We narrowed to 10,309 results for: yeast
-
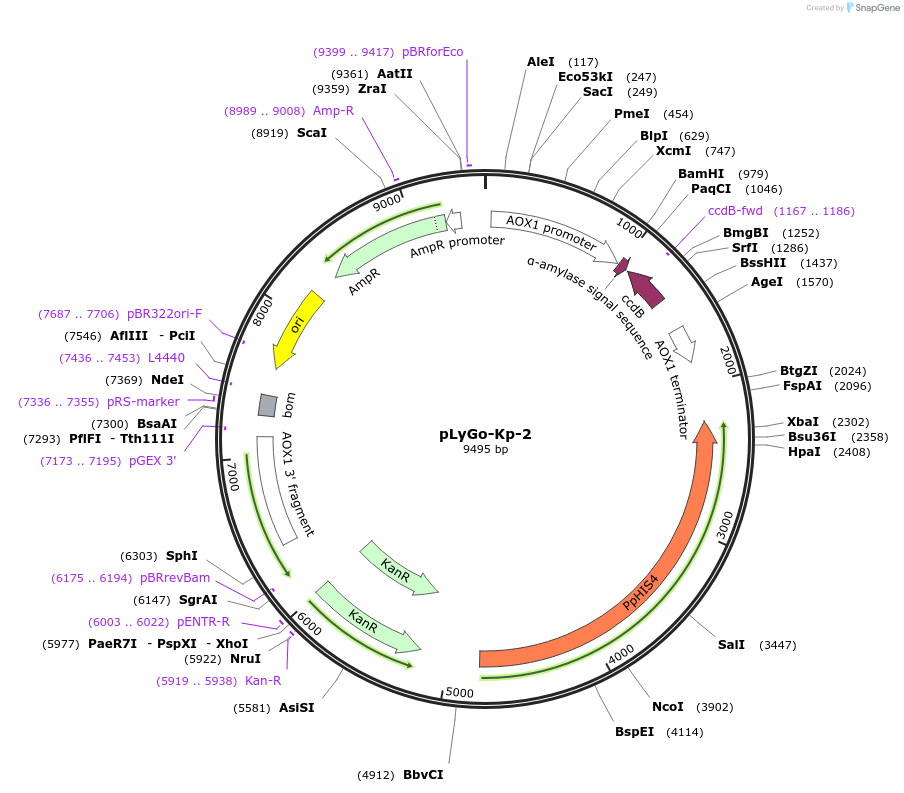
Plasmid#163144PurposepLyGo cloning vector for a sequence of interest (LPMO) in K. phaffii. Vector encoding the alfa-amylase signal peptide and the LyGo cassette (SapI-ccdB-SapI)DepositorTypeEmpty backboneUseSynthetic BiologyExpressionYeastPromoterAOX1Available SinceMay 7, 2021AvailabilityAcademic Institutions and Nonprofits only
-
pAG416GPD-SEC7-Cerulean
Plasmid#18851DepositorAvailable SinceNov. 21, 2008AvailabilityAcademic Institutions and Nonprofits only -
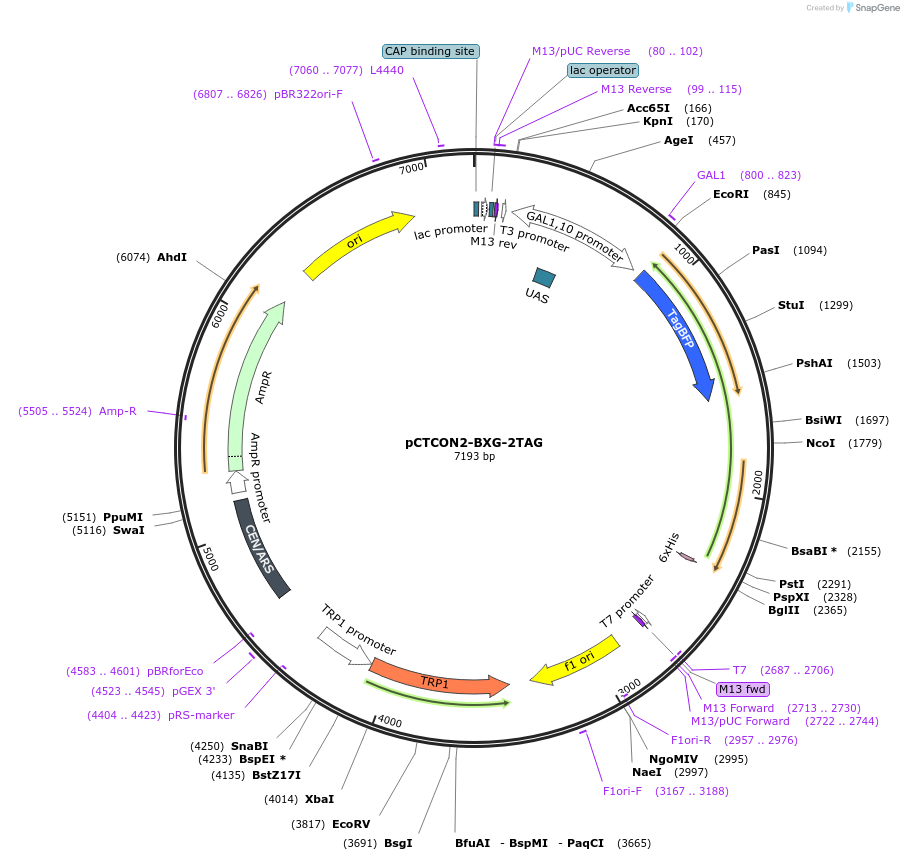
pCTCON2-BXG-2TAG
Plasmid#158138PurposeDual fluorescent protein reporter (BFP-GFP) for ncAA incorporation (2 TAG construct)DepositorInsertBFP-GFP fragment
