We narrowed to 14,277 results for: GFP expression plasmids
-
Plasmid#51137PurposeExpress turbo GFP tagged three SH3 domains of Cin85 (truncation) in mammalian cellsDepositorInsertCin85 (SH3KBP1 Human)
TagsTurboGFPExpressionMammalianMutationcontains 3 SH3 domainsPromoterCMVAvailable SinceNov. 21, 2018AvailabilityAcademic Institutions and Nonprofits only -
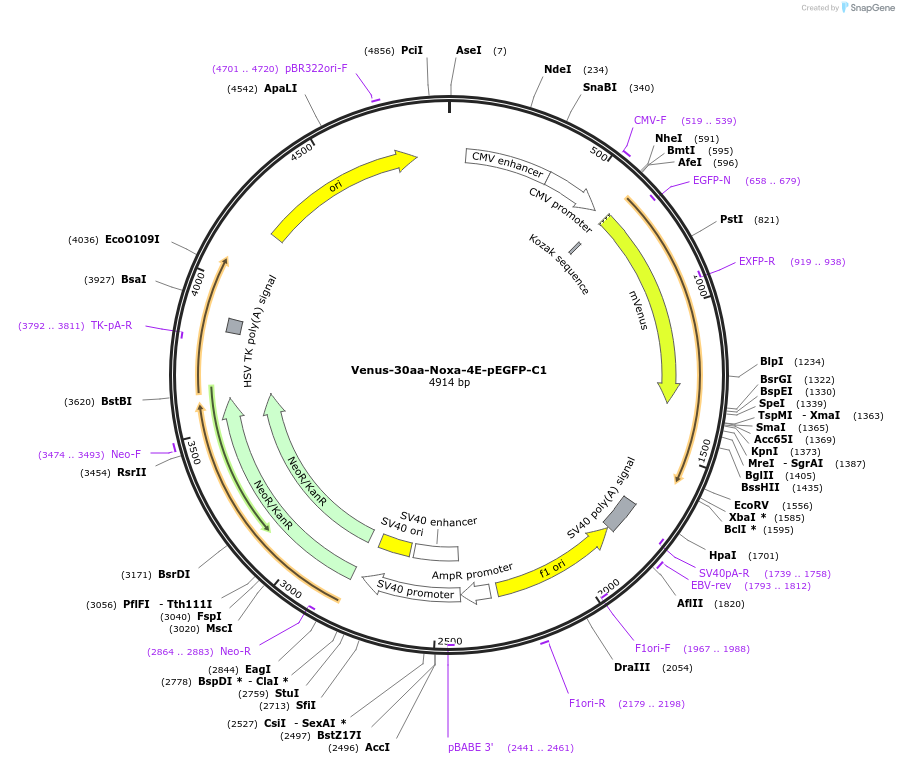
Venus-30aa-Noxa-4E-pEGFP-C1
Plasmid#177424PurposeTo express Venus fused to Noxa-4E, with a longer linker sequence between compared to Addgene #166745.DepositorInsertNoxa, PMA-induced protein 1 (PMAIP1 Human)
TagsVenusExpressionMammalianMutation4 hydrophobic residues in the BH3 region mutated …PromoterCMVAvailable SinceApril 1, 2025AvailabilityAcademic Institutions and Nonprofits only -
Venus-30aa-Noxa-4E-pEGFP-C1
Plasmid#177424PurposeTo express Venus fused to Noxa-4E, with a longer linker sequence between compared to Addgene #166745.DepositorInsertNoxa, PMA-induced protein 1 (PMAIP1 Human)
TagsVenusExpressionMammalianMutation4 hydrophobic residues in the BH3 region mutated …PromoterCMVAvailable SinceApril 1, 2025AvailabilityAcademic Institutions and Nonprofits only -
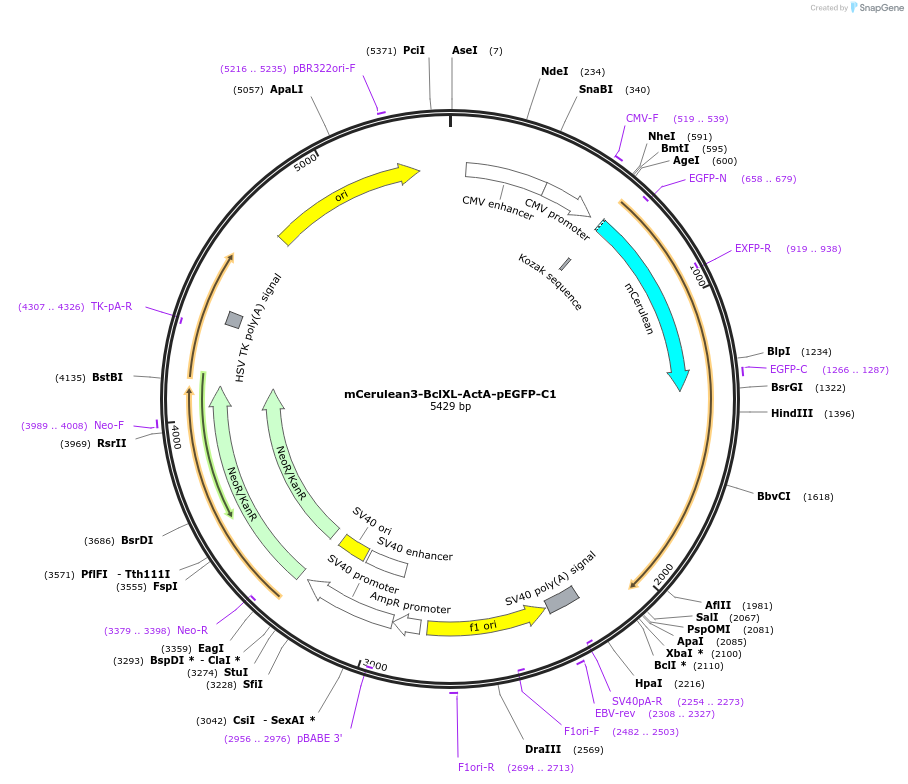
mCerulean3-BclXL-ActA-pEGFP-C1
Plasmid#177412PurposeTo express mCerulean3 fused to the N-terminus of the Bcl-2 family chimera protein, Bcl-XL-ActA: Bcl-XL with its membrane binding region swapped for ActA (mitochondrial targeting sequence).DepositorInsertBcl-XL-ActA, BclX-acta
TagsmCerulean3ExpressionMammalianPromoterCMVAvailable SinceJan. 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
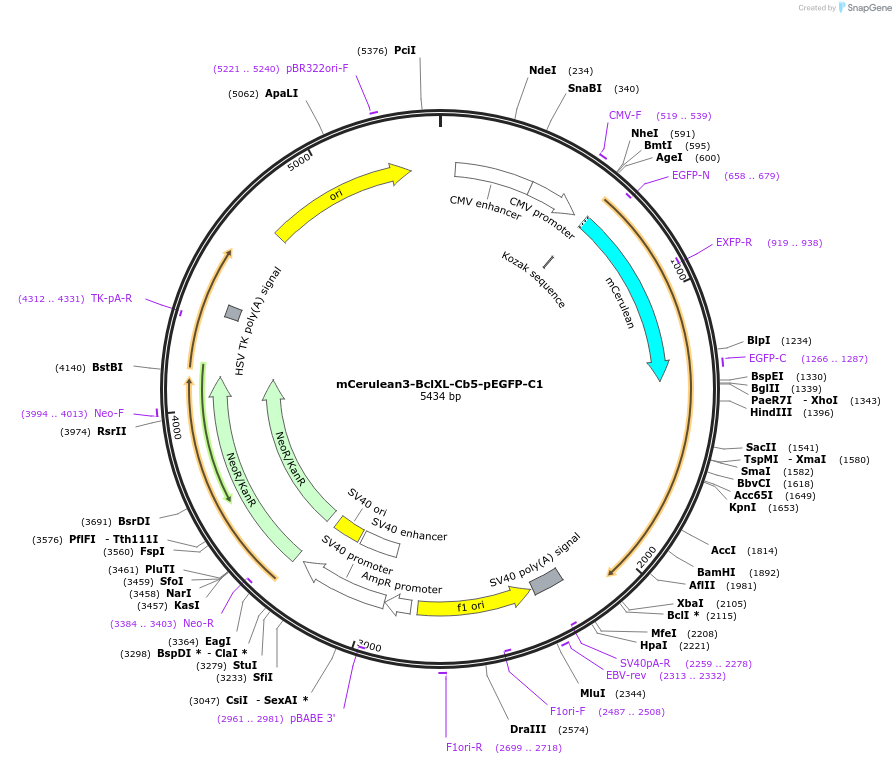
mCerulean3-BclXL-Cb5-pEGFP-C1
Plasmid#177414PurposeTo express mCerulean3 fused to the N-terminus of the Bcl-2 family chimera protein, Bcl-XL-Cb5: Bcl-XL with its membrane binding region swapped for Cb5 (ER targeting sequence).DepositorInsertBcl-XL-Cb5, BclX-cb5
TagsmCerulean3ExpressionMammalianPromoterCMVAvailable SinceJan. 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
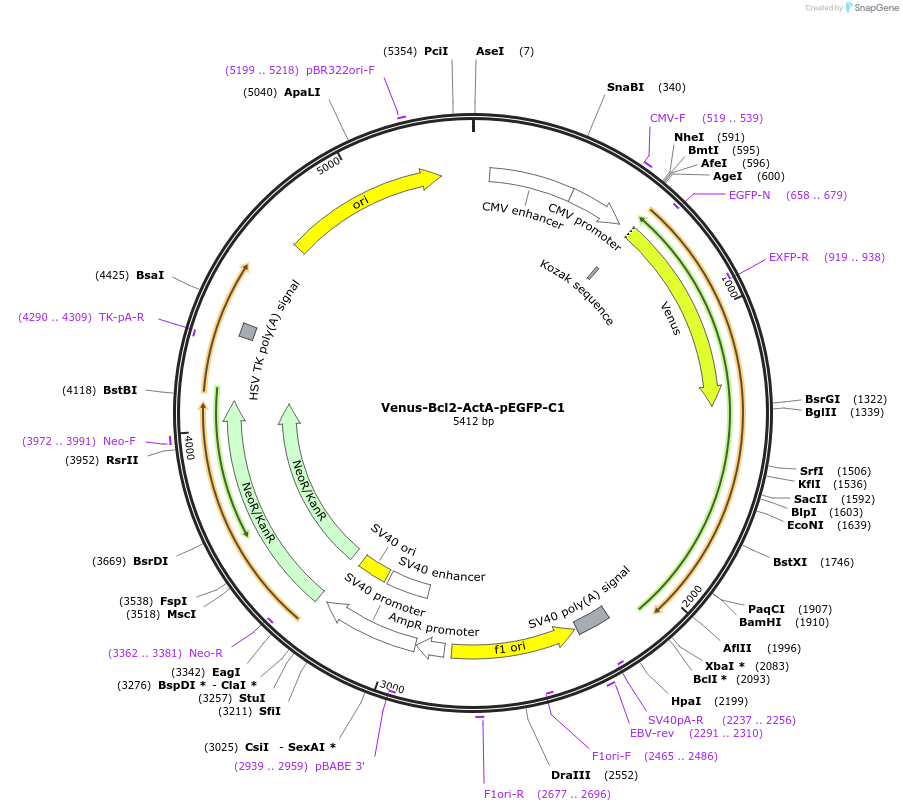
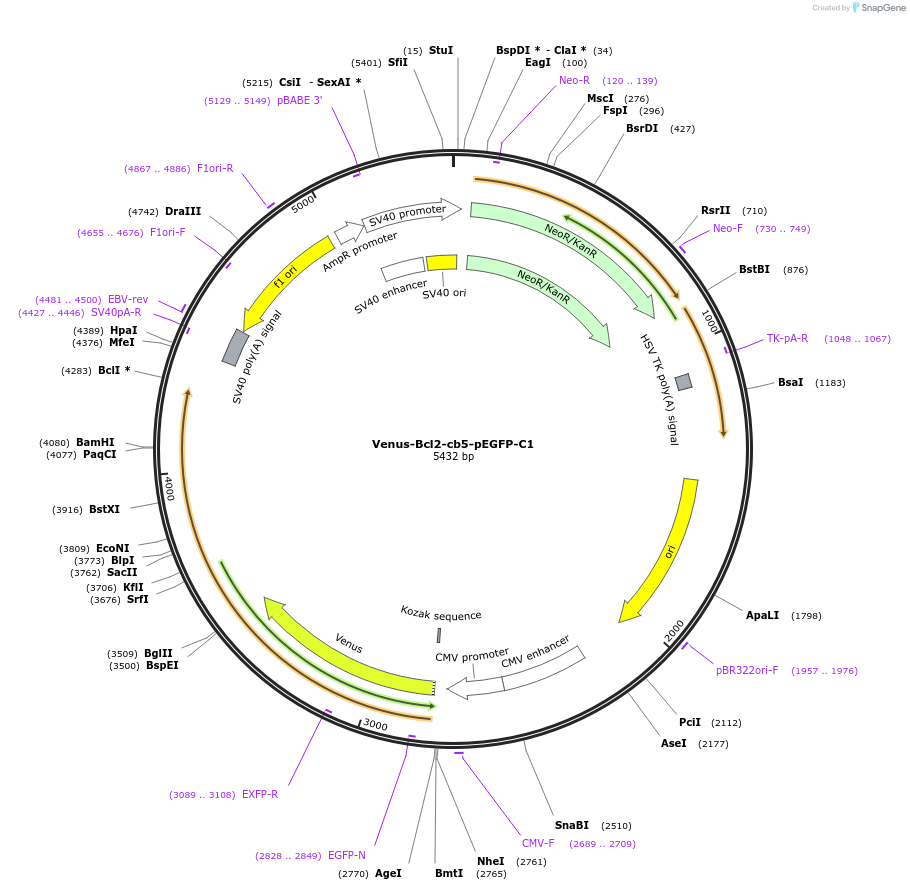
Venus-Bcl2-ActA-pEGFP-C1
