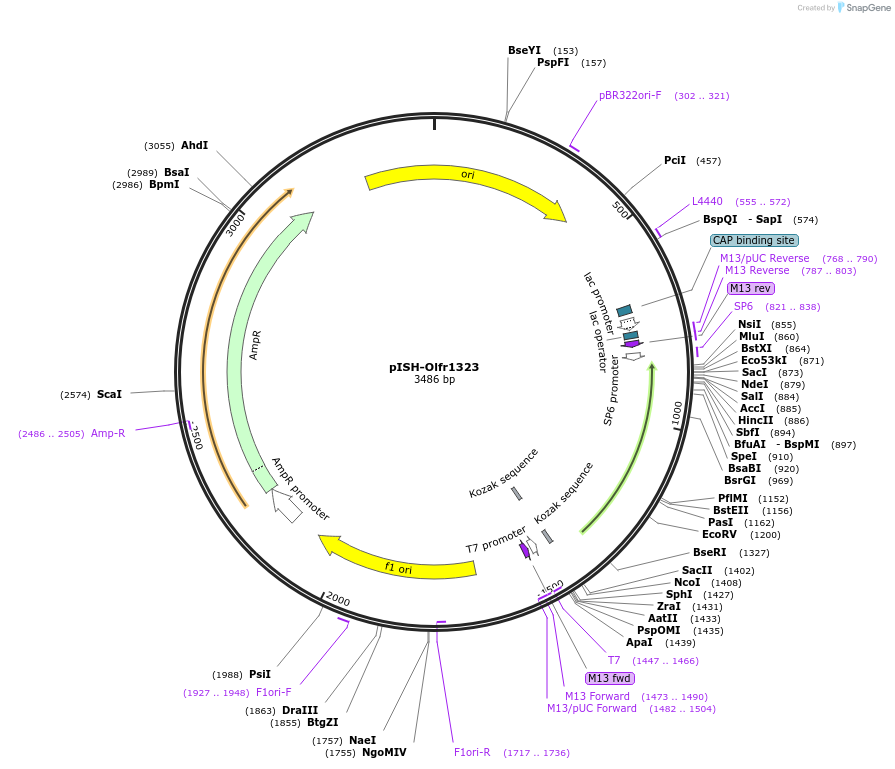
pISH-Olfr1323
Plasmid#136738PurposeTemplate to synthesize riboprobes for in situ hybridization (ISH) for Olfr1323DepositorInsertOlfr1323
UseIn situ probeMutationISH Probe insertAvailable SinceOct. 14, 2020AvailabilityAcademic Institutions and Nonprofits only -
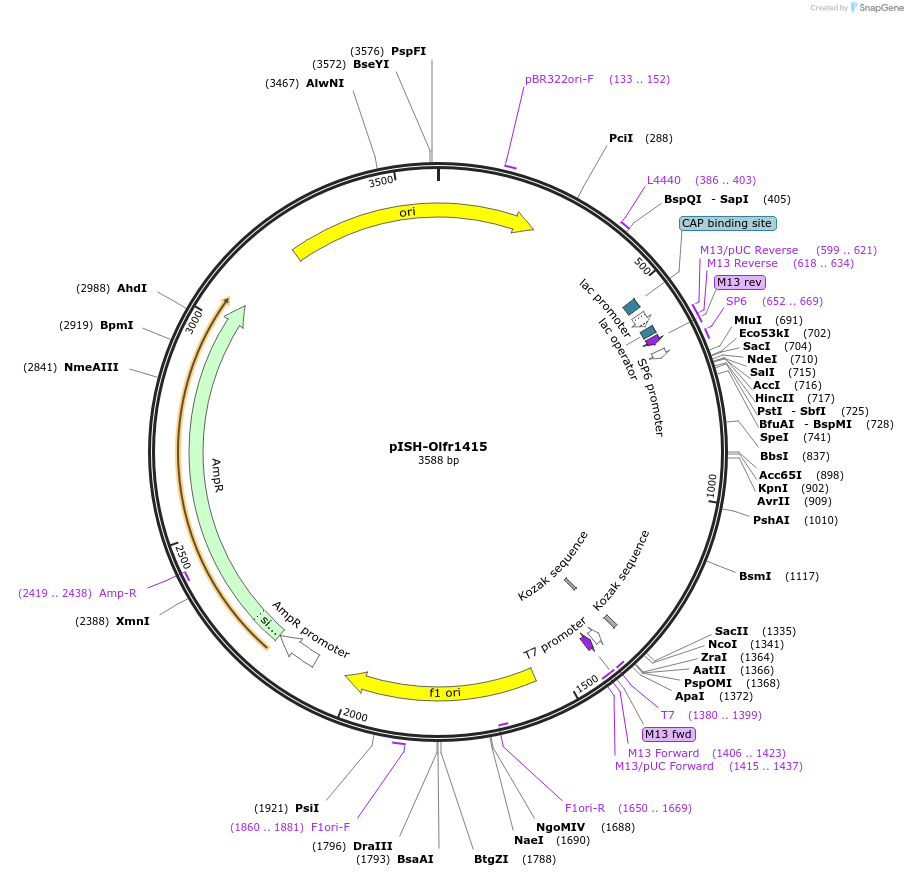
pISH-Olfr1415
Plasmid#136739PurposeTemplate to synthesize riboprobes for in situ hybridization (ISH) for Olfr1415DepositorInsertOlfr1415
UseIn situ probeMutationISH Probe insertAvailable SinceOct. 14, 2020AvailabilityAcademic Institutions and Nonprofits only -
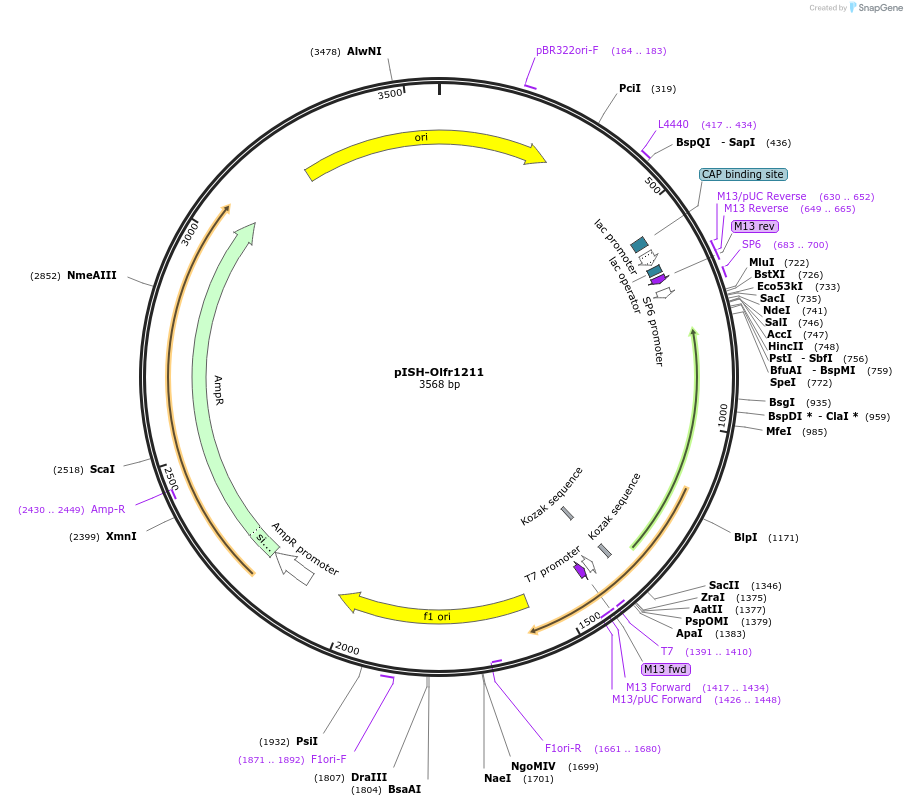
pISH-Olfr1211
Plasmid#136725PurposeTemplate to synthesize riboprobes for in situ hybridization (ISH) for Olfr1211DepositorInsertOlfr1211
UseIn situ probeMutationISH Probe insertAvailable SinceOct. 14, 2020AvailabilityAcademic Institutions and Nonprofits only -
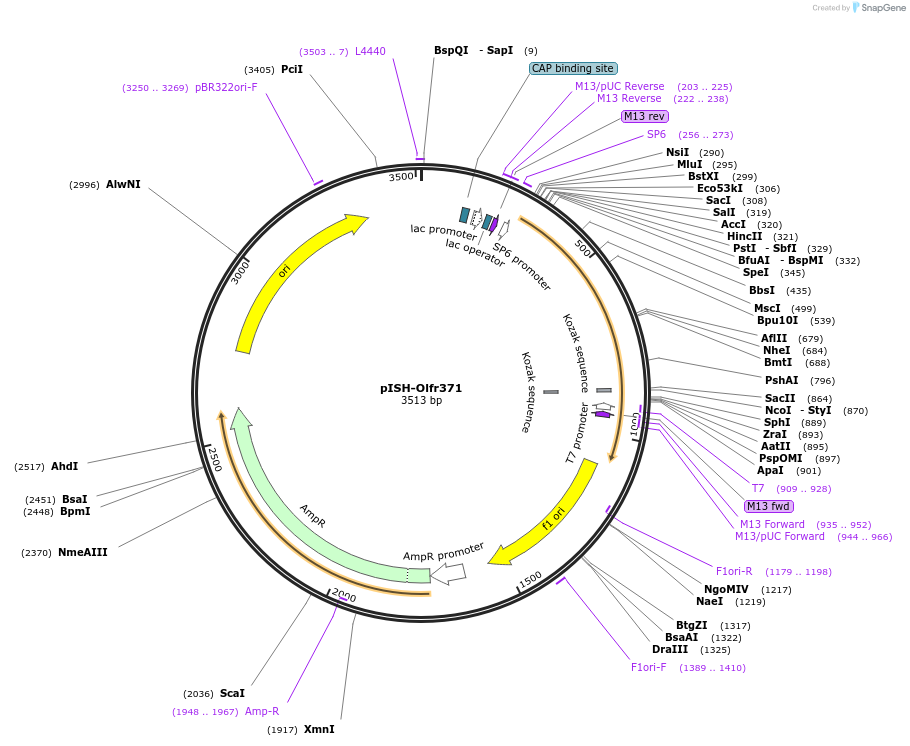
pISH-Olfr371
Plasmid#136709PurposeTemplate to synthesize riboprobes for in situ hybridization (ISH) for Olfr371DepositorInsertOlfr371
