We narrowed to 325 results for: tbp
-
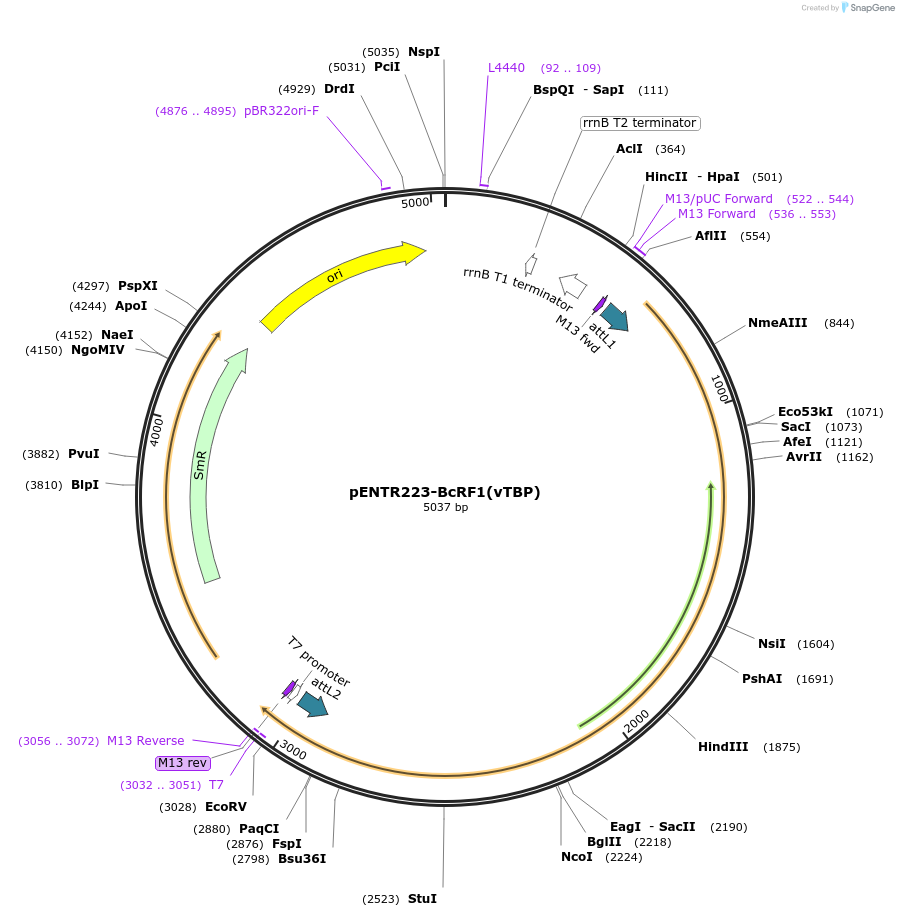
Plasmid#195832PurposeGateway Entry Clone of Epstein-Barr virus BcRF1DepositorInsertBcRF1 (BcRF1 Epstein-Barr virus)
UseGateway entry vectorAvailable SinceMay 19, 2023AvailabilityAcademic Institutions and Nonprofits only -
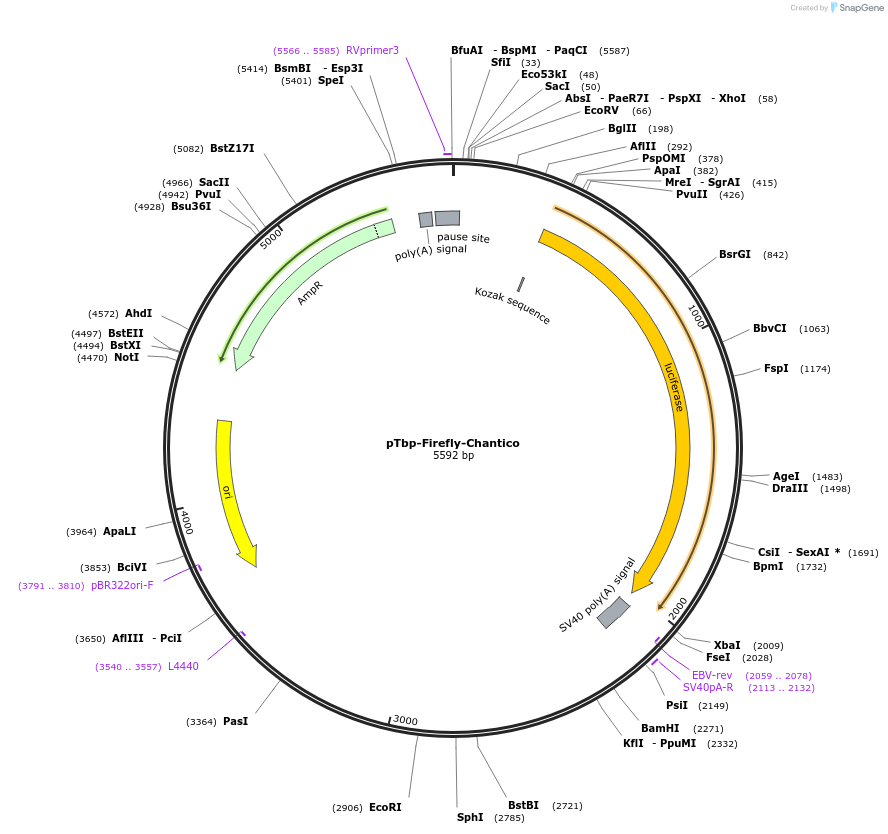
pTbp-Firefly-Chantico
Plasmid#177836PurposeLuciferase reporter for mouse Tbp with Chantico promoterDepositorInsertLuciferase
UseLuciferasePromoterTbpAvailable SinceAug. 29, 2022AvailabilityAcademic Institutions and Nonprofits only -
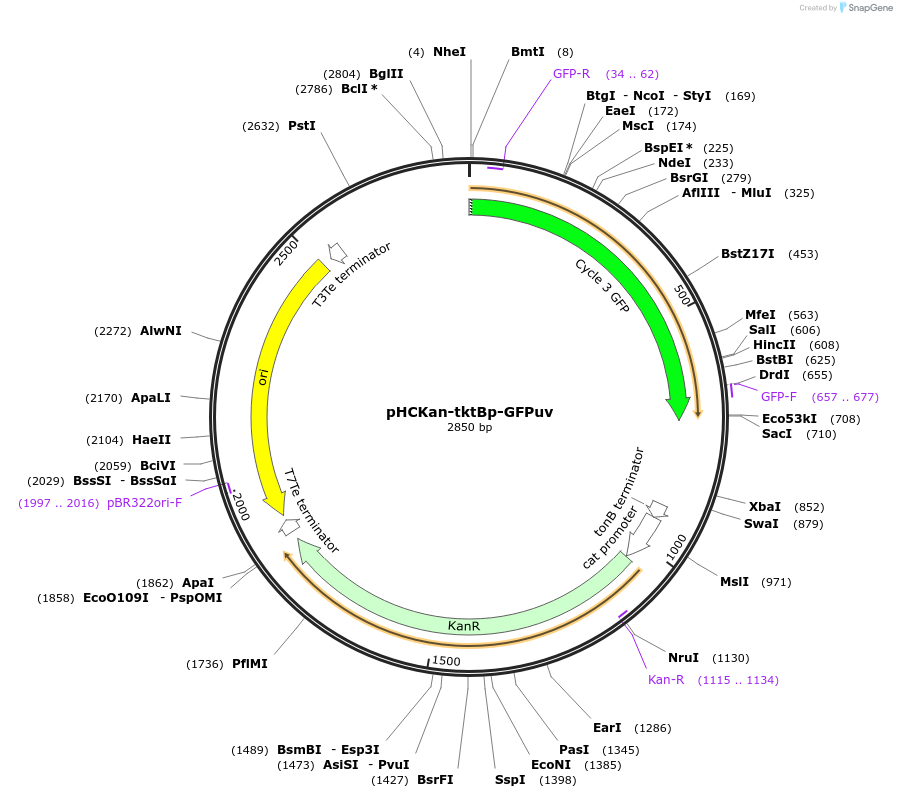
pHCKan-tktBp-GFPuv
Plasmid#139081PurposeLow phosphate inducible promoter tktBp for GFP expressionDepositorInserttktBp-GFPuv
ExpressionBacterialAvailable SinceSept. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