Plasmid#177419PurposeTo express Venus fused to the N-terminus of the Bcl-2 family chimera protein, Bcl-2-ActA: Bcl-2 with its membrane binding region swapped for ActA (mitochondrial targeting sequence).DepositorInsertBcl-2-ActA, Bcl2-acta, Bcl-acta
TagsVenusExpressionMammalianPromoterCMVAvailable SinceJan. 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
Venus-Bcl2-cb5-pEGFP-C1
Plasmid#177422PurposeTo express Venus fused to the N-terminus of the Bcl-2 family chimera protein, Bcl-2-Cb5: Bcl-2 with its membrane binding region swapped for Cb5 (ER targeting sequence).DepositorInsertBcl-2-Cb5, Bcl2-cb5, Bcl-cb5
TagsVenusExpressionMammalianPromoterCMVAvailable SinceJan. 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
mCerulean3-BclXL-ActA-pEGFP-C1
Plasmid#177412PurposeTo express mCerulean3 fused to the N-terminus of the Bcl-2 family chimera protein, Bcl-XL-ActA: Bcl-XL with its membrane binding region swapped for ActA (mitochondrial targeting sequence).DepositorInsertBcl-XL-ActA, BclX-acta
TagsmCerulean3ExpressionMammalianPromoterCMVAvailable SinceJan. 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
mCerulean3-BclXL-Cb5-pEGFP-C1
Plasmid#177414PurposeTo express mCerulean3 fused to the N-terminus of the Bcl-2 family chimera protein, Bcl-XL-Cb5: Bcl-XL with its membrane binding region swapped for Cb5 (ER targeting sequence).DepositorInsertBcl-XL-Cb5, BclX-cb5
TagsmCerulean3ExpressionMammalianPromoterCMVAvailable SinceJan. 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
Venus-Bcl2-ActA-pEGFP-C1
Plasmid#177419PurposeTo express Venus fused to the N-terminus of the Bcl-2 family chimera protein, Bcl-2-ActA: Bcl-2 with its membrane binding region swapped for ActA (mitochondrial targeting sequence).DepositorInsertBcl-2-ActA, Bcl2-acta, Bcl-acta
TagsVenusExpressionMammalianPromoterCMVAvailable SinceJan. 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
Venus-Bcl2-cb5-pEGFP-C1
Plasmid#177422PurposeTo express Venus fused to the N-terminus of the Bcl-2 family chimera protein, Bcl-2-Cb5: Bcl-2 with its membrane binding region swapped for Cb5 (ER targeting sequence).DepositorInsertBcl-2-Cb5, Bcl2-cb5, Bcl-cb5
TagsVenusExpressionMammalianPromoterCMVAvailable SinceJan. 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