UseIn situ probeMutationISH Probe insertAvailable SinceOct. 14, 2020AvailabilityAcademic Institutions and Nonprofits only -
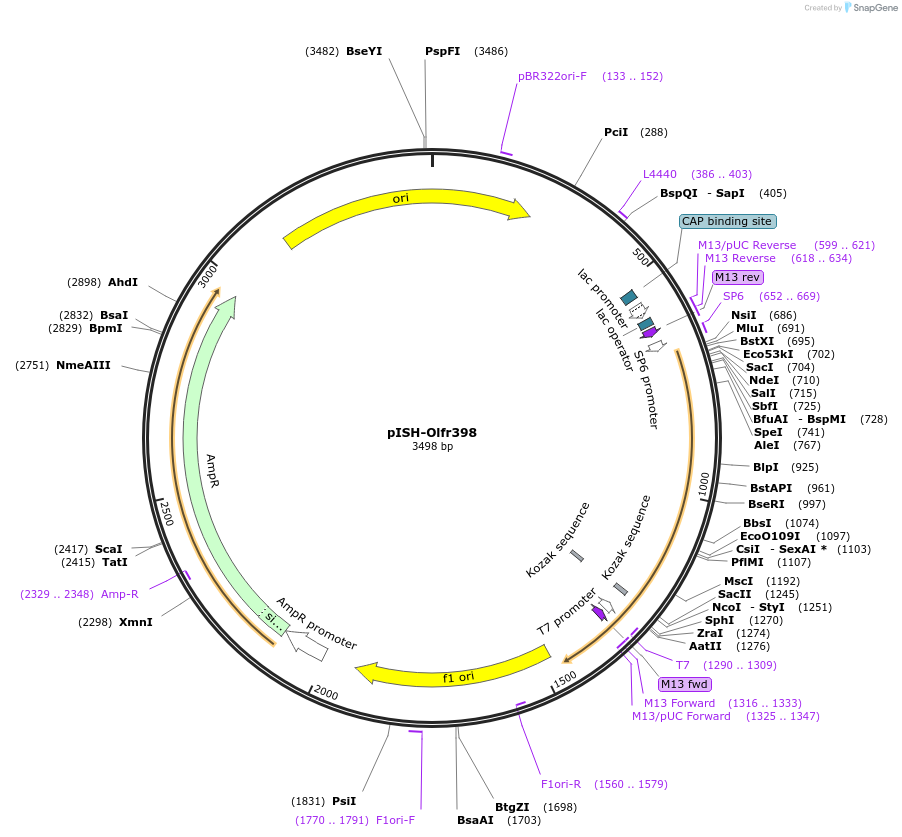
pISH-Olfr398
Plasmid#136711PurposeTemplate to synthesize riboprobes for in situ hybridization (ISH) for Olfr821DepositorInsertOlfr821
UseIn situ probeMutationISH Probe insertAvailable SinceOct. 14, 2020AvailabilityAcademic Institutions and Nonprofits only -
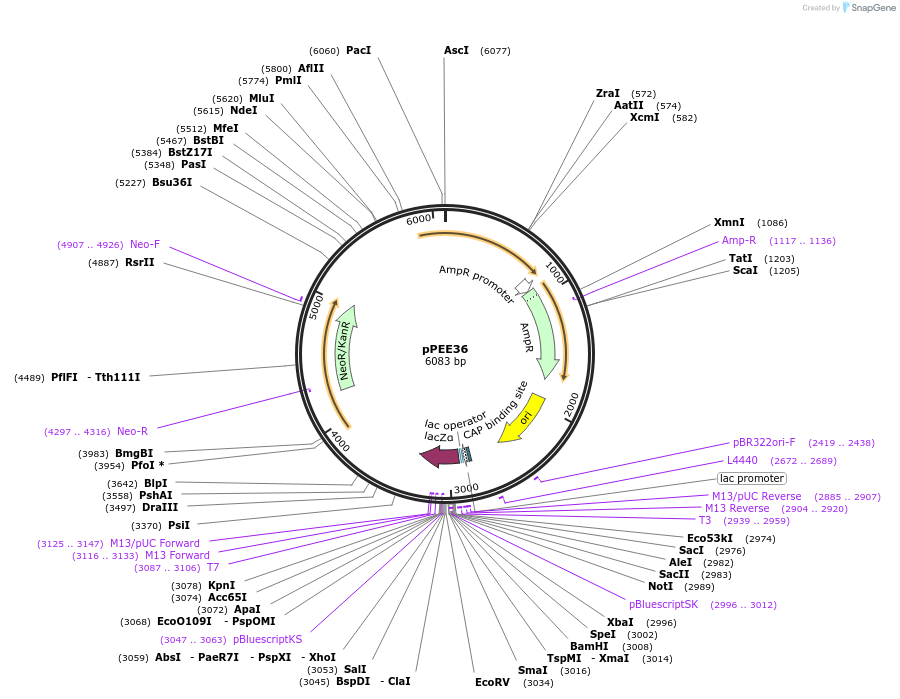
pPEE36
Plasmid#128376PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 7, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceApril 28, 2020AvailabilityAcademic Institutions and Nonprofits only -
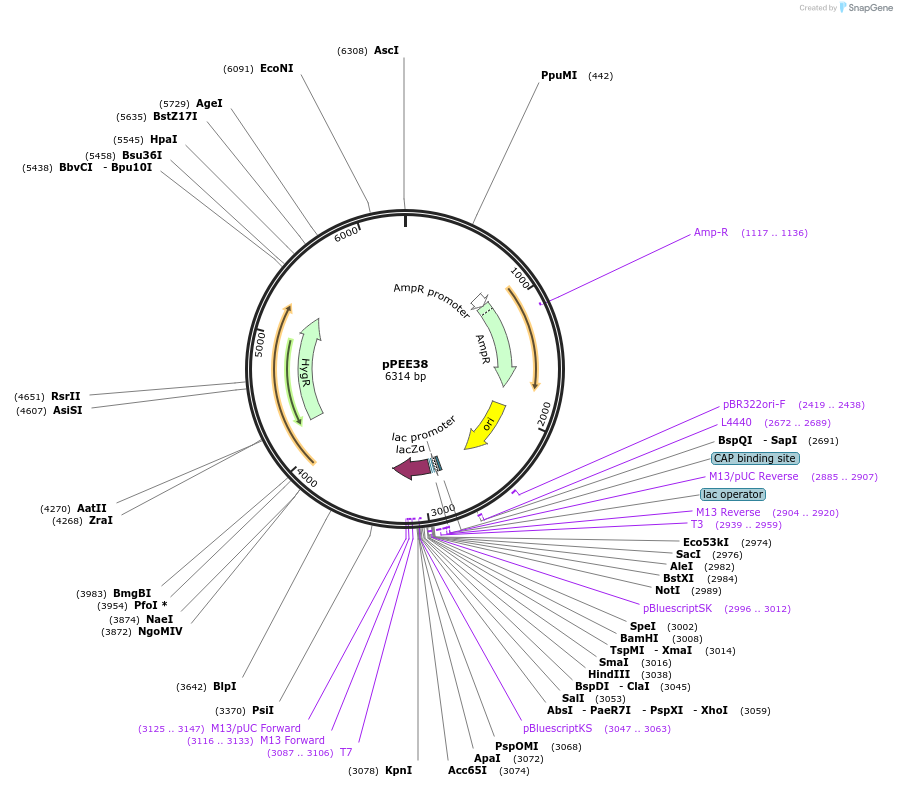
pPEE38
Plasmid#128378PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 4, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceJan. 8, 2020AvailabilityAcademic Institutions and Nonprofits only -
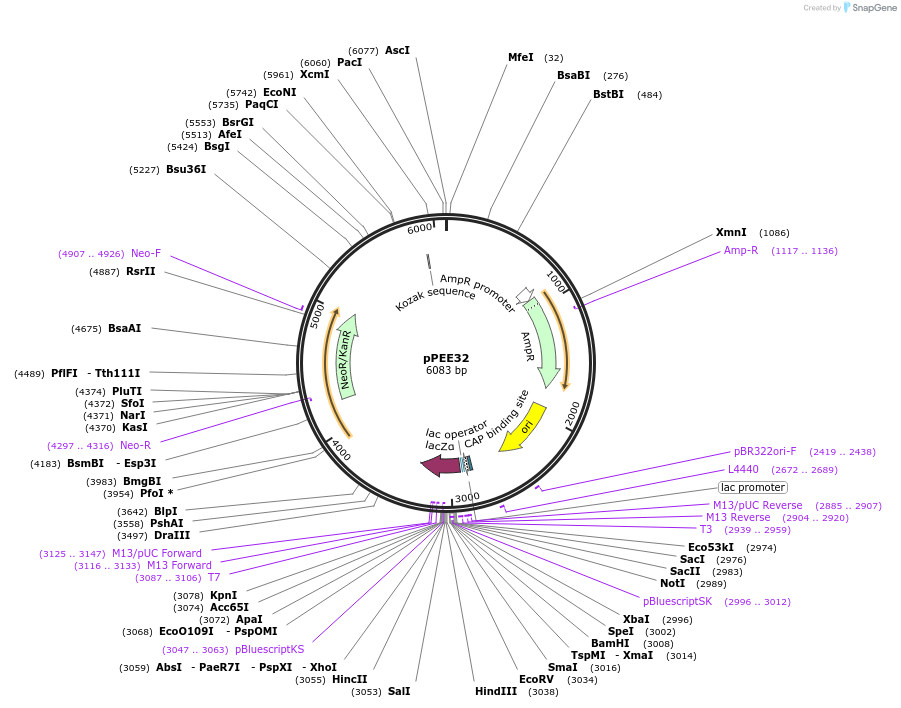
pPEE32
Plasmid#128372PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 3, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceJan. 8, 2020AvailabilityAcademic Institutions and Nonprofits only -