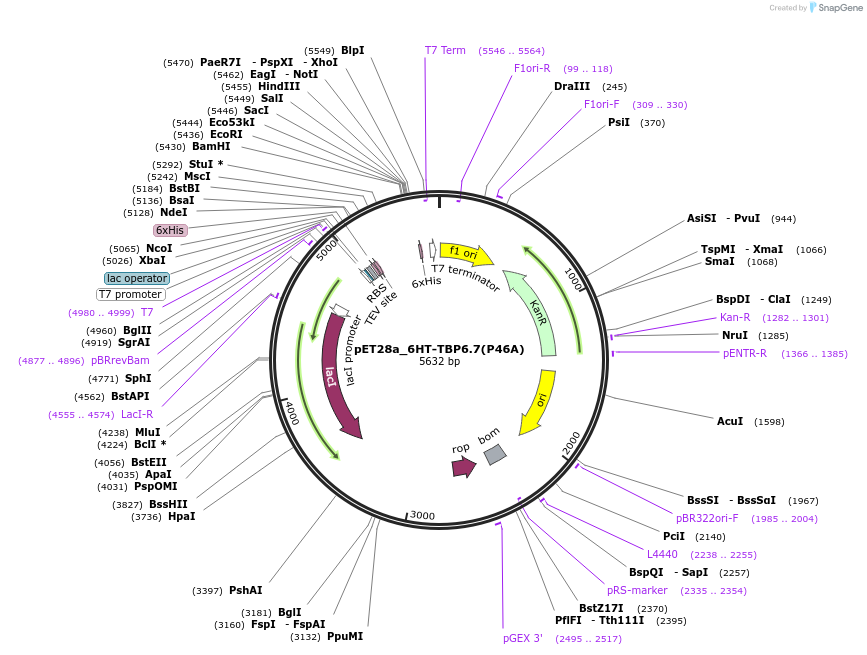
pET28a_6HT-TBP6.7(P46A)
Plasmid#112721PurposeThis mutation (P46A) marks the first residue within the evolved region of U1A.DepositorInsert6His-TEV-TBP6.7(P46A)
Tags6His-TEVExpressionBacterialMutationP46APromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
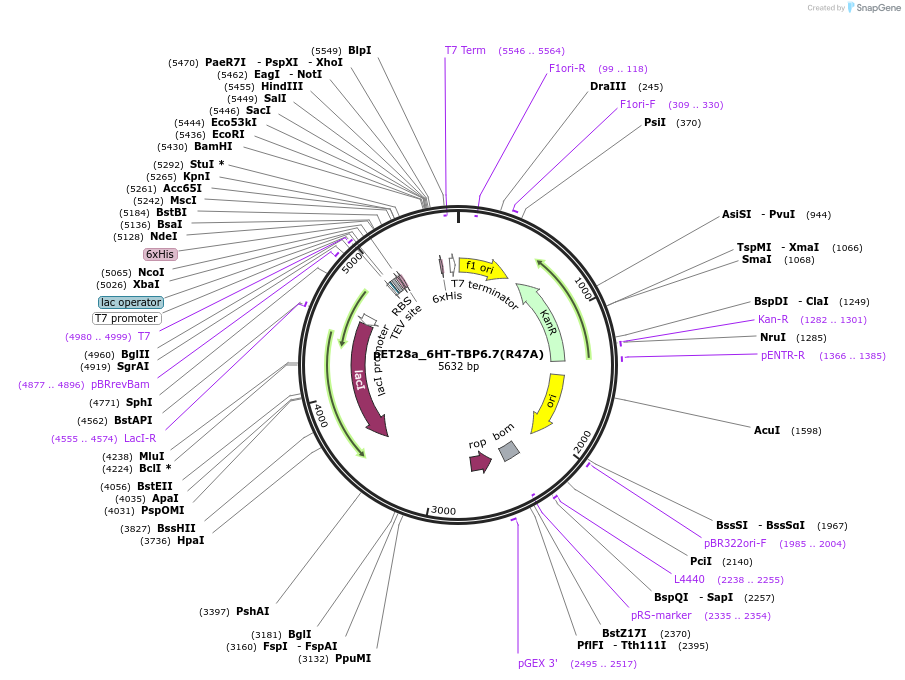
pET28a_6HT-TBP6.7(R47A)
Plasmid#112722PurposeArg47 within TBP6.7 is the most critical residue in terms of forming interactions with WT TAR. Mutating this amino acid to Ala results in ~600-fold reduction in Kd.DepositorInsert6His-TEV-TBP6.7(R47A)
Tags6His-TEVExpressionBacterialMutationR47APromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
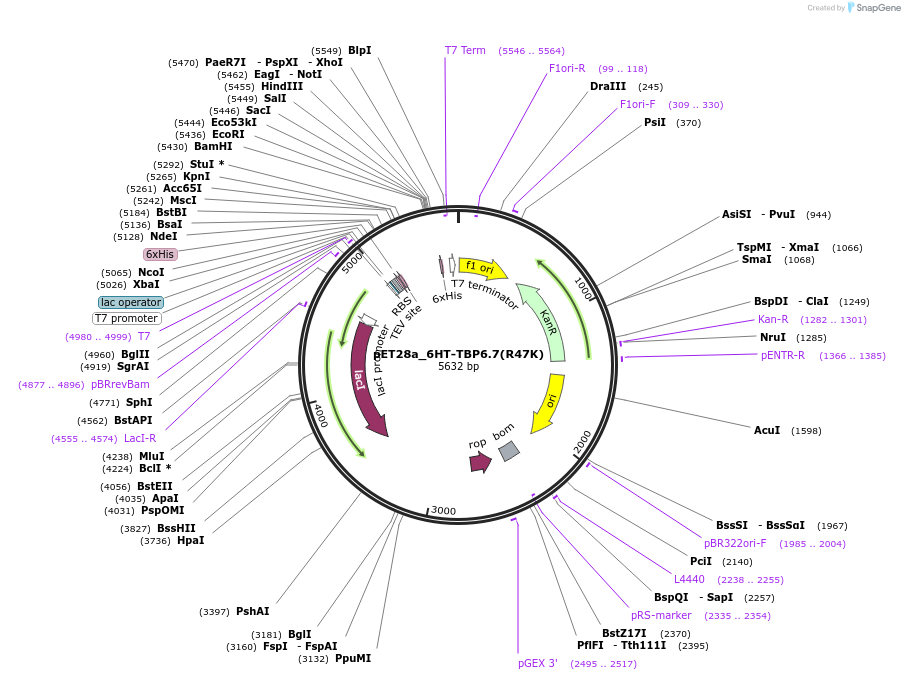