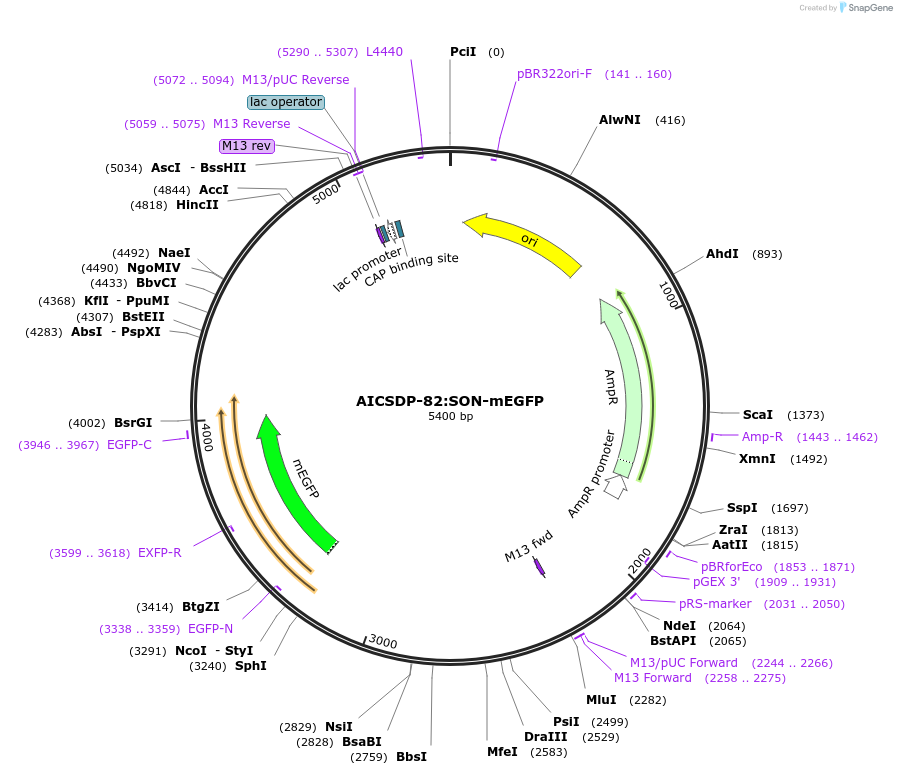
AICSDP-82:SON-mEGFP
Plasmid#133964PurposeHomology arms and mEGFP-linker sequence for N-terminus tagging of human SONDepositorInsertSON Homology Arms with mEGFP-linker (SON Human)
UseCRISPRTagsmEGFP-linkerExpressionMammalianMutationhomology arms contain point mutations to disrupt …Available SinceNov. 22, 2019AvailabilityIndustry, Academic Institutions, and Nonprofits -
AICSDP-82:SON-mEGFP
Plasmid#133964PurposeHomology arms and mEGFP-linker sequence for N-terminus tagging of human SONDepositorInsertSON Homology Arms with mEGFP-linker (SON Human)
UseCRISPRTagsmEGFP-linkerExpressionMammalianMutationhomology arms contain point mutations to disrupt …Available SinceNov. 22, 2019AvailabilityIndustry, Academic Institutions, and Nonprofits -
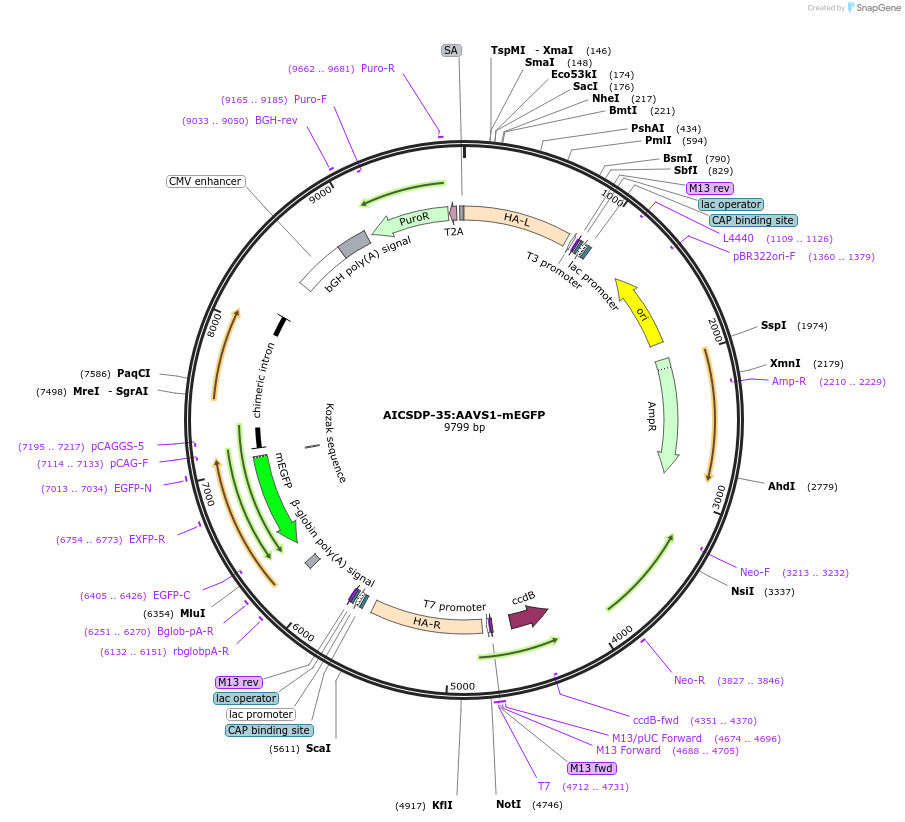
AICSDP-35:AAVS1-mEGFP
Plasmid#91565PurposeHomology arms and linker-mEGFP sequence for internal insertion of CAGGS-mEGFP at the AAVS1 safe harbor location in human cells as a reporter for cytoplasmic mEGFPDepositorInsertAAVS1 Homology Arms with CAGGS driven mEGFP (AAVS1 Human)
UseCRISPR; Donor templateExpressionMammalianPromoterCAGAvailable SinceJune 7, 2017AvailabilityIndustry, Academic Institutions, and Nonprofits -
AICSDP-35:AAVS1-mEGFP
Plasmid#91565PurposeHomology arms and linker-mEGFP sequence for internal insertion of CAGGS-mEGFP at the AAVS1 safe harbor location in human cells as a reporter for cytoplasmic mEGFPDepositorInsertAAVS1 Homology Arms with CAGGS driven mEGFP (AAVS1 Human)
UseCRISPR; Donor templateExpressionMammalianPromoterCAGAvailable SinceJune 7, 2017AvailabilityIndustry, Academic Institutions, and Nonprofits -
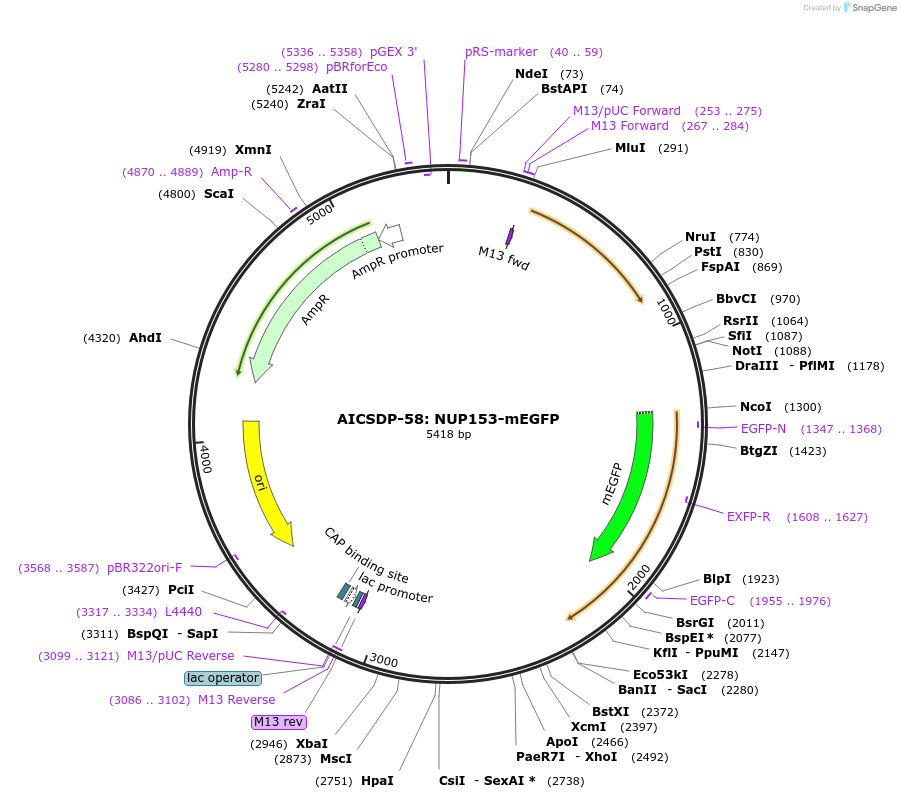
AICSDP-58: NUP153-mEGFP
Plasmid#114407PurposeHomology arms and mEGFP-linker sequence for N-terminus tagging of human NUP153DepositorInsertNUP153 Homology Arms with mEGFP-linker (NUP153 Human)
UseCRISPR; Donor templateTagsmEGFP-linkerExpressionMammalianMutationhomology arms contain point mutations to disrupt …Available SinceSept. 5, 2018AvailabilityIndustry, Academic Institutions, and Nonprofits -
AICSDP-58: NUP153-mEGFP
