We narrowed to 14,335 results for: ung
-
Plasmid#59840PurposeConstitutively expresses AmiE with proK9 promoterDepositorAvailable SinceOct. 20, 2014AvailabilityAcademic Institutions and Nonprofits only
-
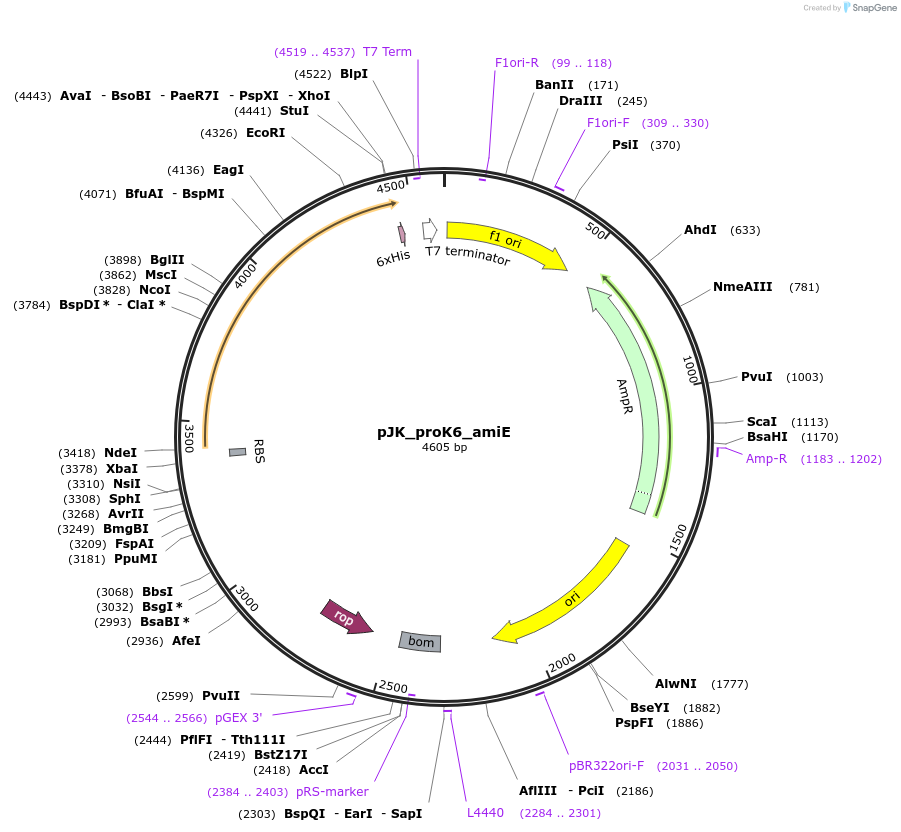
pJK_proK6_amiE
Plasmid#59841PurposeConstitutively expresses AmiE with proK6 promoterDepositorAvailable SinceOct. 20, 2014AvailabilityAcademic Institutions and Nonprofits only -
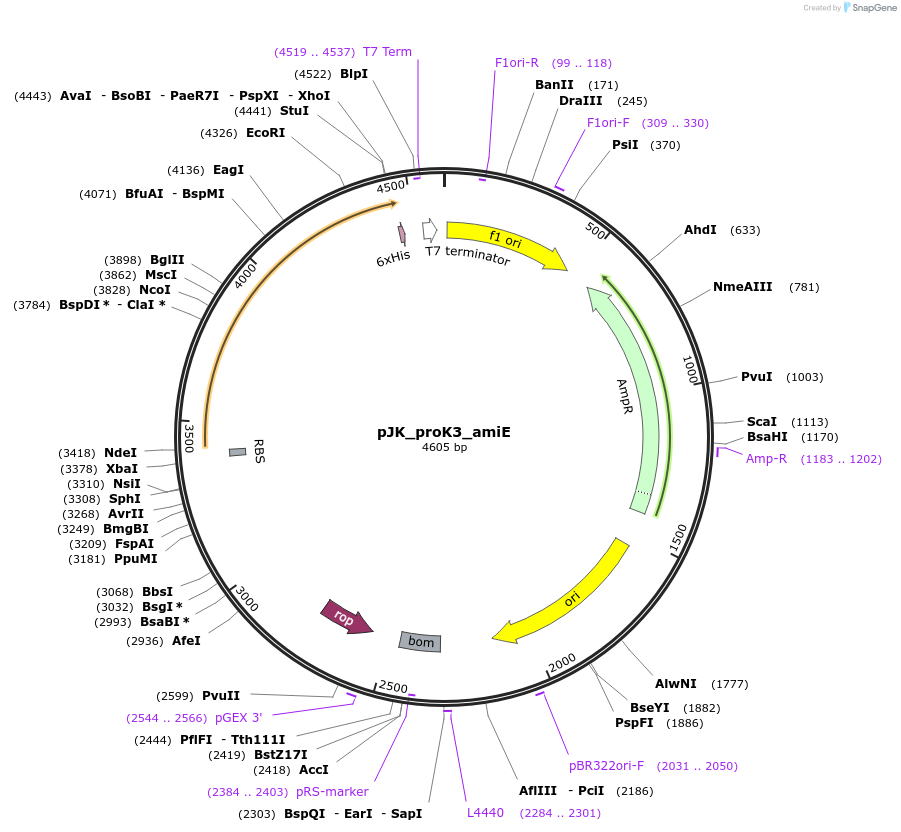
pJK_proK3_amiE
Plasmid#59842PurposeConstitutively expresses AmiE with proK3 promoterDepositorAvailable SinceOct. 20, 2014AvailabilityAcademic Institutions and Nonprofits only -
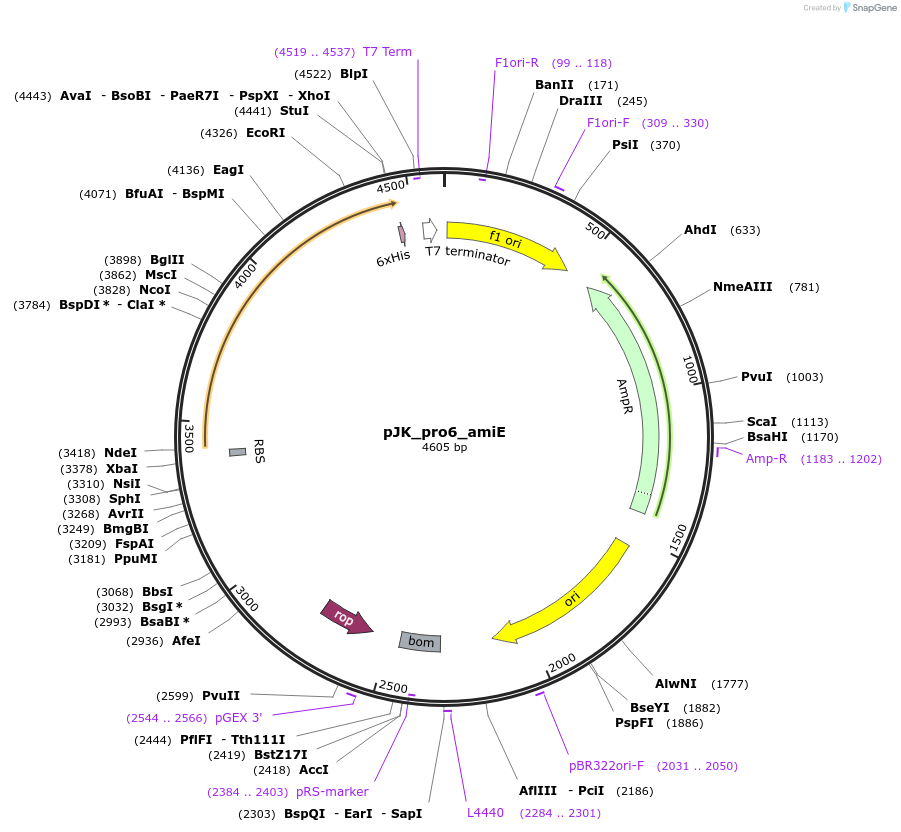
pJK_pro6_amiE
Plasmid#59844PurposeConstitutively expresses AmiE with pro6 promoterDepositorAvailable SinceOct. 20, 2014AvailabilityAcademic Institutions and Nonprofits only -
pAS-7 (mm12-21)
Plasmid#40842PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm12-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-6 (mm17-21)
Plasmid#40841PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm17-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-5 (mm18-21)