ExpressionYeastMutationCloned in a TAG codon at the first serine residue…PromoterGAL1-10Available SinceJan. 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
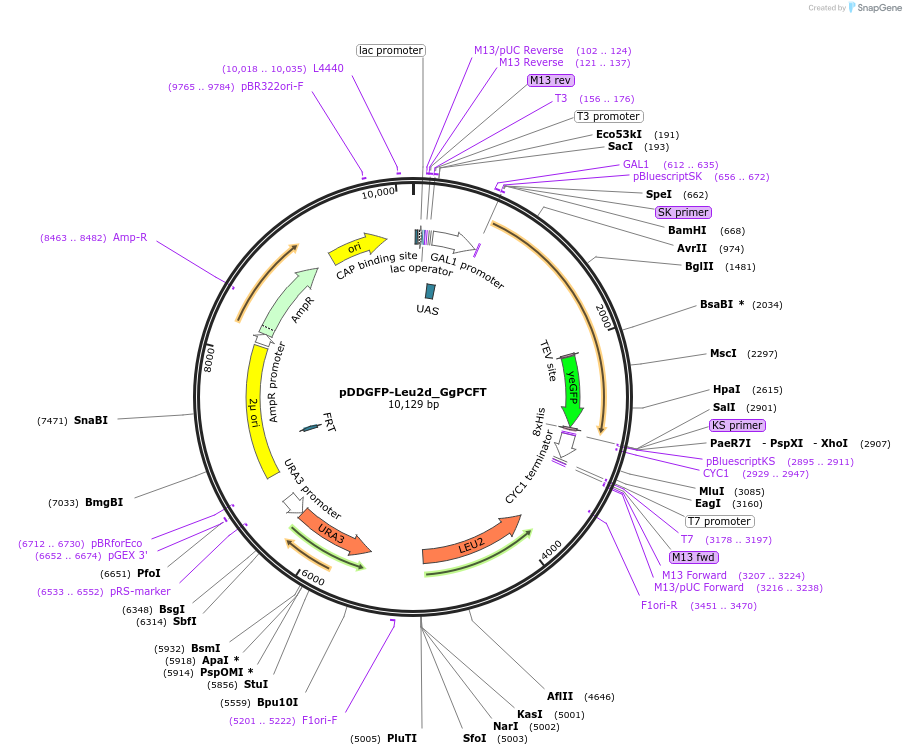
pDDGFP-Leu2d_GgPCFT
Plasmid#165414Purposeexpression GgPCFT protein with C terminally tagged GFP-His tag cleavable with TEV protease.DepositorAvailable SinceJuly 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
pAG416GPD-Cerulean-CHC1
Plasmid#18850DepositorAvailable SinceNov. 21, 2008AvailabilityAcademic Institutions and Nonprofits only -
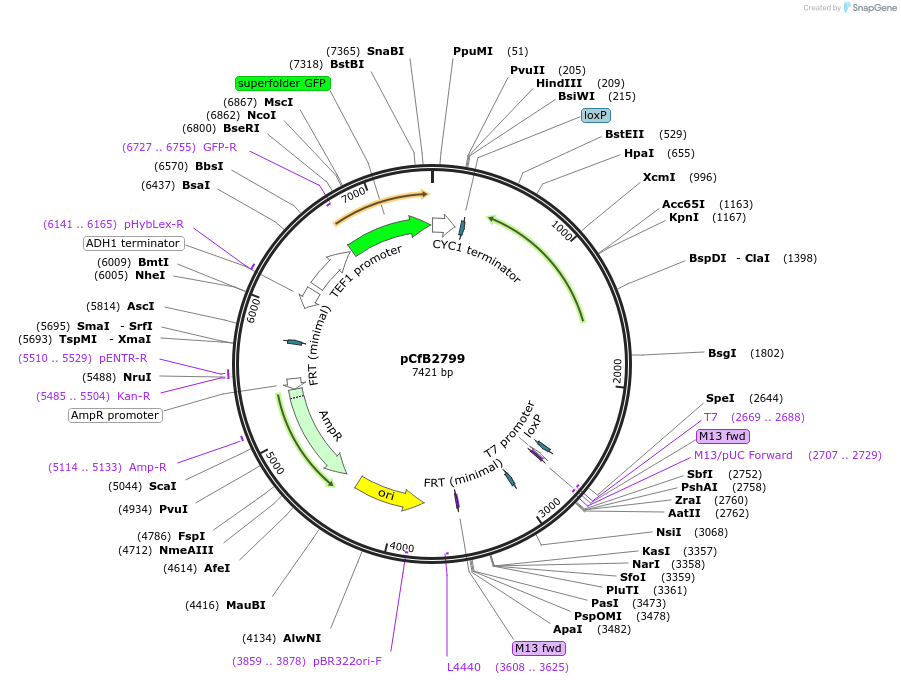
pCfB2799
Plasmid#63658PurposepCfB2799 is a vector for multiple integrations at sites sharing homology with Ty3Cons. The selective marker is Kl.LEU2. Additionally it contains a PTEF1-GFP as reporter.DepositorInsertGFP