Plasmid#114407PurposeHomology arms and mEGFP-linker sequence for N-terminus tagging of human NUP153DepositorInsertNUP153 Homology Arms with mEGFP-linker (NUP153 Human)
UseCRISPR; Donor templateTagsmEGFP-linkerExpressionMammalianMutationhomology arms contain point mutations to disrupt …Available SinceSept. 5, 2018AvailabilityIndustry, Academic Institutions, and Nonprofits -
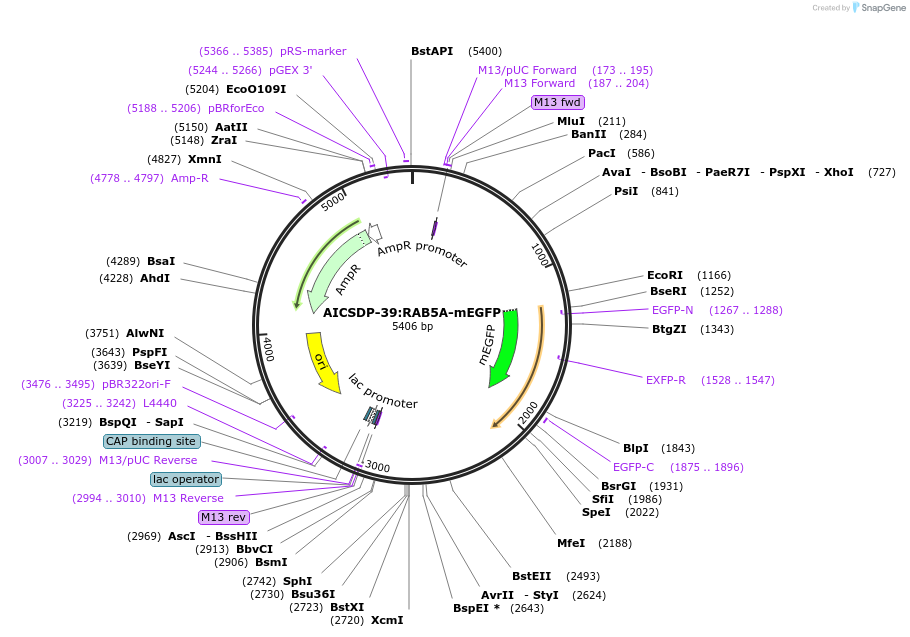
AICSDP-39:RAB5A-mEGFP
Plasmid#107579PurposeHomology arms and mEGFP-linker sequence for N-terminus tagging of human RAB5ADepositorInsertRAB5A Homology Arms with mEGFP-linker (RAB5A Human)
UseCRISPR; Donor templateTagsmEGFP-linkerExpressionMammalianMutationhomology arms contain point mutations to disrupt …Available SinceMay 9, 2018AvailabilityIndustry, Academic Institutions, and Nonprofits -
AICSDP-39:RAB5A-mEGFP
Plasmid#107579PurposeHomology arms and mEGFP-linker sequence for N-terminus tagging of human RAB5ADepositorInsertRAB5A Homology Arms with mEGFP-linker (RAB5A Human)
UseCRISPR; Donor templateTagsmEGFP-linkerExpressionMammalianMutationhomology arms contain point mutations to disrupt …Available SinceMay 9, 2018AvailabilityIndustry, Academic Institutions, and Nonprofits -
lgp120+linker-pEGFP N1
Plasmid#62963Purposerat lgp120 + 10 amino acid linker cloned into pEGFP N1. The linker separates the targetting motif in lgp120 from EGFP.DepositorInsertlgp120 (Lamp1 Rat)
TagsEGFP and linkerExpressionMammalianMutationhas 2 amino acid mutations compared to the coding…PromoterCMVAvailable SinceMay 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
lgp120+linker-pEGFP N1
Plasmid#62963Purposerat lgp120 + 10 amino acid linker cloned into pEGFP N1. The linker separates the targetting motif in lgp120 from EGFP.DepositorInsertlgp120 (Lamp1 Rat)
TagsEGFP and linkerExpressionMammalianMutationhas 2 amino acid mutations compared to the coding…PromoterCMVAvailable SinceMay 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
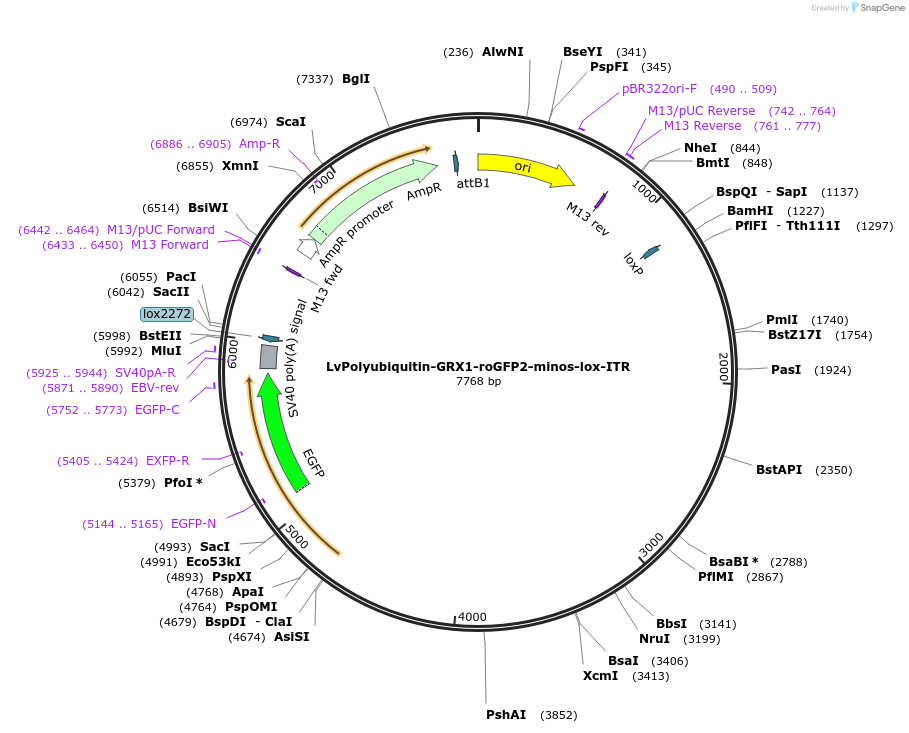
LvPolyubiquitin-GRX1-roGFP2-minos-lox-ITR
Plasmid#247292PurposePolyubiquitin promoter driving expression of glutathione biosensor GRX1-roGFP2. Flanked by ITRs compatible with minos-mediated transgenesisDepositorInsertGRX1-roGFP2