pET28a_6HT-TBP6.7(R47K)
Plasmid#112723PurposeArg47 of TBP6.7 interacts with Hoogsteen edge of TAR's Gua26. Mutating this amino acid to Lys reduces binding to the WT TAR RNA ~330-fold.DepositorInsert6His-TEV-TBP6.7(R47K)
Tags6His-TEVExpressionBacterialMutationR47KPromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
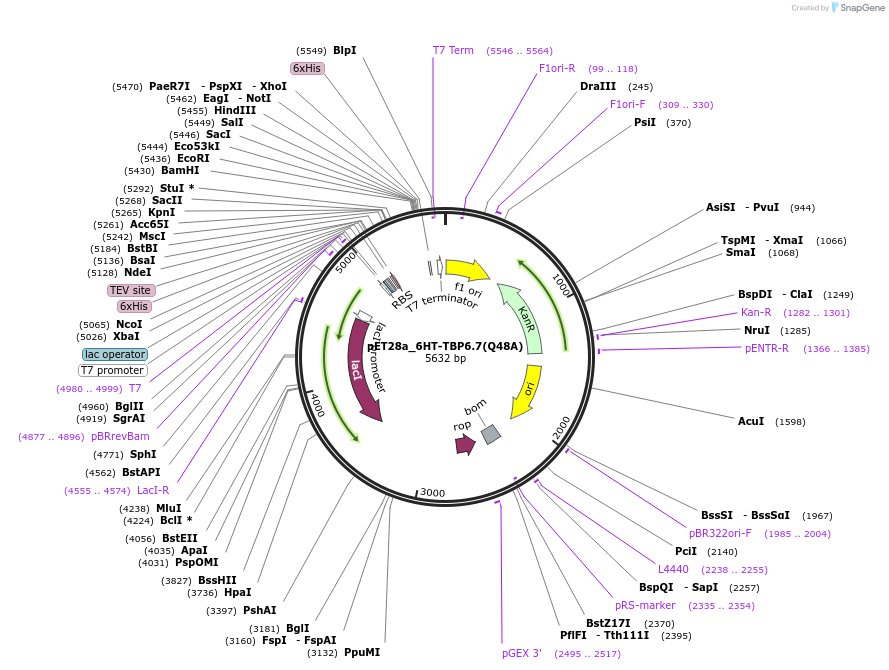
pET28a_6HT-TBP6.7(Q48A)
Plasmid#112724PurposeGln48 contacts the backbone of TAR RNA as well as mediates intramoecular interaction to stabilize the conformation of beta2-beta3 loop within TBP6.7.DepositorInsert6His-TEV-TBP6.7(Q48A)
Tags6His-TEVExpressionBacterialMutationQ48APromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
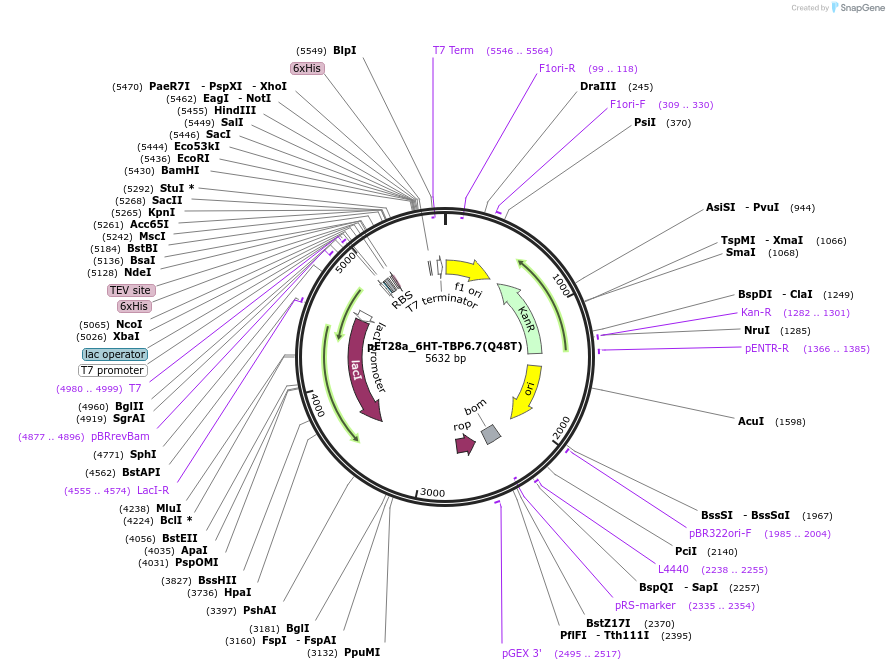
pET28a_6HT-TBP6.7(Q48T)