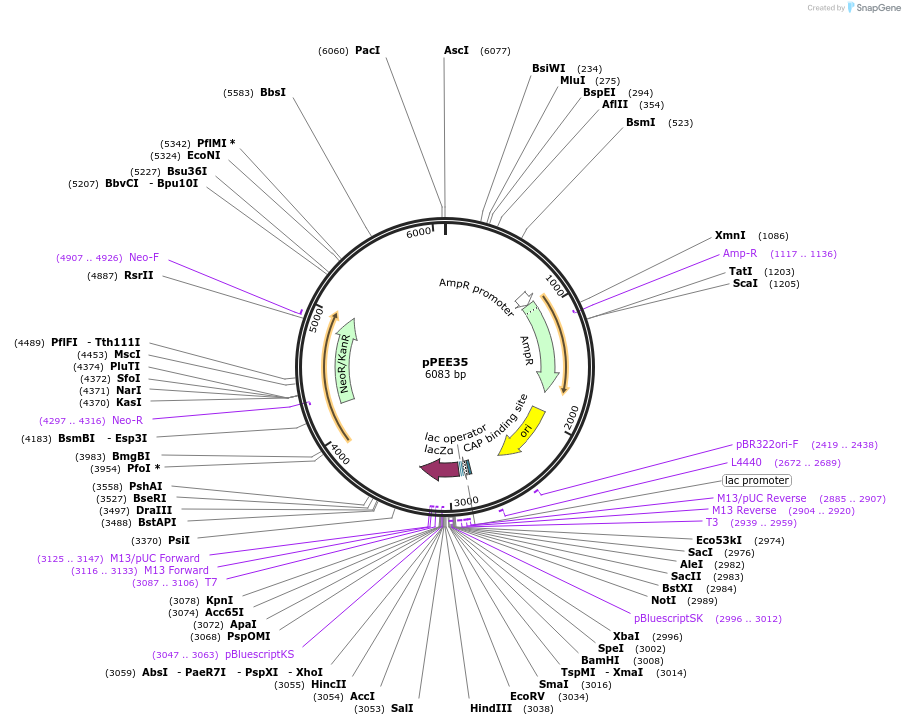
pPEE35
Plasmid#128375PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 6, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
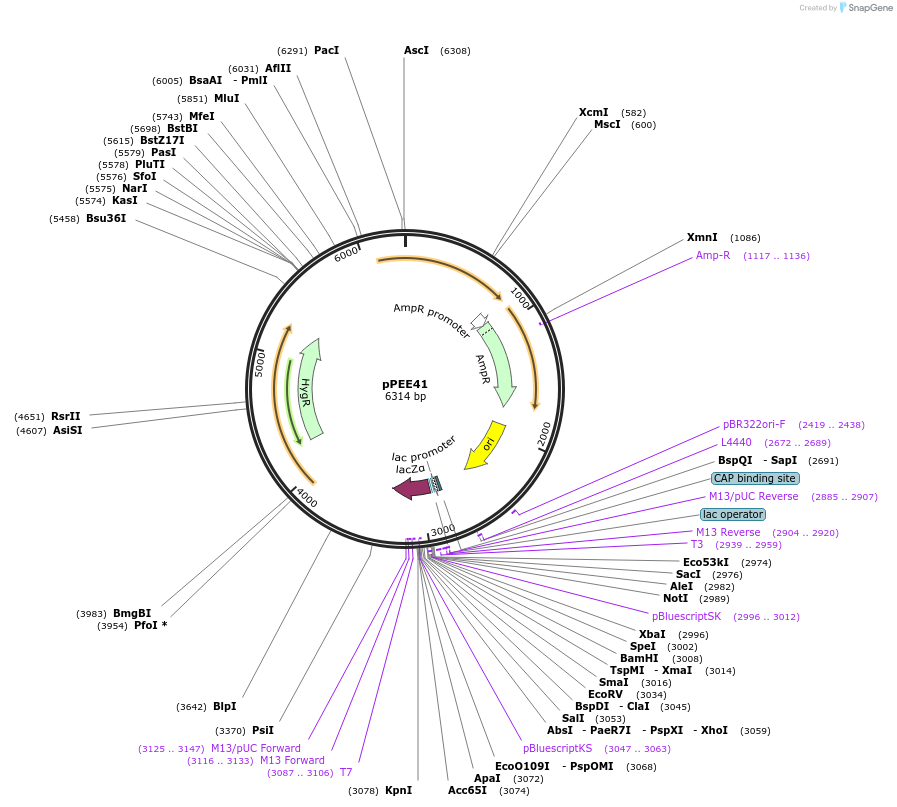
pPEE41
Plasmid#128381PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 7, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
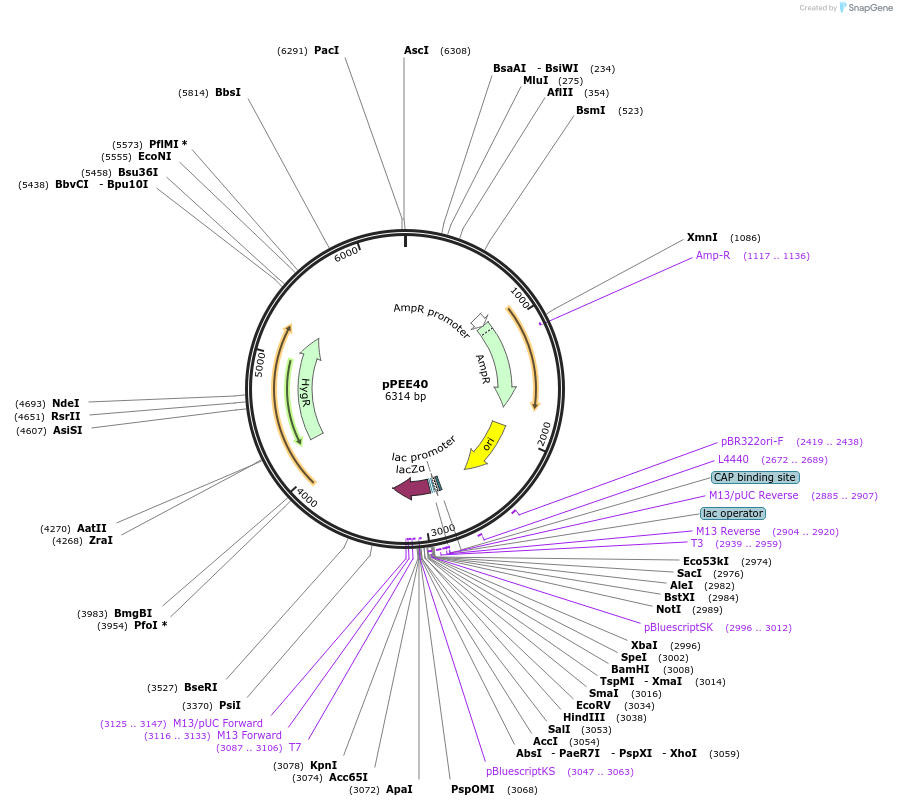
pPEE40
Plasmid#128380PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 6, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
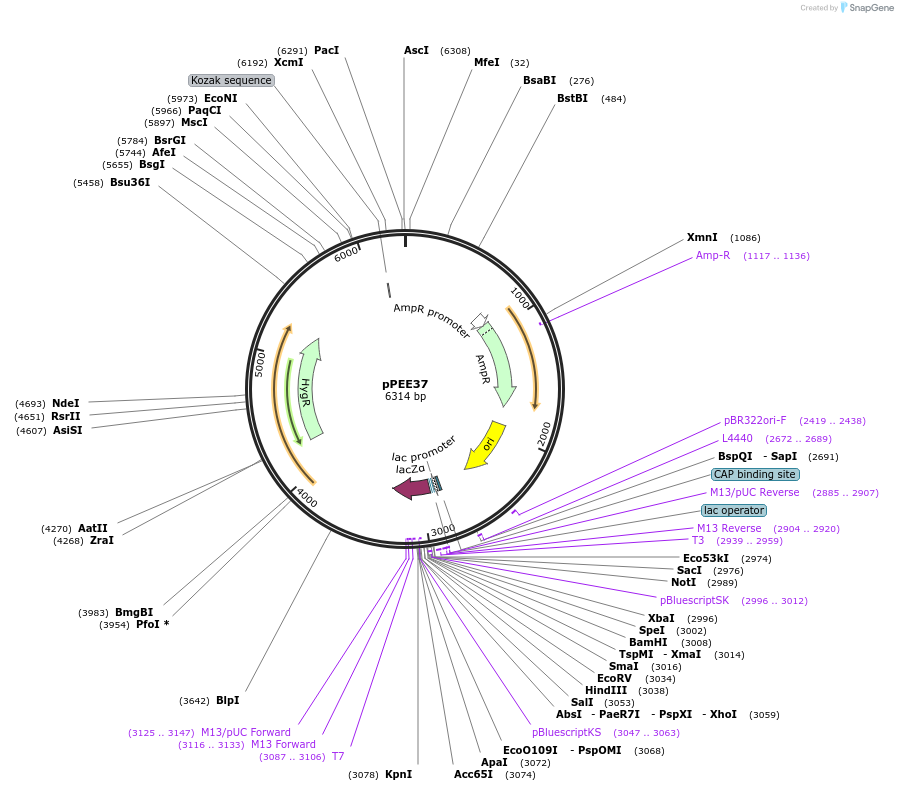
pPEE37
Plasmid#128377PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 3, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
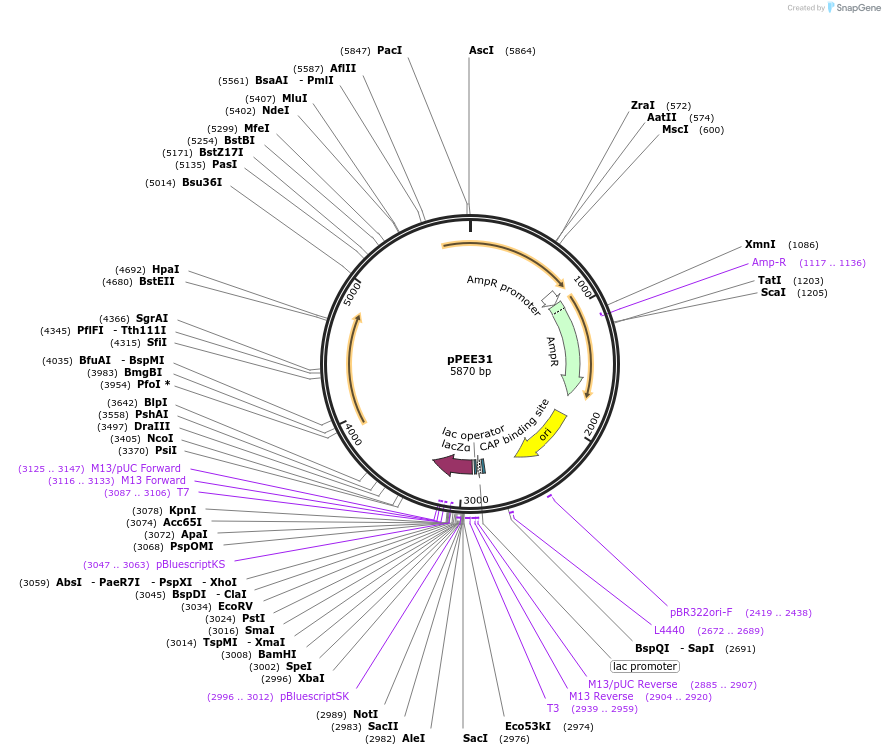
pPEE31
Plasmid#128371PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 7, a small gene-free region. Contains the nourseothricin resistance marker (NAT)DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
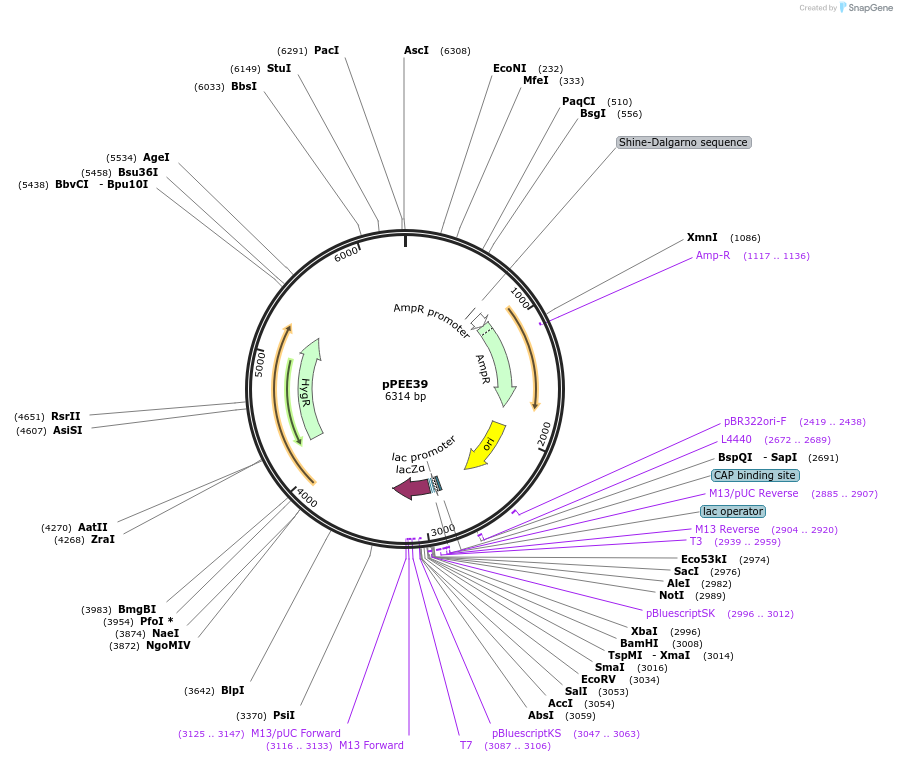
pPEE39