UseSynthetic Biology; Sea urchin expressionPromoterPolyubiquitinAvailable SinceNov. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
LvPolyubiquitin-GRX1-roGFP2-minos-lox-ITR
Plasmid#247292PurposePolyubiquitin promoter driving expression of glutathione biosensor GRX1-roGFP2. Flanked by ITRs compatible with minos-mediated transgenesisDepositorInsertGRX1-roGFP2
UseSynthetic Biology; Sea urchin expressionPromoterPolyubiquitinAvailable SinceNov. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
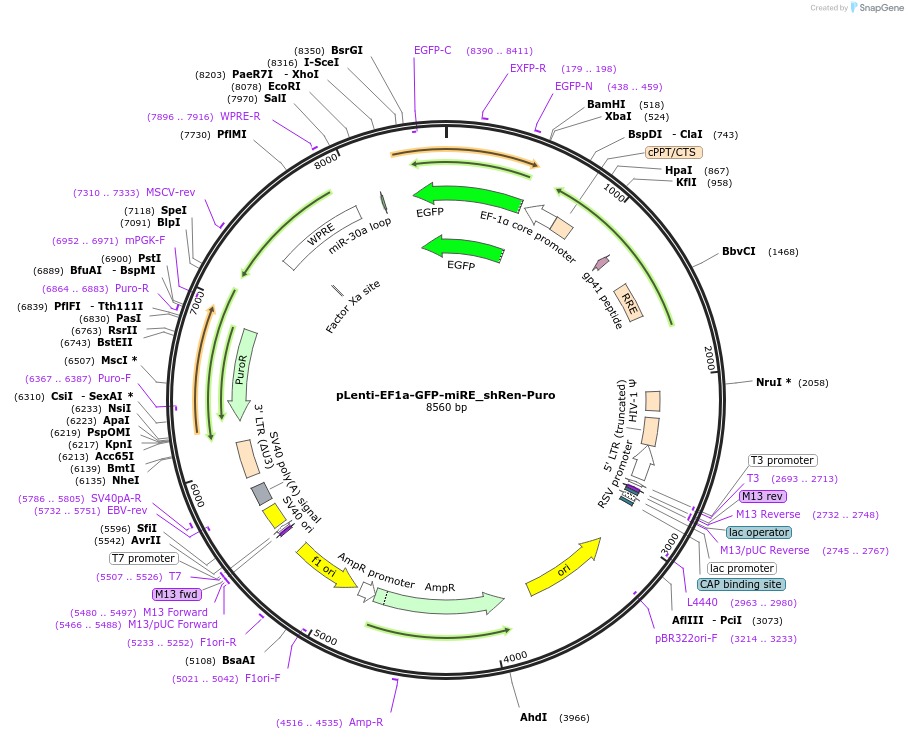
pLenti-EF1a-GFP-miRE_shRen-Puro
Plasmid#135683PurposeConstitutive expression (EF1a promoter) of GFP cDNA plus miRE-embedded shRenilla (control shRNA), Puromycin selectionDepositorInsertshRenilla
UseLentiviralAvailable SinceJan. 13, 2022AvailabilityAcademic Institutions and Nonprofits only -
pLenti-EF1a-GFP-miRE_shRen-Puro
Plasmid#135683PurposeConstitutive expression (EF1a promoter) of GFP cDNA plus miRE-embedded shRenilla (control shRNA), Puromycin selectionDepositorInsertshRenilla
UseLentiviralAvailable SinceJan. 13, 2022AvailabilityAcademic Institutions and Nonprofits only -
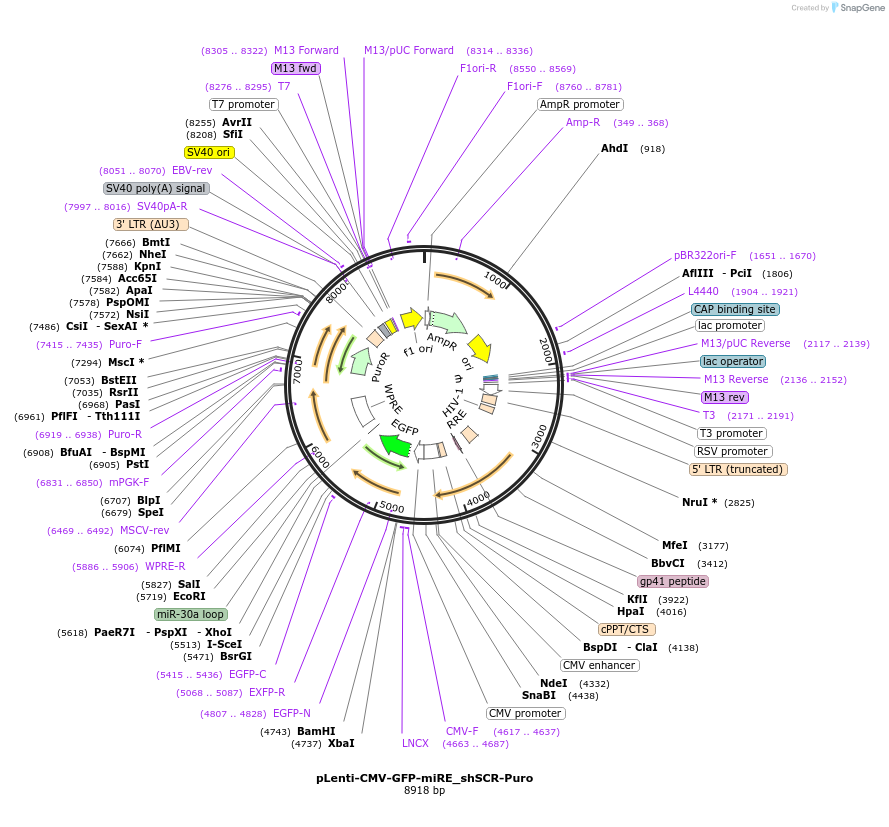
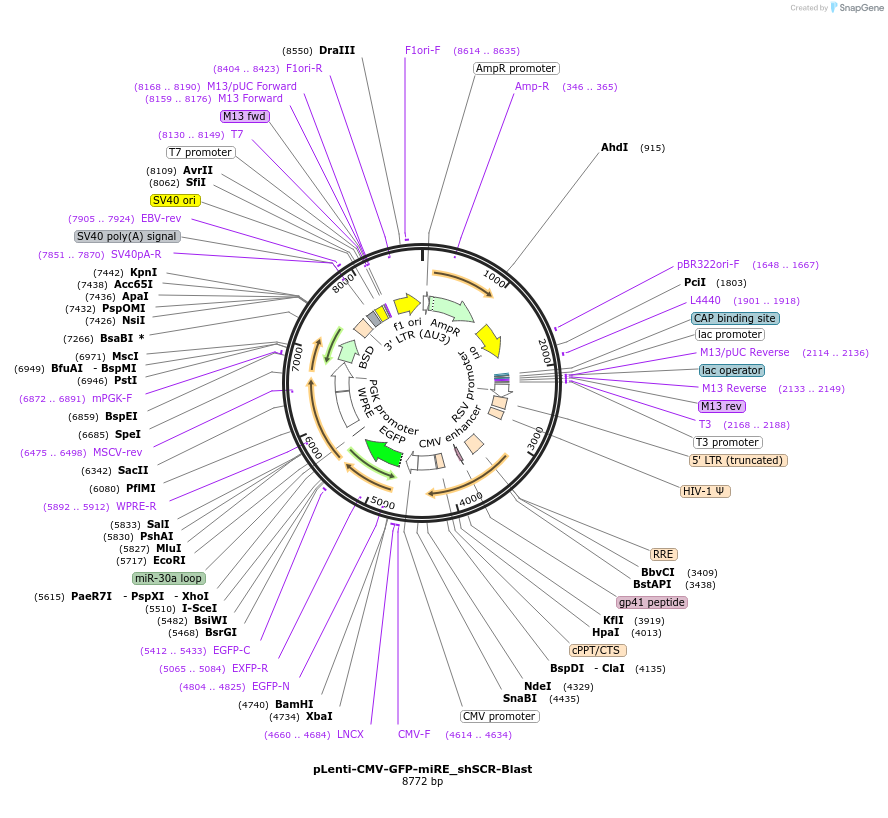
pLenti-CMV-GFP-miRE_shSCR-Puro
Plasmid#135689PurposeConstitutive expression (CMV promoter) of GFP cDNA plus miRE-embedded scrambled control shRNA, Puromycin selectionDepositorInsertshScrambled
UseLentiviralAvailable SinceJan. 13, 2022AvailabilityAcademic Institutions and Nonprofits only -
pLenti-CMV-GFP-miRE_shSCR-Puro
Plasmid#135689PurposeConstitutive expression (CMV promoter) of GFP cDNA plus miRE-embedded scrambled control shRNA, Puromycin selectionDepositorInsertshScrambled
UseLentiviralAvailable SinceJan. 13, 2022AvailabilityAcademic Institutions and Nonprofits only -
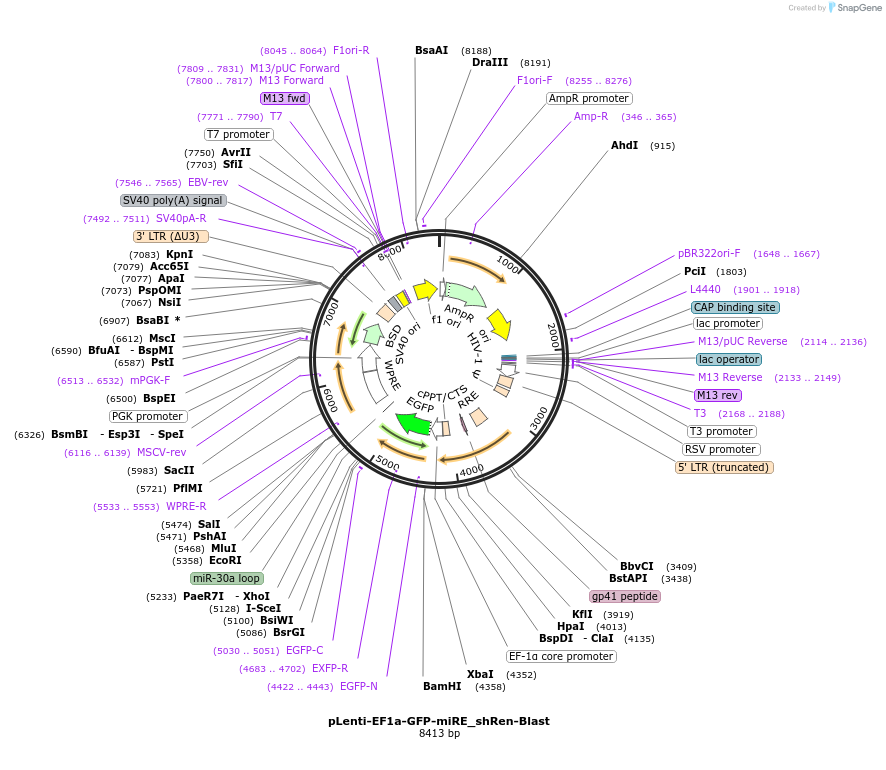
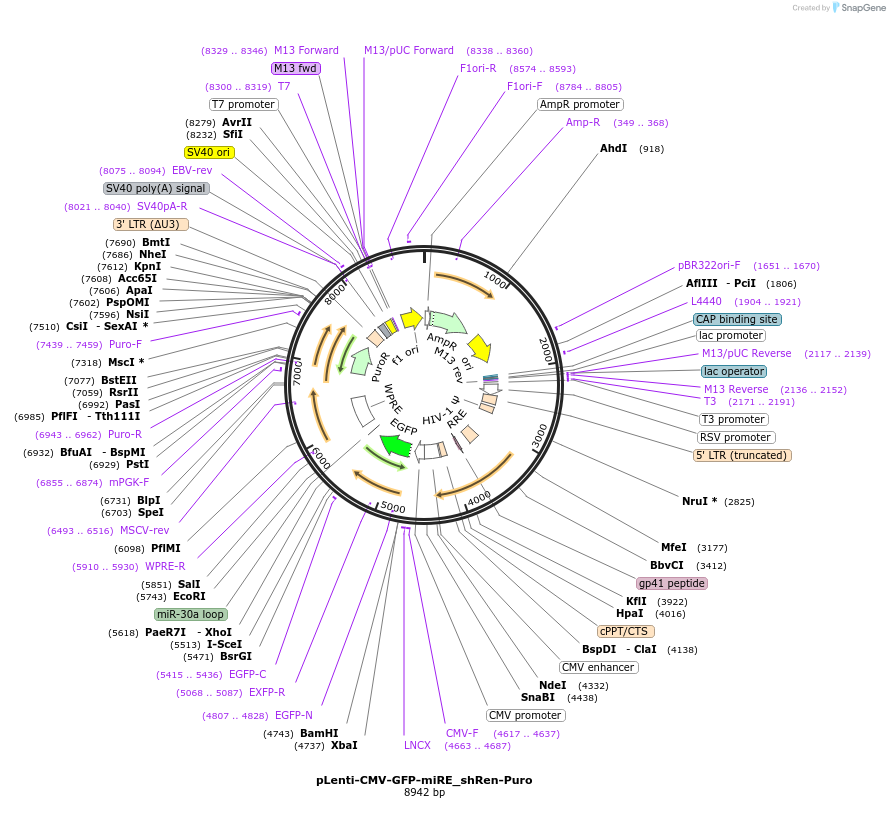
pLenti-EF1a-GFP-miRE_shRen-Blast
Plasmid#135682PurposeConstitutive expression (EF1a promoter) of GFP cDNA plus miRE-embedded shRenilla (control shRNA), Blasticidin selectionDepositorInsertshRenilla
UseLentiviralAvailable SinceMarch 15, 2021AvailabilityAcademic Institutions and Nonprofits only -