Plasmid#112725PurposeThis mutant of TBP6.7 binds to TAR very similarly to TBP6.7(WT), with only 1.4-fold reduced Kd.DepositorInsert6His-TEV-TBP6.7(Q48T)
Tags6His-TEVExpressionBacterialMutationQ48TPromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
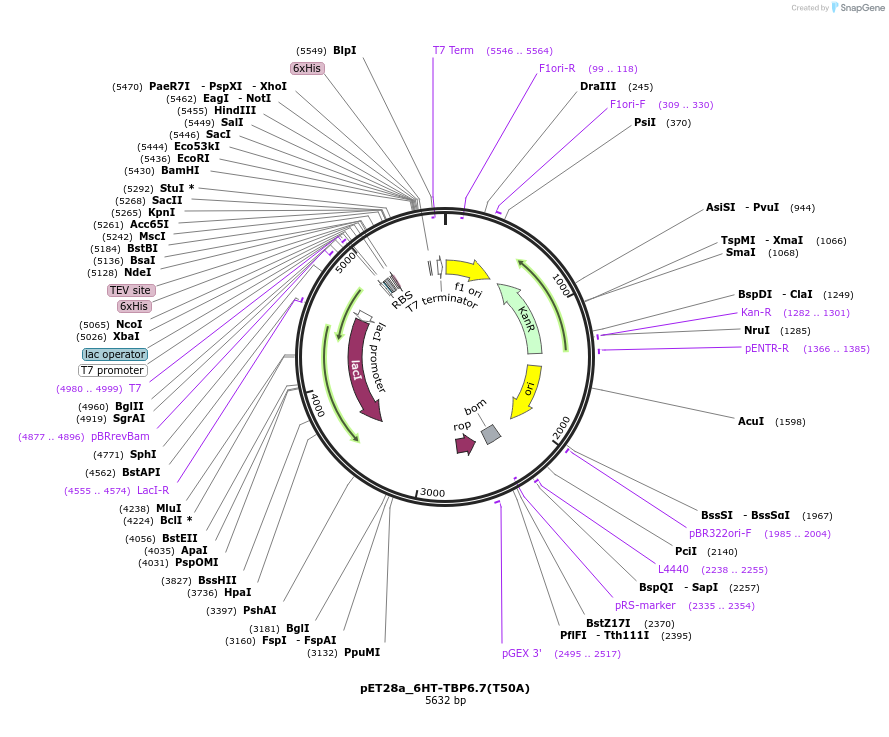
pET28a_6HT-TBP6.7(T50A)
Plasmid#112727PurposeLav-evolved amino acid T50 within TBPs is important in engaging intramolecularly to steer also lab-evolved R49 into conformation compatible binding to RNA.DepositorInsert6His-TEV-TBP6.7(T50A)
Tags6His-TEVExpressionBacterialMutationT50APromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
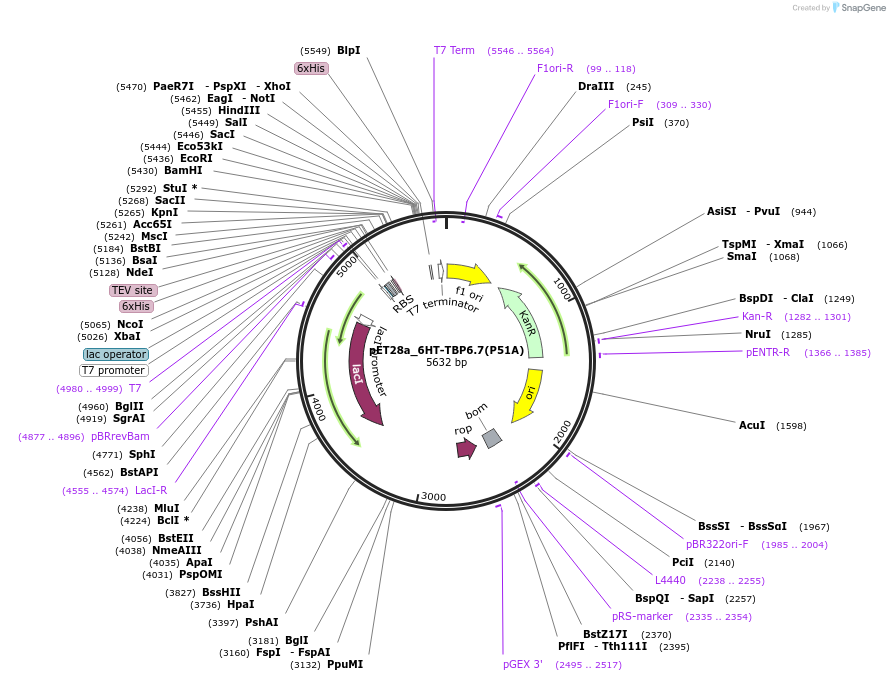
pET28a_6HT-TBP6.7(P51A)
Plasmid#112728PurposeSimilar to P46, P51 was selected and is present in all evolved TBPs. Mutating this position into Ala has a modest effect on binding.DepositorInsert6His-TEV-TBP6.7(P51A)