UseCre/LoxExpressionYeastPromoterTEF1 promoterAvailable SinceAug. 15, 2016AvailabilityAcademic Institutions and Nonprofits only -
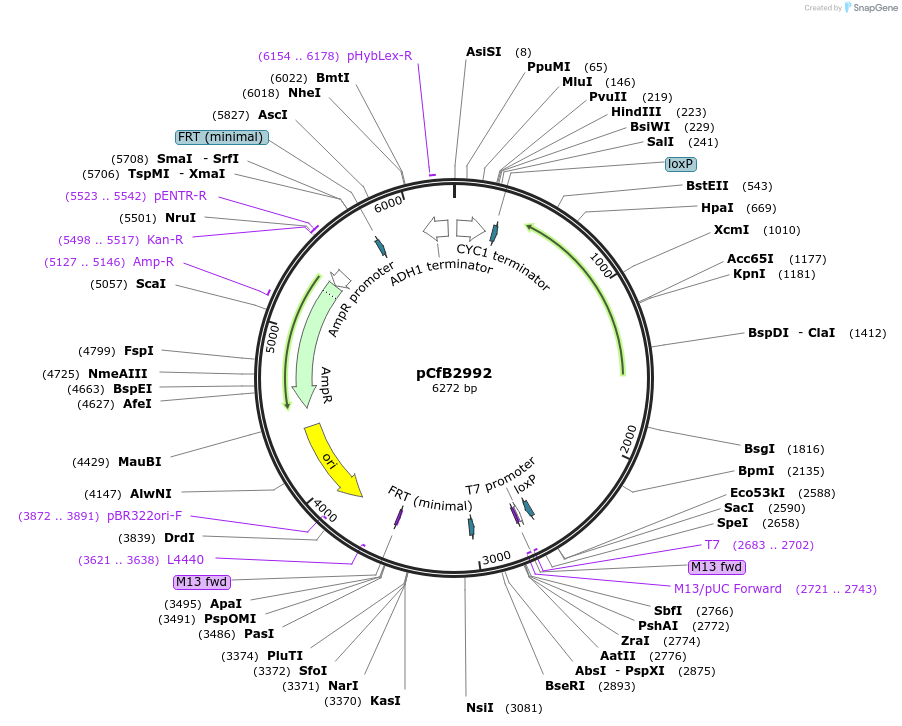
pCfB2992
Plasmid#63642PurposepCfB2992 is a vector for multiple integrations at sites sharing homology with Ty1Cons1. The selective marker is Kl.LEU2. Additionally it contains a USER cassette.DepositorTypeEmpty backboneUseCre/LoxExpressionYeastPromoternoneAvailable SinceAug. 15, 2016AvailabilityAcademic Institutions and Nonprofits only -
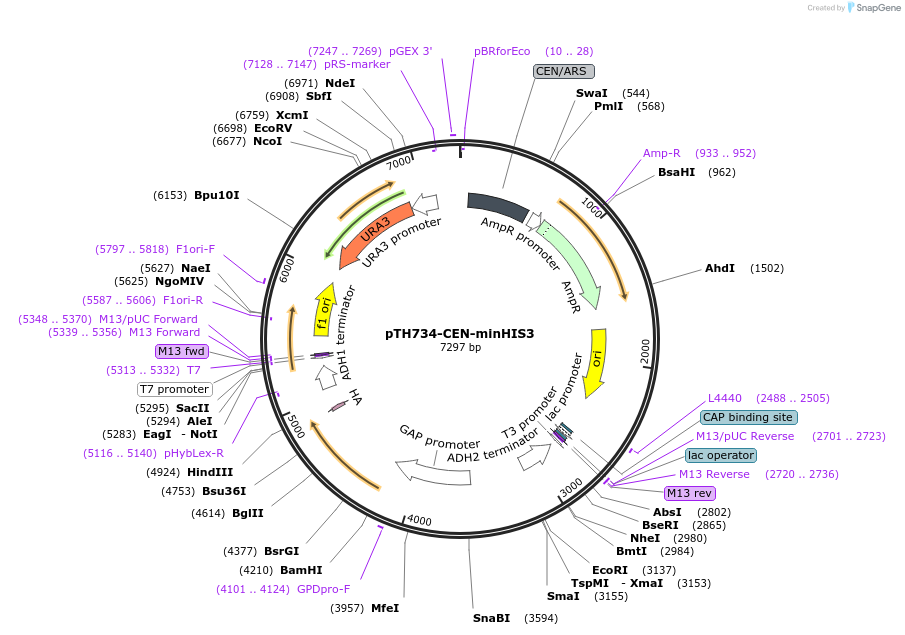
pTH734-CEN-minHIS3
Plasmid#38227DepositorInsertHIS3 (HIS3 Synthetic, Budding Yeast)
TagsHAExpressionYeastMutationEvery codon of the coding sequence has been repla…PromoterTDH3Available SinceDec. 19, 2012AvailabilityAcademic Institutions and Nonprofits only -