pLenti-EF1a-GFP-miRE_shRen-Blast
Plasmid#135682PurposeConstitutive expression (EF1a promoter) of GFP cDNA plus miRE-embedded shRenilla (control shRNA), Blasticidin selectionDepositorInsertshRenilla
UseLentiviralAvailable SinceMarch 15, 2021AvailabilityAcademic Institutions and Nonprofits only -
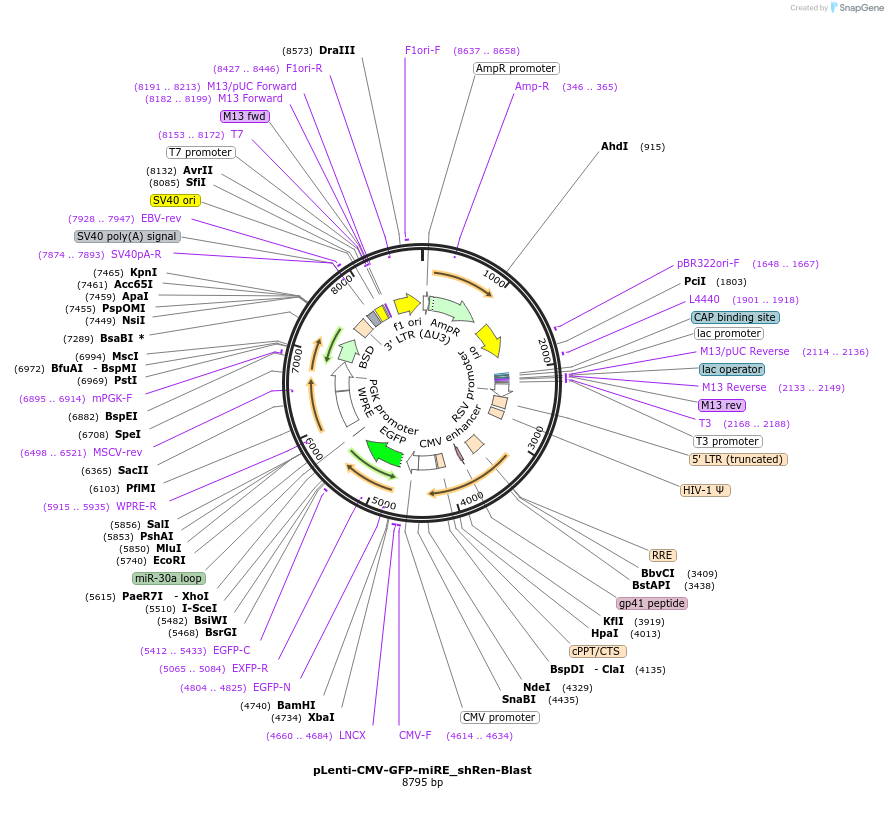
pLenti-CMV-GFP-miRE_shRen-Blast
Plasmid#135684PurposeConstitutive expression (CMV promoter) of GFP cDNA plus miRE-embedded shRenilla (control shRNA), Blasticidin selectionDepositorInsertshRenilla
UseLentiviralAvailable SinceMarch 16, 2021AvailabilityAcademic Institutions and Nonprofits only -
pLenti-CMV-GFP-miRE_shRen-Blast
Plasmid#135684PurposeConstitutive expression (CMV promoter) of GFP cDNA plus miRE-embedded shRenilla (control shRNA), Blasticidin selectionDepositorInsertshRenilla
UseLentiviralAvailable SinceMarch 16, 2021AvailabilityAcademic Institutions and Nonprofits only -
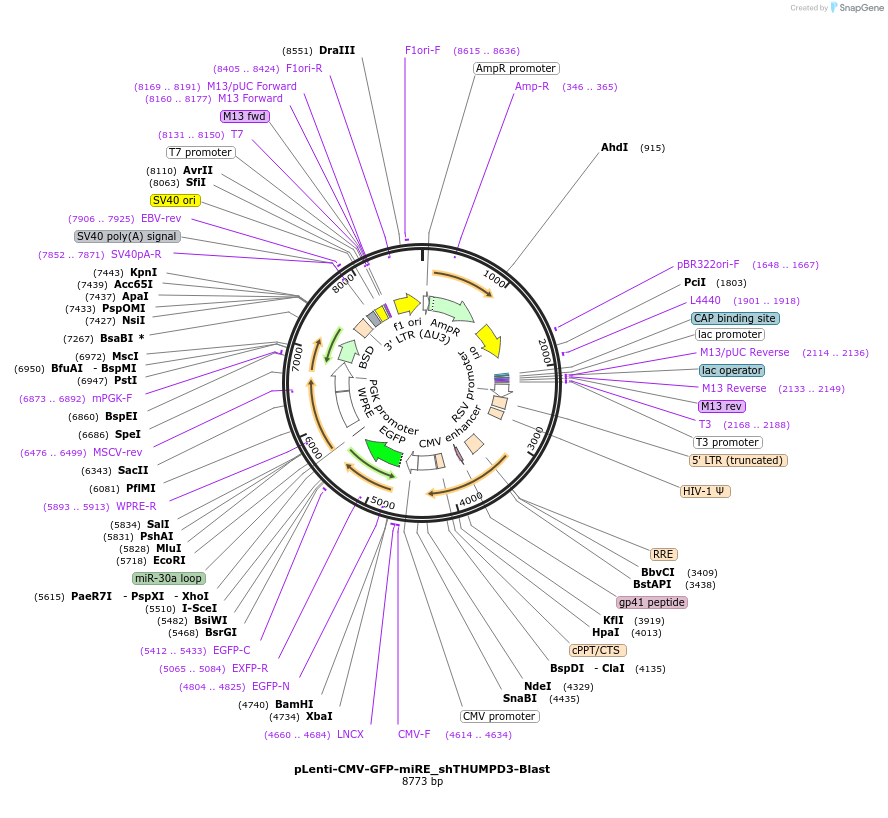
pLenti-CMV-GFP-miRE_shTHUMPD3-Blast
Plasmid#135690PurposeConstitutive expression (CMV promoter) of GFP cDNA plus miRE-embedded shROSA26 (control shRNA), Blasticidin selectionDepositorInsertshRosa26
UseLentiviralAvailable SinceJan. 13, 2022AvailabilityAcademic Institutions and Nonprofits only -
pLenti-CMV-GFP-miRE_shTHUMPD3-Blast
Plasmid#135690PurposeConstitutive expression (CMV promoter) of GFP cDNA plus miRE-embedded shROSA26 (control shRNA), Blasticidin selectionDepositorInsertshRosa26
UseLentiviralAvailable SinceJan. 13, 2022AvailabilityAcademic Institutions and Nonprofits only -
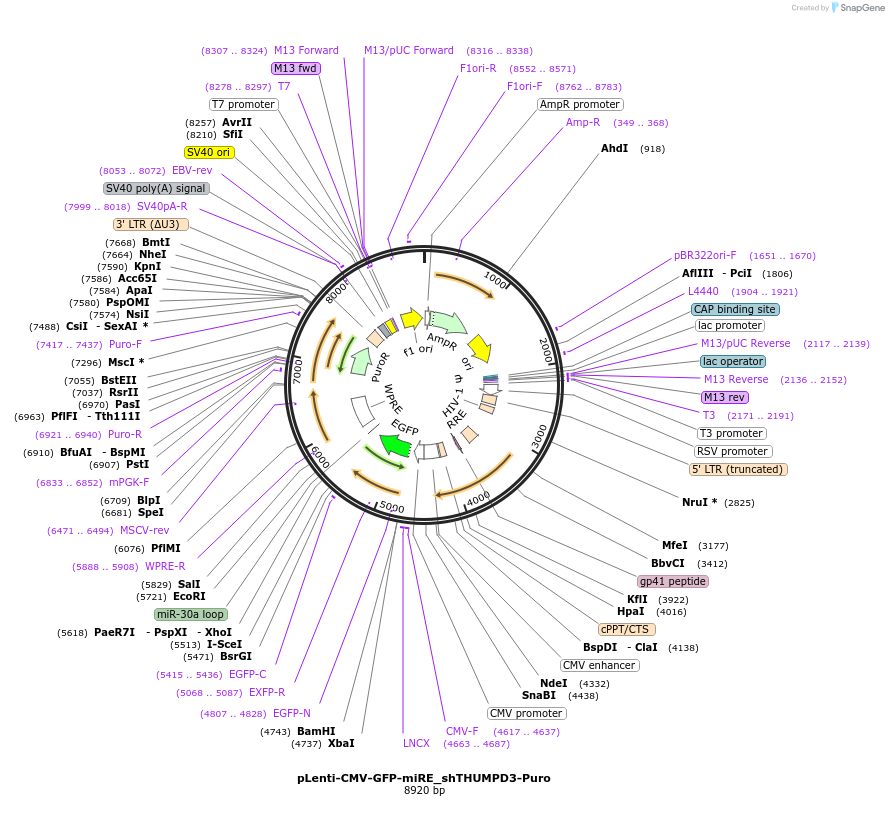
pLenti-CMV-GFP-miRE_shTHUMPD3-Puro
Plasmid#135691PurposeConstitutive expression (CMV promoter) of GFP cDNA plus miRE-embedded shROSA26 (control shRNA), Puromycin selectionDepositorInsertshRosa26
UseLentiviralAvailable SinceJan. 13, 2022AvailabilityAcademic Institutions and Nonprofits only -
pLenti-CMV-GFP-miRE_shTHUMPD3-Puro
Plasmid#135691PurposeConstitutive expression (CMV promoter) of GFP cDNA plus miRE-embedded shROSA26 (control shRNA), Puromycin selectionDepositorInsertshRosa26
UseLentiviralAvailable SinceJan. 13, 2022AvailabilityAcademic Institutions and Nonprofits only -