Tags6His-TEVExpressionBacterialMutationP51APromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
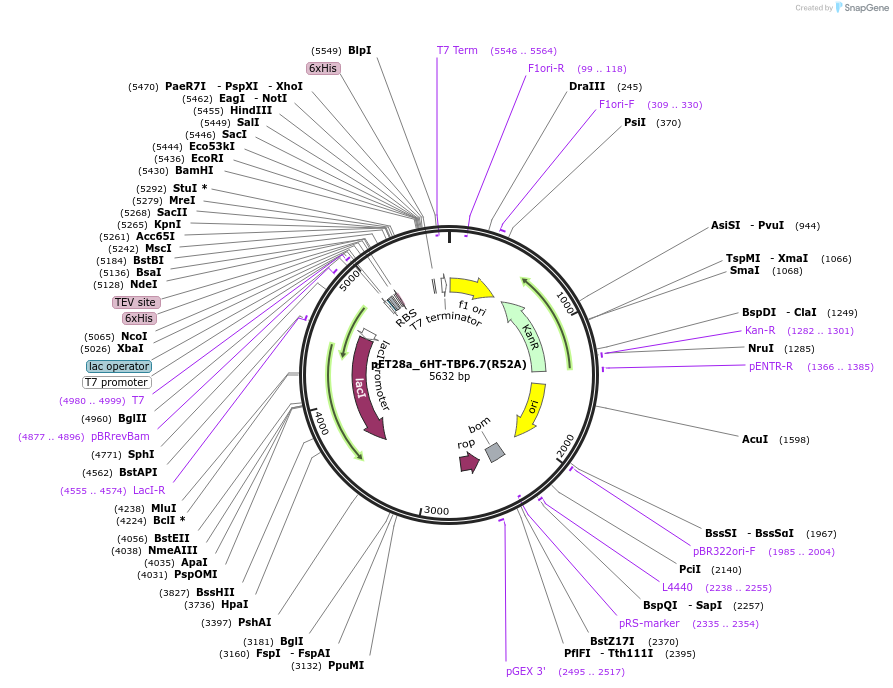
pET28a_6HT-TBP6.7(R52A)
Plasmid#112729Purpose52nd position is Arg in both wild-type U1A and TBPs, but the RNA recognition by this residue in both types of proteins is fundamentally different.DepositorInsert6His-TEV-TBP6.7(R52A)
Tags6His-TEVExpressionBacterialMutationR52APromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
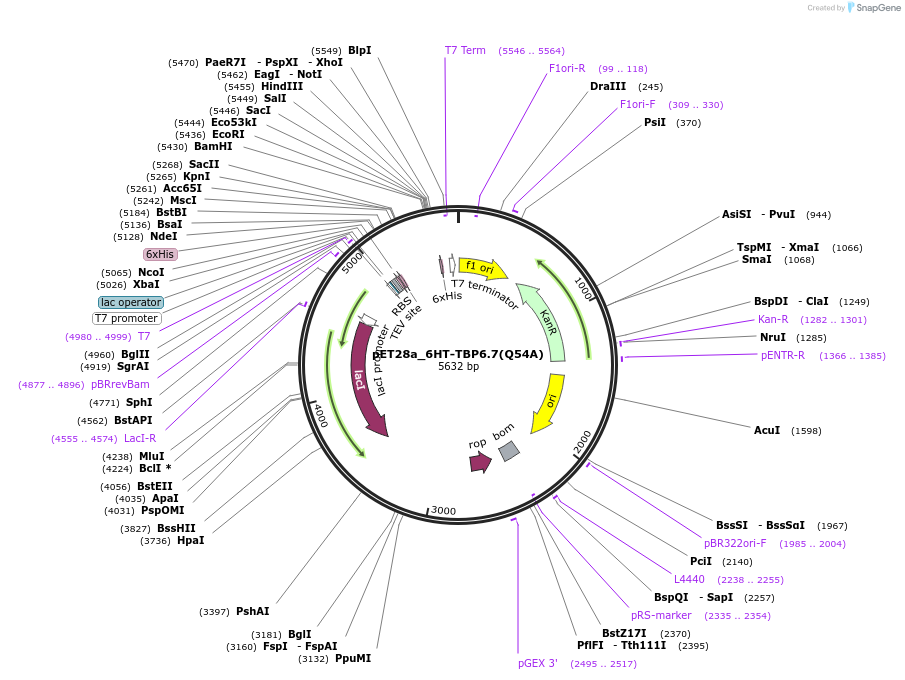
pET28a_6HT-TBP6.7(Q54A)
Plasmid#112730PurposeAlthough not evolved, this residue within TBPs participates in stabilizing the polypeptide loop responsible for RNA recognition.DepositorInsert6His-TEV-TBP6.7(Q54A)