Plasmid#128379PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 5, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
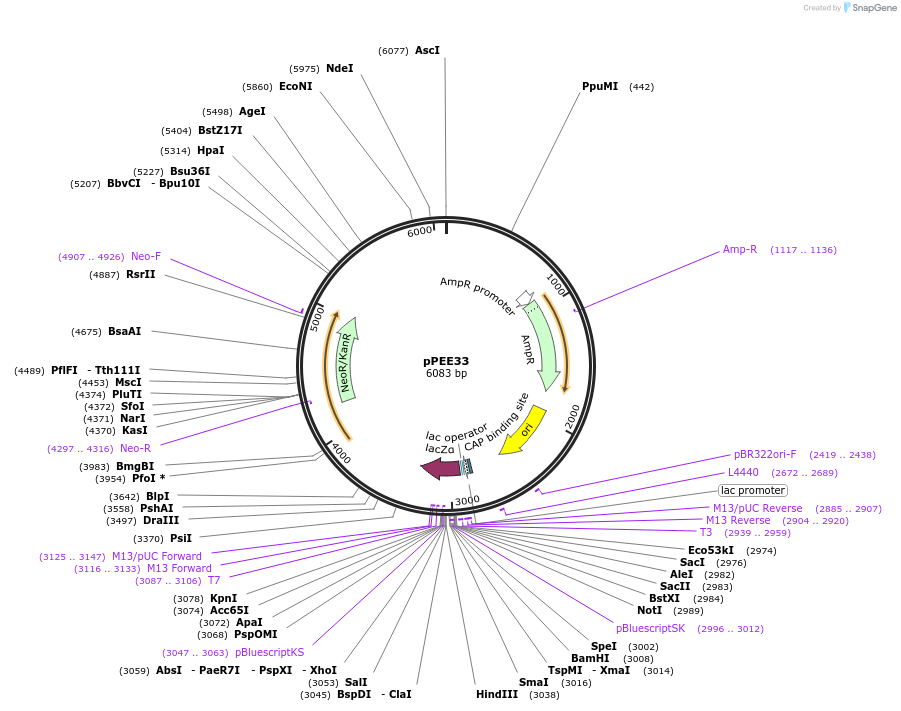
pPEE33
Plasmid#128373PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 4, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
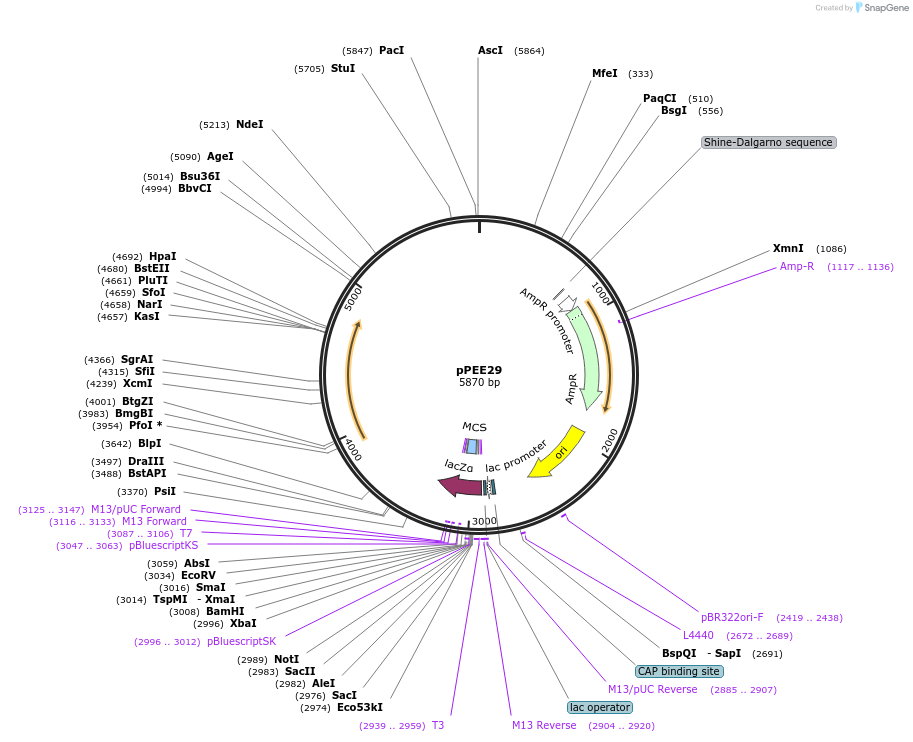
pPEE29
Plasmid#128369PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 5, a small gene-free region. Contains the nourseothricin resistance marker (NAT)DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
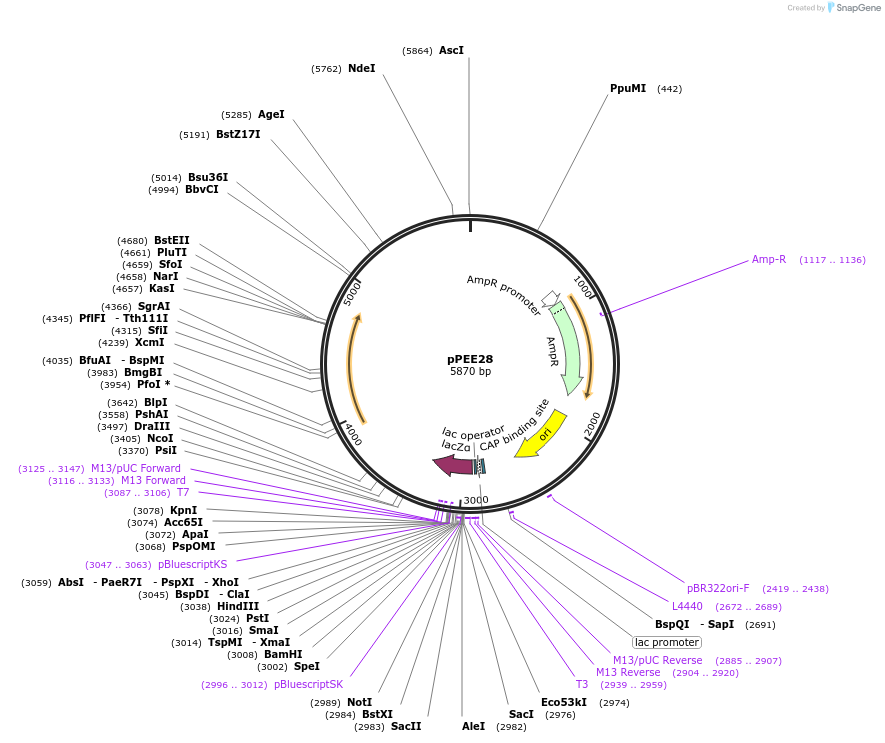
pPEE28
Plasmid#128368PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 4, a small gene-free region. Contains the nourseothricin resistance marker (NAT)DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
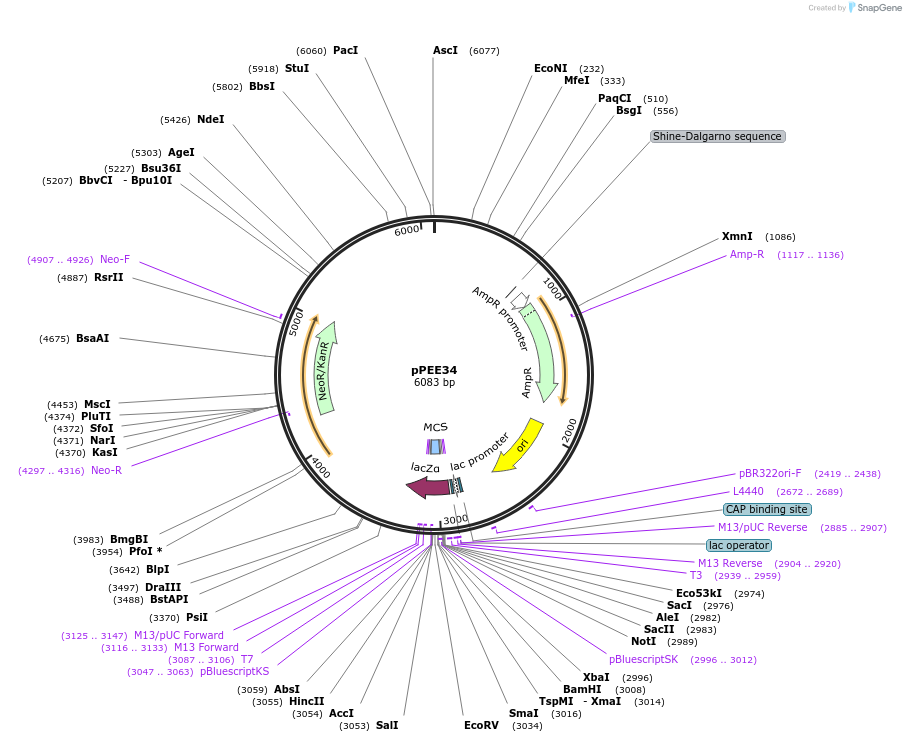
pPEE34
Plasmid#128374PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 5, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
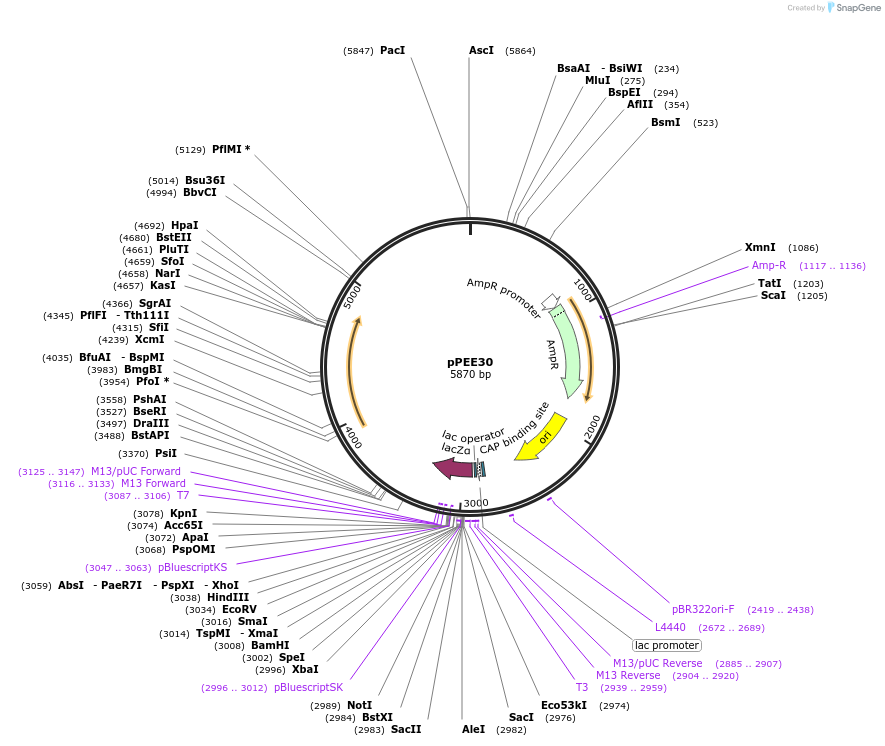
pPEE30