Plasmid#40840PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm18-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-4 (mm19-21)
Plasmid#40839PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm19-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-3 (mm20-21)
Plasmid#40838PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm20-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-2 (mm21)
Plasmid#40837PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-16 (mm14-21)
Plasmid#40851PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm14-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-15 (mm15-21)
Plasmid#40850PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm15-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS14 (mm16-21)
Plasmid#40849PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm16-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-13
Plasmid#40848PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–Bartel
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-12 (mm1-8)
Plasmid#40847PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm1-8
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-11 (bulge 9-15)
Plasmid#40846PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–bulge9-15
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-10 (bulge 9-13)
Plasmid#40845PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–bulge 9-13
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-9 (bulge 9-11)
Plasmid#40844PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–bulge 9-11
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
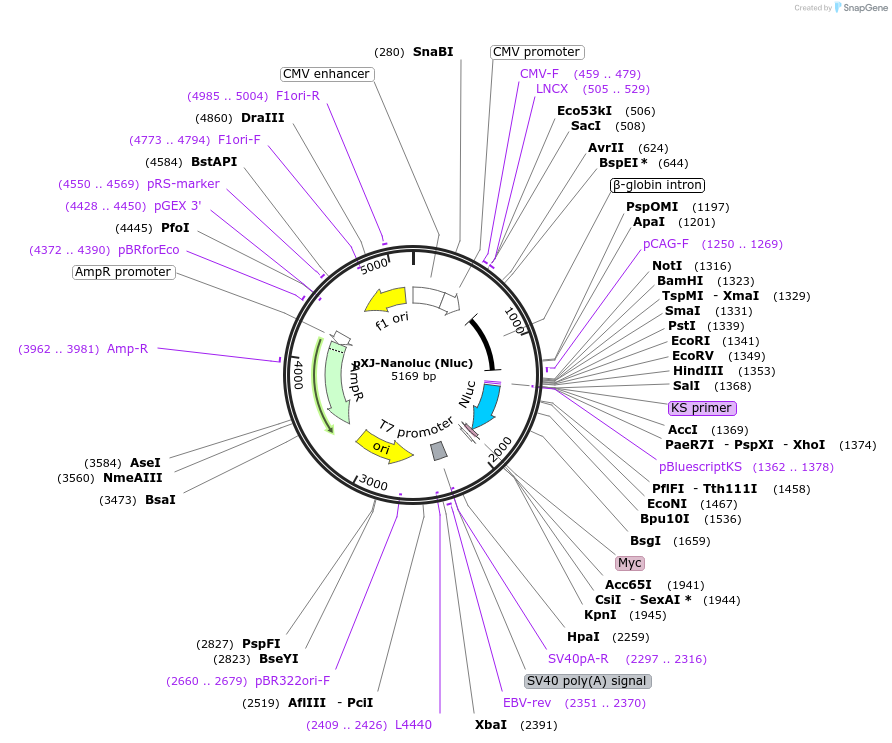
pXJ-Nanoluc (Nluc)
Plasmid#232656PurposeNanoluc luciferase expression driven CMV promoterDepositorInsertNanoluc
UseLuciferaseTagsMycExpressionMammalianPromoterCMVAvailable SinceMarch 7, 2025AvailabilityAcademic Institutions and Nonprofits only -
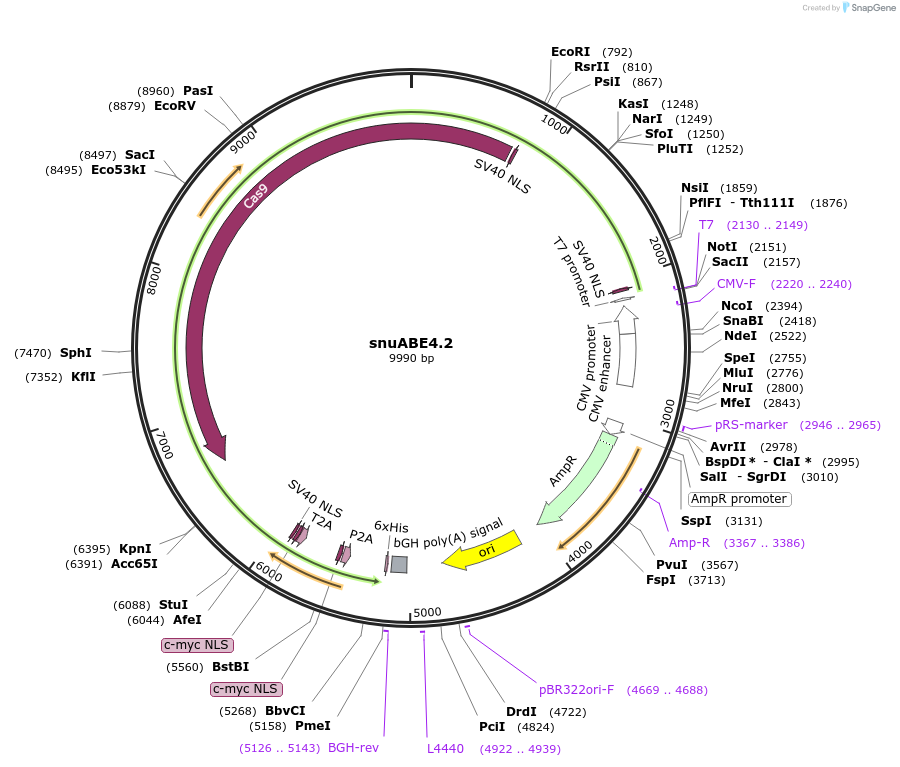
snuABE4.2
Plasmid#246014PurposesnuABE4.1 with MLH1-SB fused to the C-terminus via P2A.DepositorInsertPediculus humanus ADAR deaminase domain (E438Q, I536S, E637F mutant)
TagsMLH1-SBExpressionMammalianMutationE438Q, I536S, E637F, with 10 amino acids truncati…PromoterCMVAvailable SinceNov. 7, 2025AvailabilityAcademic Institutions and Nonprofits only