Tags6His-TEVExpressionBacterialMutationQ54APromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
XE90 pCS2P+ XCtBP
Plasmid#16704DepositorInsertXCtBP (ctbp2 Frog)
UseXenopus expressionAvailable SinceMarch 14, 2008AvailabilityAcademic Institutions and Nonprofits only -
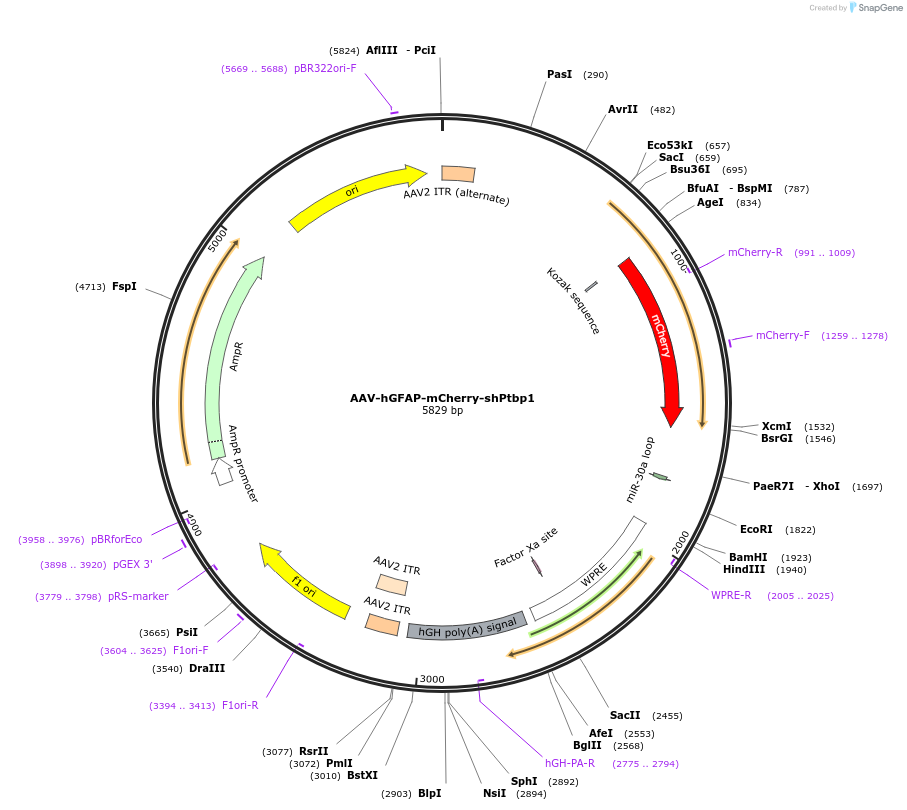
AAV-hGFAP-mCherry-shPtbp1
Plasmid#178589PurposeExpression of mCherry and a shRNA against Ptbp1 under the human GFAP promoterDepositorInsertmiRE-Ptbp1
UseAAVExpressionMammalianAvailable SinceFeb. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
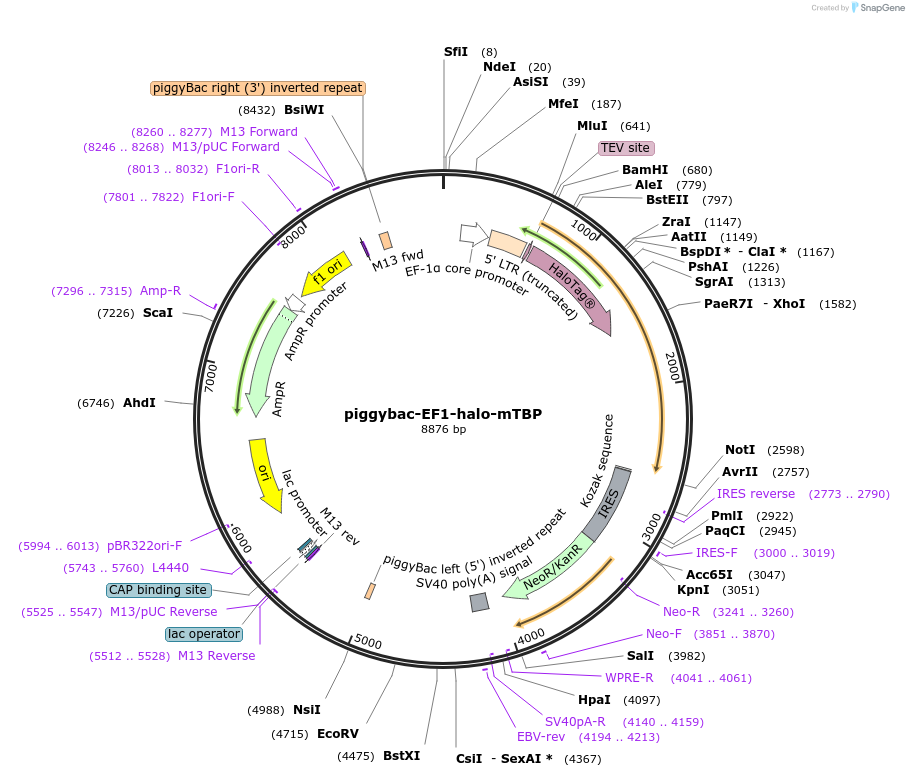
piggybac-EF1-halo-mTBP
Plasmid#197908Purposesingle-particle tracking for mTBPDepositorAvailable SinceMarch 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
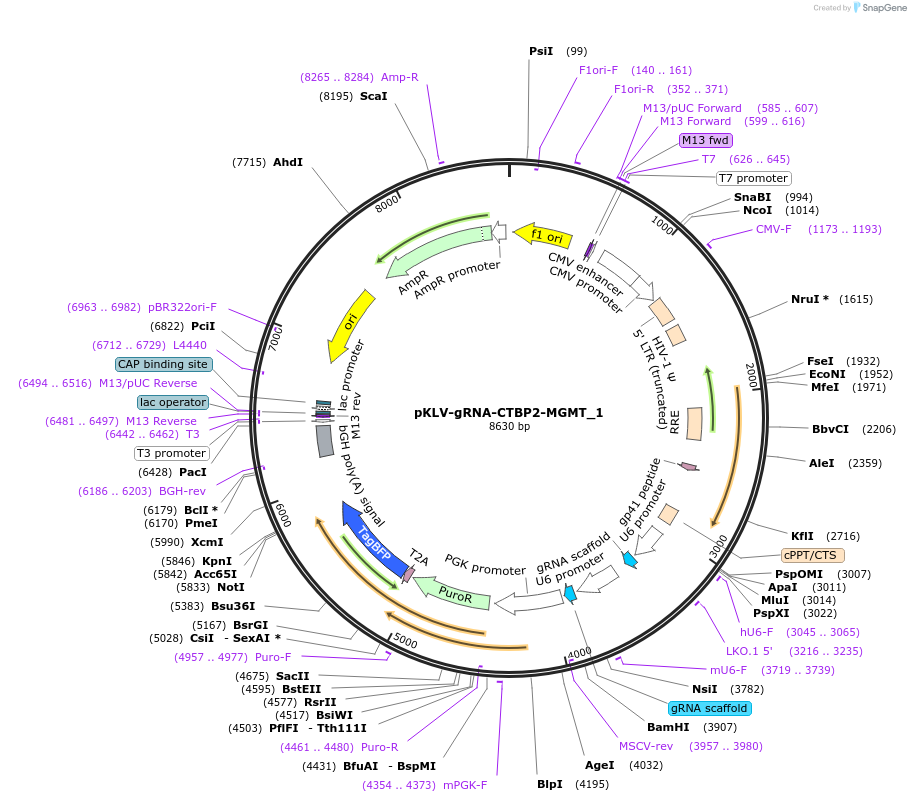