Plasmid#128370PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 6, a small gene-free region. Contains the nourseothricin resistance marker (NAT)DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
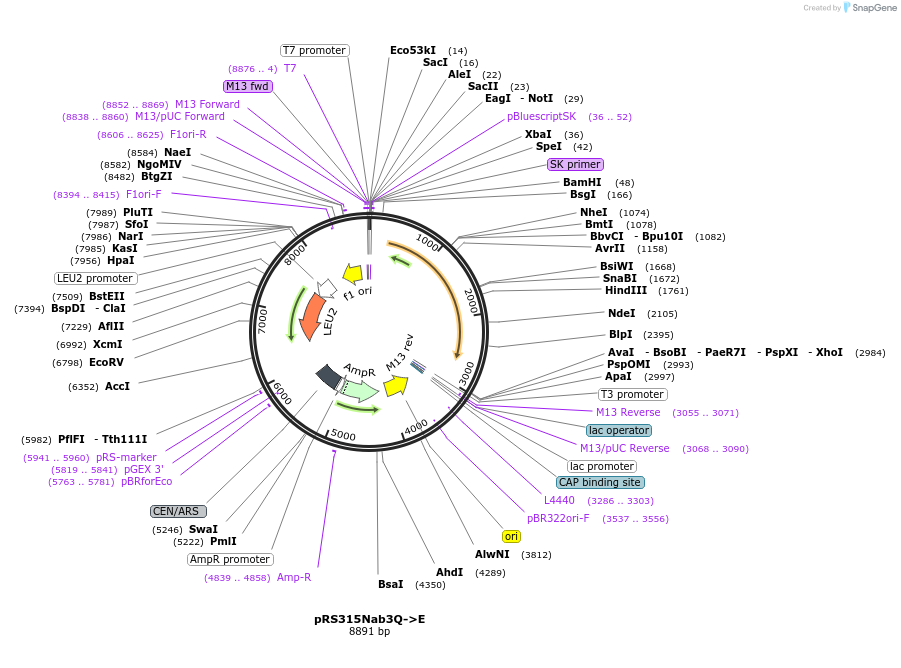
pRS315Nab3Q->E
Plasmid#110538PurposeExpresses yeast NAB3 using the endogenous promoterDepositorInsertNAB3 (NAB3 Budding Yeast)
ExpressionYeastMutationQ to E mutations in Nab3 low complexity domainAvailable SinceJune 19, 2018AvailabilityAcademic Institutions and Nonprofits only -
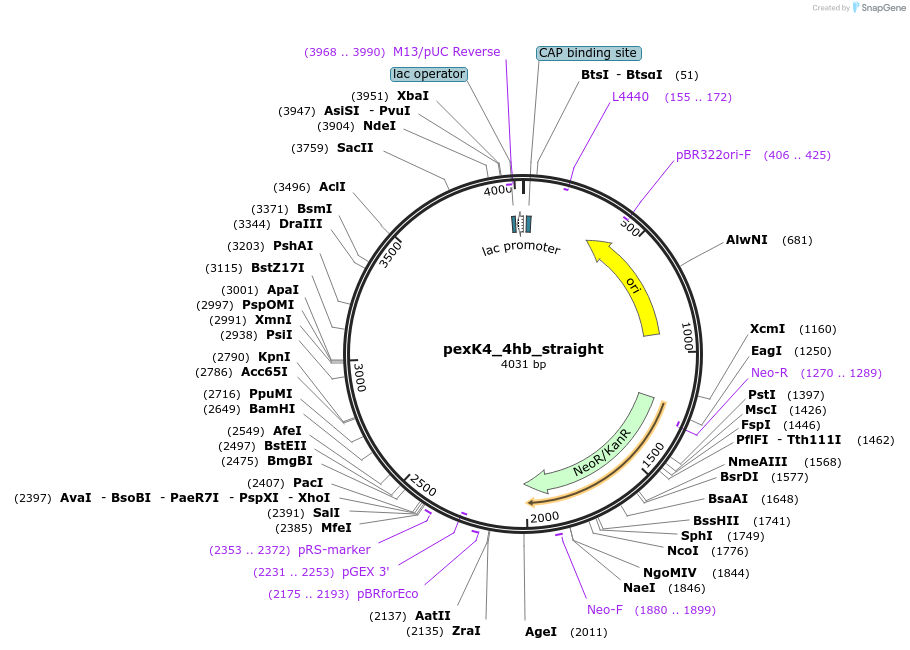
pexK4_4hb_straight
Plasmid#89678PurposePlasmid for the construction of DNA-protein hybrid nanostructures. Template for the straight four-helix-bundle shown in figure 4A of associated article.DepositorInsert4hb_straight
UseUnspecifiedAvailable SinceAug. 23, 2017AvailabilityAcademic Institutions and Nonprofits only -
pexK4_4hb_bent
Plasmid#89679PurposePlasmid for the construction of DNA-protein hybrid nanostructures. Template for the bent four-helix-bundle shown in figure 4E of associated article.DepositorInsert4hb_bent
UseUnspecifiedAvailable SinceAug. 23, 2017AvailabilityAcademic Institutions and Nonprofits only -