pLenti-CMV-GFP-miRE_shSCR-Blast
Plasmid#135688PurposeConstitutive expression (CMV promoter) of GFP cDNA plus miRE-embedded scrambled control shRNA, Blasticidin selectionDepositorInsertshScrambled
UseLentiviralAvailable SinceJan. 13, 2022AvailabilityAcademic Institutions and Nonprofits only -
pLenti-CMV-GFP-miRE_shSCR-Blast
Plasmid#135688PurposeConstitutive expression (CMV promoter) of GFP cDNA plus miRE-embedded scrambled control shRNA, Blasticidin selectionDepositorInsertshScrambled
UseLentiviralAvailable SinceJan. 13, 2022AvailabilityAcademic Institutions and Nonprofits only -
pLenti-CMV-GFP-miRE_shRen-Puro
Plasmid#135685PurposeConstitutive expression (CMV promoter) of GFP cDNA plus miRE-embedded shRenilla (control shRNA), Puromycin selectionDepositorInsertshRenilla
UseLentiviralAvailable SinceMarch 15, 2021AvailabilityAcademic Institutions and Nonprofits only -
pLenti-CMV-GFP-miRE_shRen-Puro
Plasmid#135685PurposeConstitutive expression (CMV promoter) of GFP cDNA plus miRE-embedded shRenilla (control shRNA), Puromycin selectionDepositorInsertshRenilla
UseLentiviralAvailable SinceMarch 15, 2021AvailabilityAcademic Institutions and Nonprofits only -
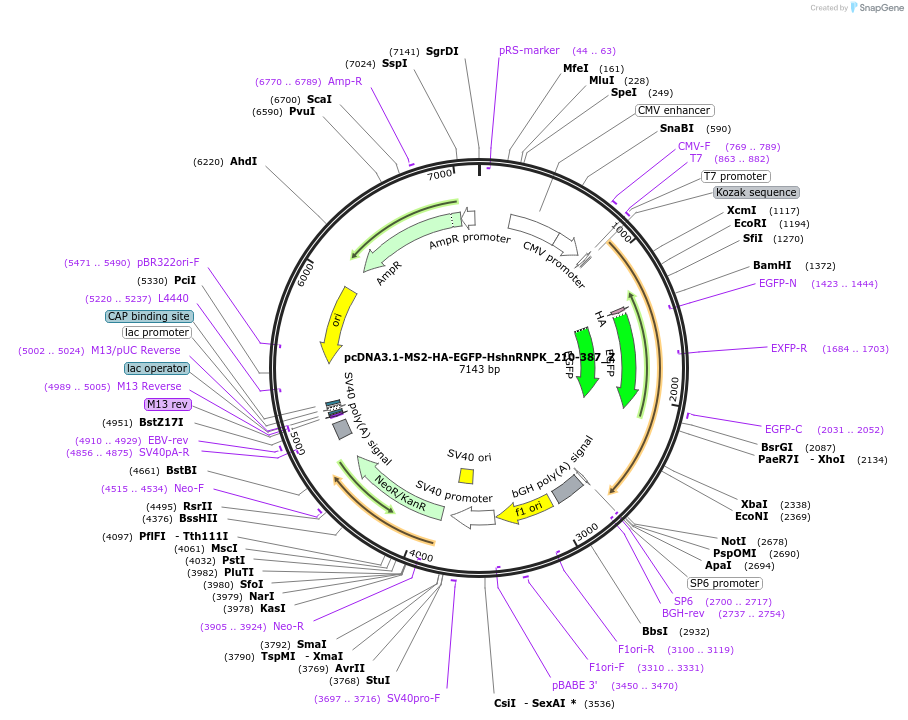
pcDNA3.1-MS2-HA-EGFP-HshnRNPK_210-387_Z
Plasmid#148110PurposeMammalian Expression of HshnRNPK_210-387DepositorInsertHshnRNPK_210-387 (HNRNPK Human)
ExpressionMammalianAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3.1-MS2-HA-EGFP-HshnRNPK_210-387_Z
Plasmid#148110PurposeMammalian Expression of HshnRNPK_210-387DepositorInsertHshnRNPK_210-387 (HNRNPK Human)
ExpressionMammalianAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
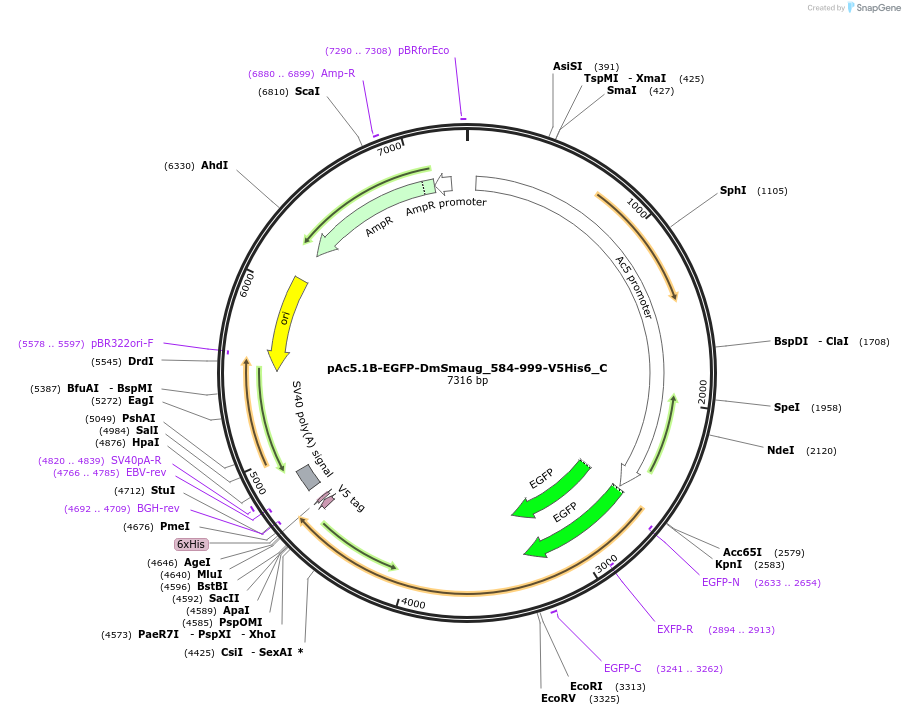
pAc5.1B-EGFP-DmSmaug_584-999-V5His6_C
Plasmid#146003PurposeInsect Expression of DmSmaug_584-999DepositorInsertDmSmaug_584-999 (smg Fly)
ExpressionInsectMutation5 silent mutations and a deletion of the AA 961-9…Available SinceFeb. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
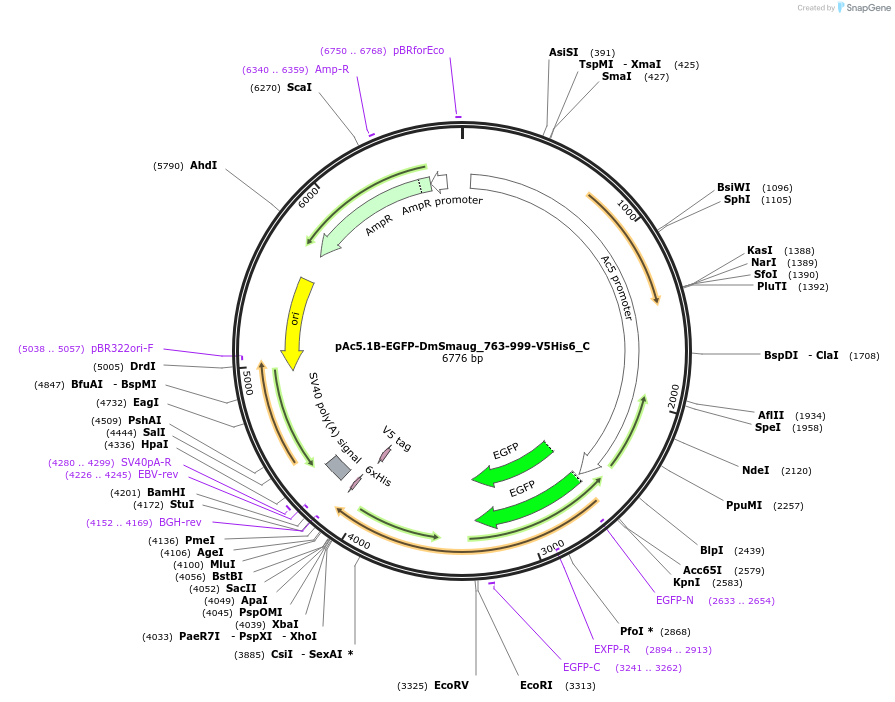
pAc5.1B-EGFP-DmSmaug_763-999-V5His6_C
Plasmid#146004PurposeInsect Expression of DmSmaug_763-999DepositorInsertDmSmaug_763-999 (smg Fly)