pKLV-gRNA-CTBP2-MGMT_1
Plasmid#136419PurposeLentiviral expression of gRNAs targeting intron 1 of human CTBP2 and intron 1 of human MGMT. Also constitutively expresses Puromycin fused to TagBFP.DepositorInsertU6_sgRNA(CTBP2)_U6_sgRNA(MGMT)
UseCRISPR and LentiviralExpressionMammalianPromoterU6Available SinceAug. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
P6598 MSCV-IP N-HAonly TBPL1
Plasmid#34896DepositorAvailable SinceFeb. 8, 2012AvailabilityAcademic Institutions and Nonprofits only -
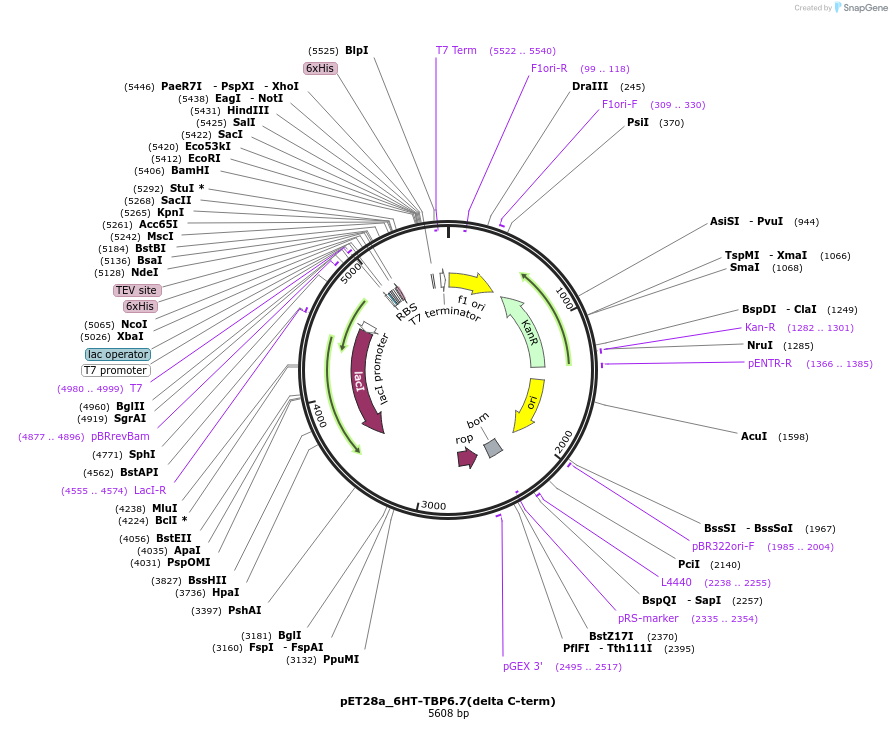
pET28a_6HT-TBP6.7(delta C-term)
Plasmid#112731PurposeThis mutant of TBP6.7 is lacking residues 91-98 at C-terminus, and was used to test the involvement of those residues in TAR recognition.DepositorInsert6His-TEV-TBP6.7(delta C-term)