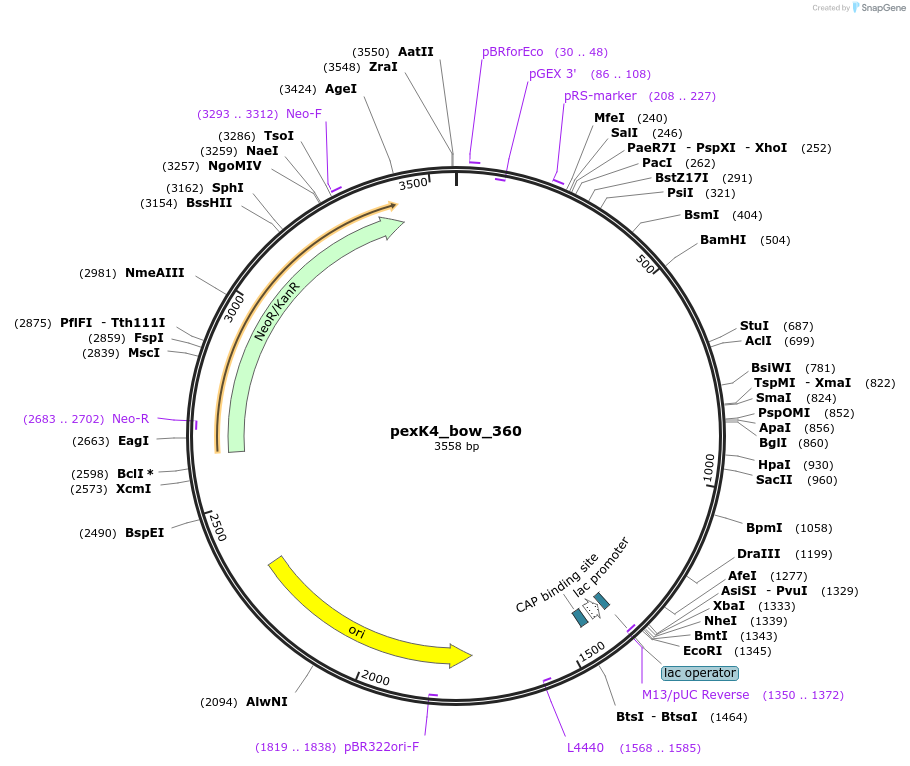
pexK4_bow_360
Plasmid#89673PurposePlasmid for the construction of DNA-protein hybrid nanostructures. Template for the bent two-helix-bundle shown in figure 3B of associated article.DepositorInsertbow_360
UseUnspecifiedAvailable SinceAug. 23, 2017AvailabilityAcademic Institutions and Nonprofits only -
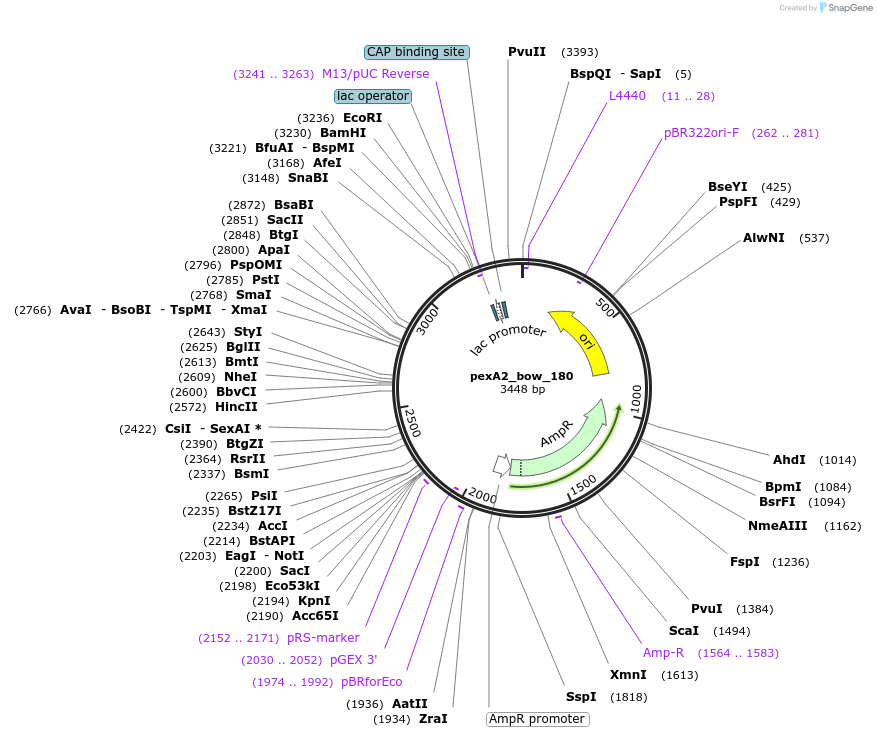
pexA2_bow_180
Plasmid#89671PurposePlasmid for the construction of DNA-protein hybrid nanostructures. Template for the bent two-helix-bundle shown in figure 3A of associated article.DepositorInsertbow_180
UseUnspecifiedAvailable SinceAug. 23, 2017AvailabilityAcademic Institutions and Nonprofits only -
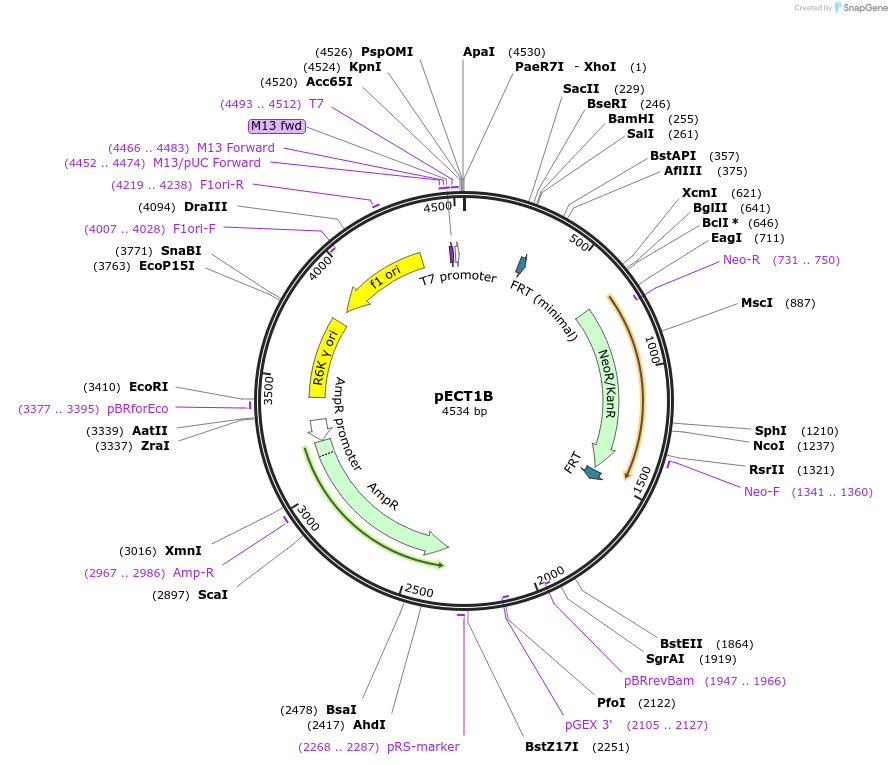
pECT1B
Plasmid#75447PurposePlasmid that serves as the PCR template to create genomic pdt1B insertions. Plasmid contains the R6K origin of replication so needs pir gene to replicateDepositorInsertpdt1B
ExpressionBacterialPromoternoneAvailable SinceOct. 5, 2016AvailabilityAcademic Institutions and Nonprofits only -
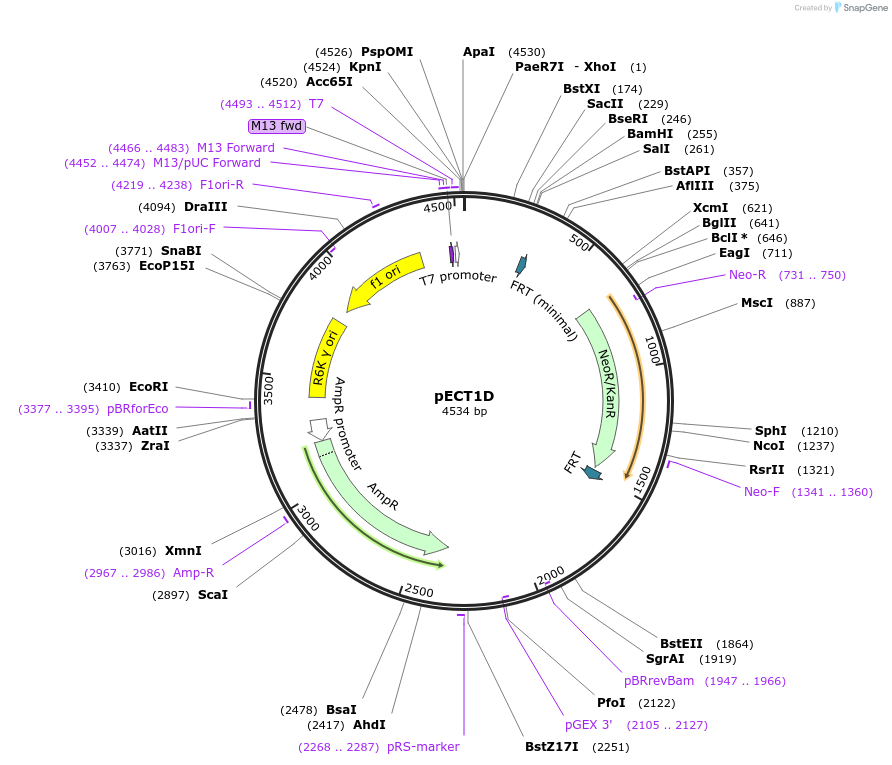