ExpressionInsectMutation2 silent mutations and a deletion of the AA 961-9…Available SinceFeb. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
pAc5.1B-EGFP-DmSmaug-A642H-V5His6_C
Plasmid#146005PurposeInsect Expression of DmSmaug-A642HDepositorInsertDmSmaug-A642H (smg Fly)
ExpressionInsectMutation10 silent mutations and a deletion of the AA 961-…Available SinceFeb. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
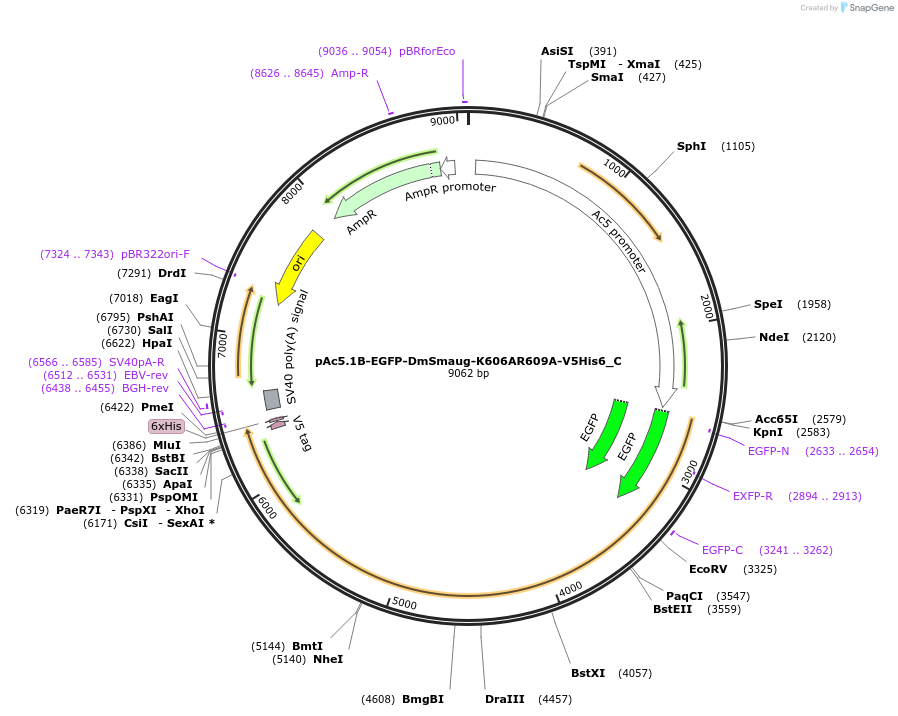
pAc5.1B-EGFP-DmSmaug-K606AR609A-V5His6_C
Plasmid#146007PurposeInsect Expression of DmSmaug-K606AR609ADepositorInsertDmSmaug-K606AR609A (smg Fly)
ExpressionInsectMutation10 silent mutations and a deletion of the AA 961-…Available SinceFeb. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
pAc5.1B-EGFP-DmSmaug_584-999-V5His6_C
Plasmid#146003PurposeInsect Expression of DmSmaug_584-999DepositorInsertDmSmaug_584-999 (smg Fly)
ExpressionInsectMutation5 silent mutations and a deletion of the AA 961-9…Available SinceFeb. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
pAc5.1B-EGFP-DmSmaug_763-999-V5His6_C
Plasmid#146004PurposeInsect Expression of DmSmaug_763-999DepositorInsertDmSmaug_763-999 (smg Fly)
ExpressionInsectMutation2 silent mutations and a deletion of the AA 961-9…Available SinceFeb. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
pAc5.1B-EGFP-DmSmaug-A642H-V5His6_C
Plasmid#146005PurposeInsect Expression of DmSmaug-A642HDepositorInsertDmSmaug-A642H (smg Fly)
ExpressionInsectMutation10 silent mutations and a deletion of the AA 961-…Available SinceFeb. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
pAc5.1B-EGFP-DmSmaug-K606AR609A-V5His6_C
Plasmid#146007PurposeInsect Expression of DmSmaug-K606AR609ADepositorInsertDmSmaug-K606AR609A (smg Fly)
ExpressionInsectMutation10 silent mutations and a deletion of the AA 961-…Available SinceFeb. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
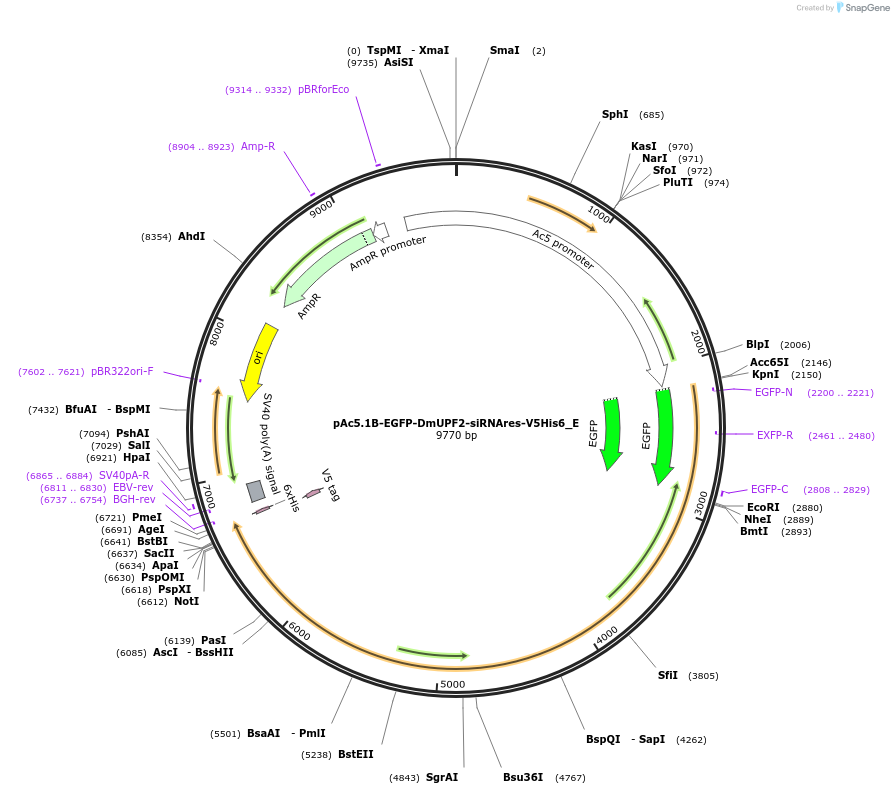
pAc5.1B-EGFP-DmUPF2-siRNAres-V5His6_E
Plasmid#146166PurposeInsect Expression of DmUPF2-siRNAresDepositorInsertDmUPF2-siRNAres (Upf2 Fly)
ExpressionInsectMutation3 non silent mutations: L1022P, V1097L and N1213S…Available SinceJune 25, 2021AvailabilityAcademic Institutions and Nonprofits only