Tags6His-TEVExpressionBacterialMutationdelta C-termPromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
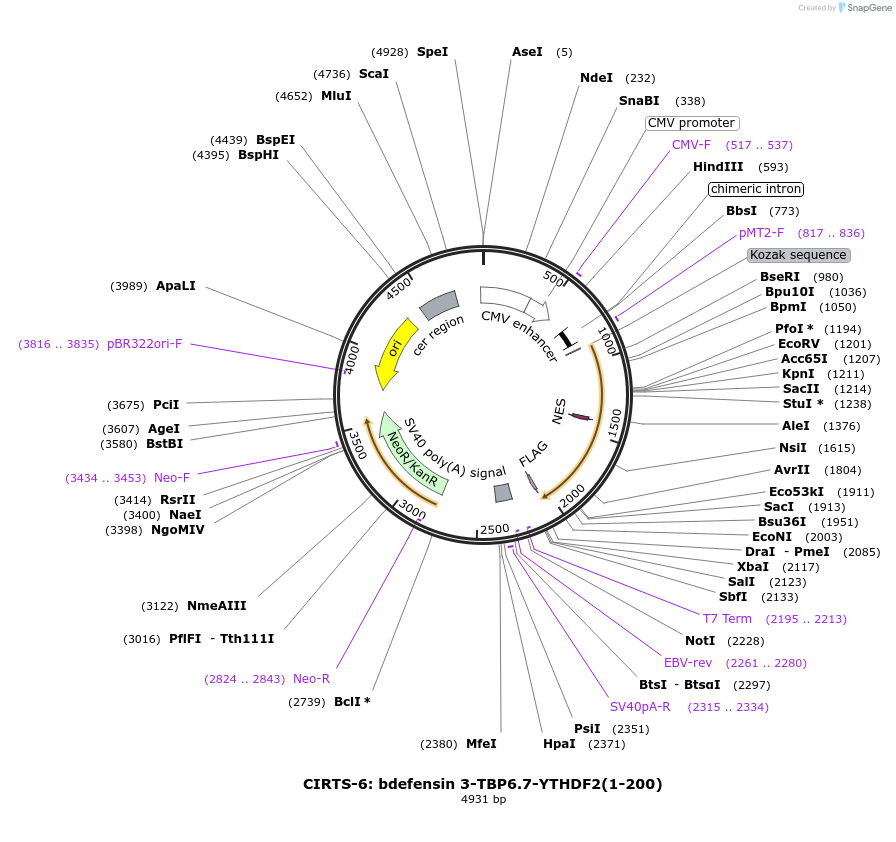
CIRTS-6: bdefensin 3-TBP6.7-YTHDF2(1-200)
Plasmid#132544PurposeExpresses CIRTS-6 (bdefensin 3-TBP6.7-YTHDF2(1-200)) in mammalian cellsDepositorInsertCIRTS-6: bdefensin 3-TBP6.7-YTHDF2(1-200)
TagsFLAG tagExpressionMammalianPromotercmv d0Available SinceSept. 30, 2019AvailabilityAcademic Institutions and Nonprofits only -
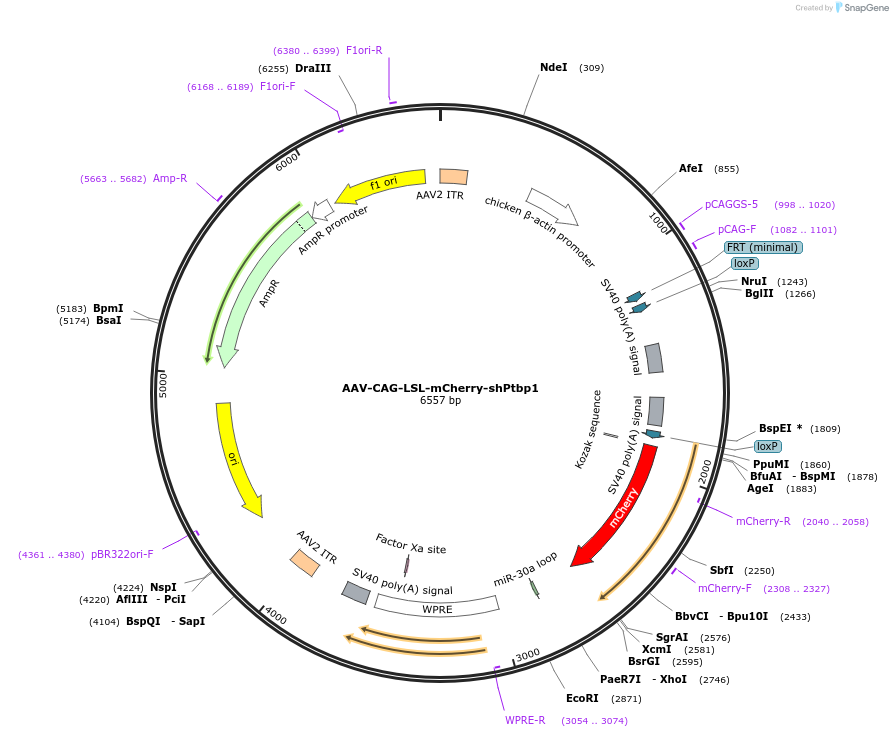
AAV-CAG-LSL-mCherry-shPtbp1
Plasmid#178591PurposeCre-dependent expression of mCherry and a shRNA against Ptbp1 under the constitutive promoterDepositorInsertmiRE-Ptbp1
UseAAVExpressionMammalianAvailable SinceFeb. 9, 2022AvailabilityAcademic Institutions and Nonprofits only