pECT1D
Plasmid#75448PurposePlasmid that serves as the PCR template to create genomic pdt1D insertions. Plasmid contains the R6K origin of replication so needs pir gene to replicateDepositorInsertpdt1D
ExpressionBacterialPromoternoneAvailable SinceOct. 5, 2016AvailabilityAcademic Institutions and Nonprofits only -
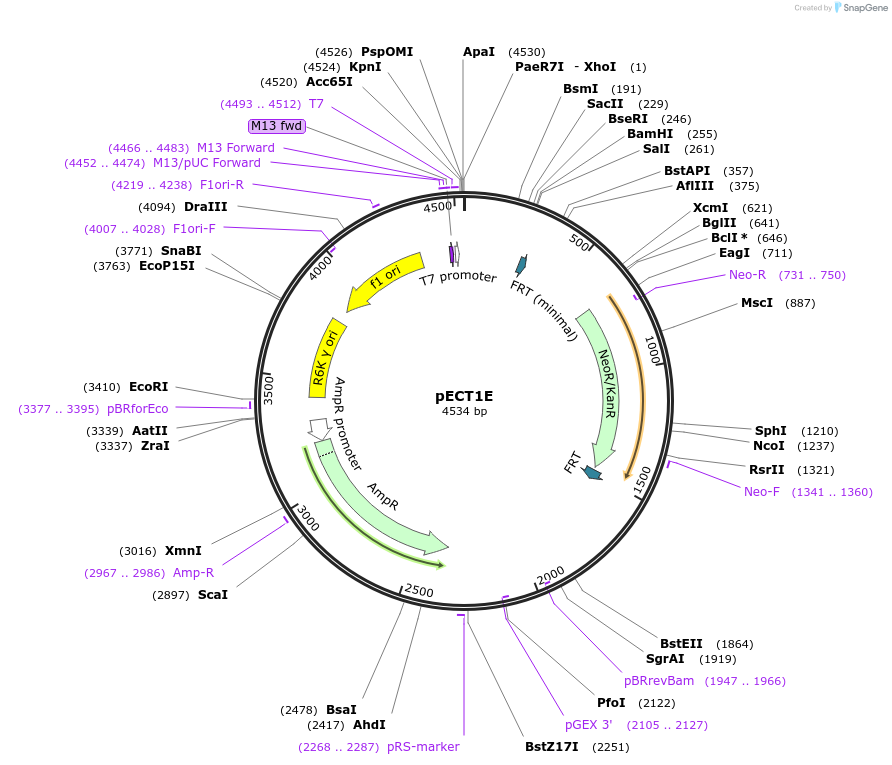
pECT1E
Plasmid#75449PurposePlasmid that serves as the PCR template to create genomic pdt1E insertions. Plasmid contains the R6K origin of replication so needs pir gene to replicateDepositorInsertpdt1E
ExpressionBacterialPromoternoneAvailable SinceOct. 5, 2016AvailabilityAcademic Institutions and Nonprofits only -
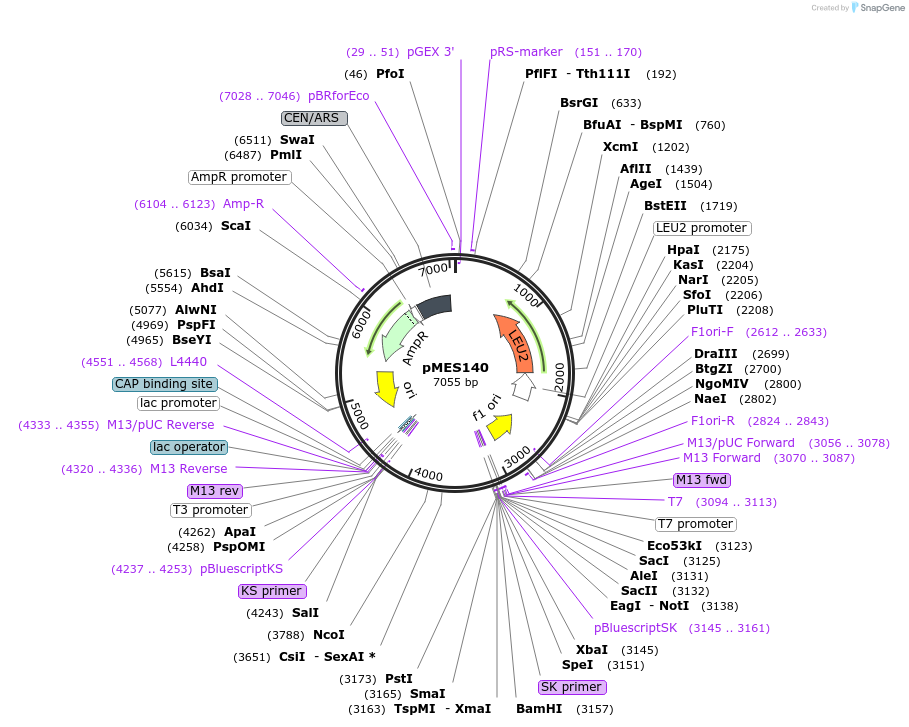
pMES140
Plasmid#71849PurposeComplementation of NME1 gene disruption or mutation in yeastDepositorAvailable SinceJan. 5, 2016AvailabilityAcademic Institutions and Nonprofits only -
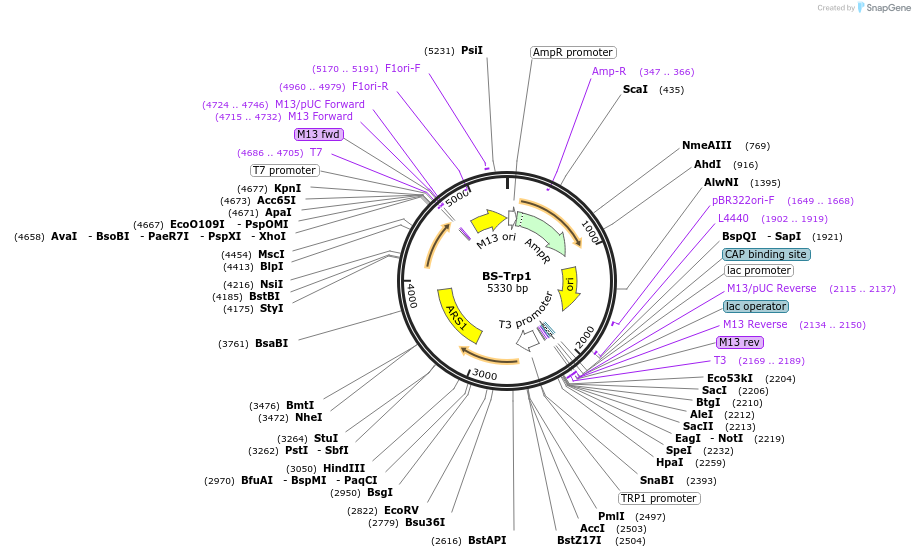
BS-Trp1
Plasmid#69196Purposecomplementation of yeast auxotrophic marker TRP1. Although this is the plasmid described in the paper, the depositor notes that it has a low integration efficiency and Plasmid 69503 should be used.DepositorInsertTRP1
ExpressionYeastAvailable SinceOct. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
-
T-B18
Bacterial Strain#159446PurposeE. coli BW25113 ΔfrmA, harboring plasmid Addgene plasmid # 129104, was adaptively evolved to grow efficiently on threonine and perform significant biosynthesis while employing methanol-derived carbonBacterial ResistanceAmpicillin and KanamycinAvailable SinceAug. 18, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
BW-Para
Bacterial Strain#172602PurposeThe BW-Para E. coli strain is used to screen the functions of chimeric AraC/XylS transcription activators using beta-galactosidase assays.DepositorBacterial ResistanceNoneAvailable SinceDec. 22, 2022AvailabilityAcademic Institutions and Nonprofits only -
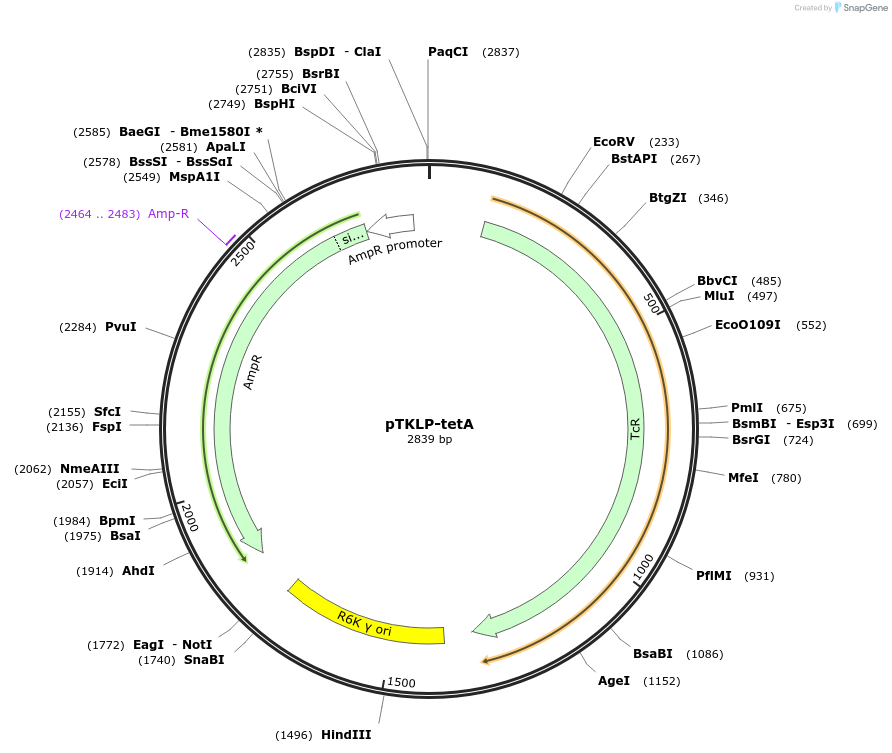
pTKLP-tetA
Plasmid#71325PurposetetA landing pad template plasmidDepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterialAvailable SinceApril 22, 2016AvailabilityAcademic Institutions and Nonprofits only