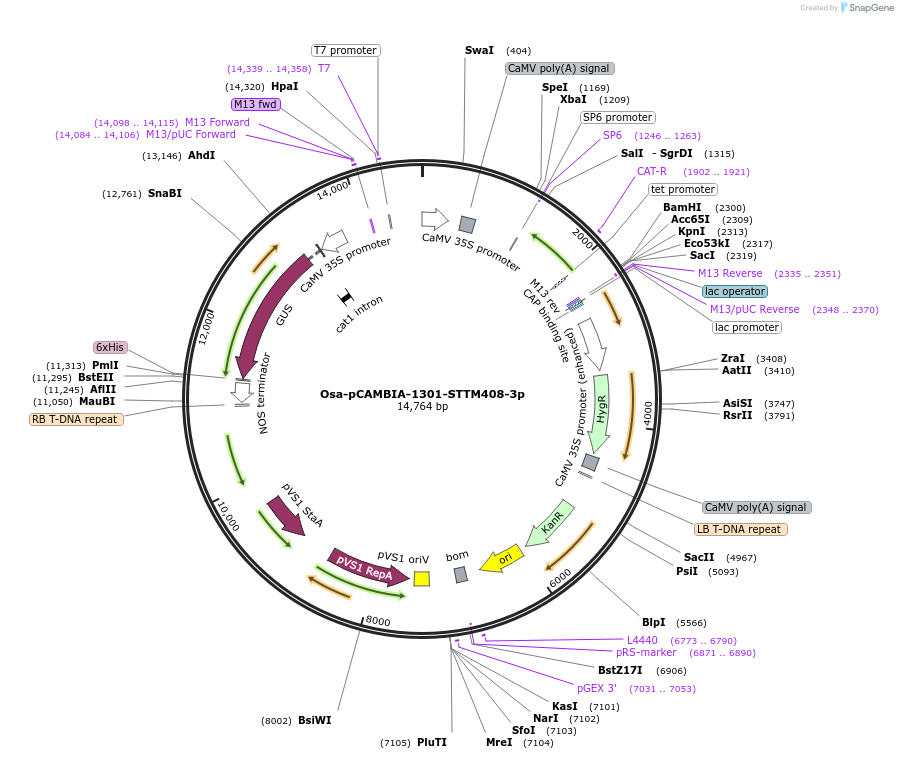
Osa-pCAMBIA-1301-STTM408-3p
Plasmid#84217PurposeDisturb the expression of miR408-3p in riceDepositorInsertmiR408-3p
ExpressionBacterial, Plant, and Yeast…Promoter2x35SAvailable SinceSept. 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
PdarC1
Plasmid#24835DepositorInsertPdarC1
TagsHA and MycExpressionYeastAvailable SinceAug. 23, 2011AvailabilityAcademic Institutions and Nonprofits only -
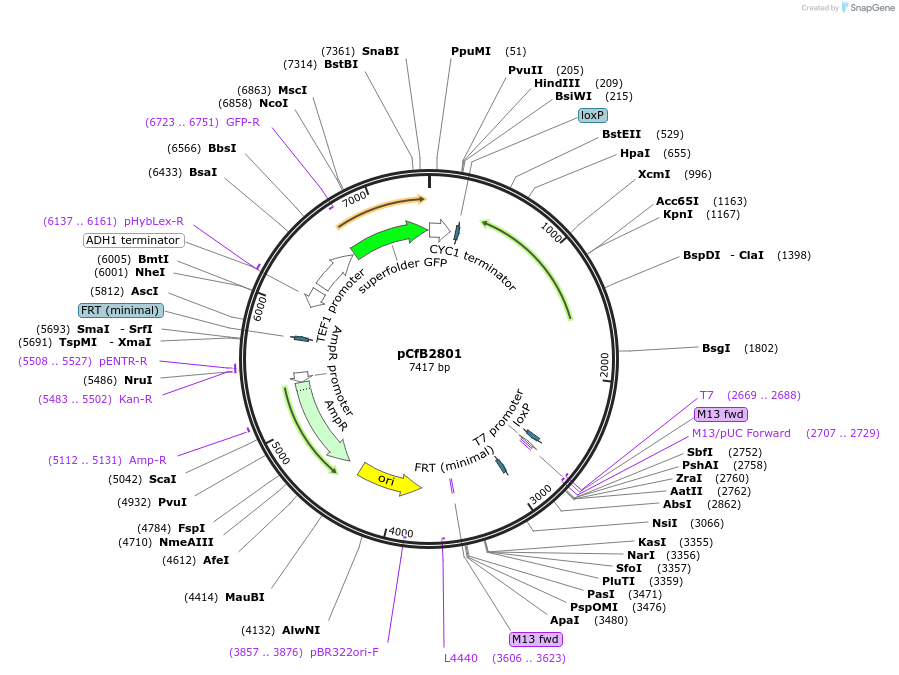
pCfB2801
Plasmid#63656PurposepCfB2801 is a vector for multiple integrations at sites sharing homology with Ty1Cons2. The selective marker is Kl.LEU2. Additionally it contains a PTEF1-GFP as reporter.DepositorInsertGFP
UseCre/LoxExpressionYeastPromoterTEF1 promoterAvailable SinceAug. 15, 2016AvailabilityAcademic Institutions and Nonprofits only -
M34 Xdsh PDZ bait-pGBT9
Plasmid#17190DepositorAvailable SinceFeb. 1, 2008AvailabilityAcademic Institutions and Nonprofits only -
Cwc25-pRSETA
Plasmid#61043PurposeEncodes the yeast Cwc25 protein for overexpression and purification using a C-terminal hexahistidine tag. A single cysteine residue is encoded after the Cwc25 sequence for fluorescent labeling.DepositorInsertCwc25
TagsTEV site, Histidine TagExpressionBacterialPromoterT7Available SinceJan. 27, 2015AvailabilityAcademic Institutions and Nonprofits only -
pAG416GPD-Cerulean-YPT52
Plasmid#18843DepositorAvailable SinceNov. 21, 2008AvailabilityAcademic Institutions and Nonprofits only -
pAG416GPD-Cerulean-YPT1
Plasmid#18848DepositorAvailable SinceNov. 21, 2008AvailabilityAcademic Institutions and Nonprofits only -
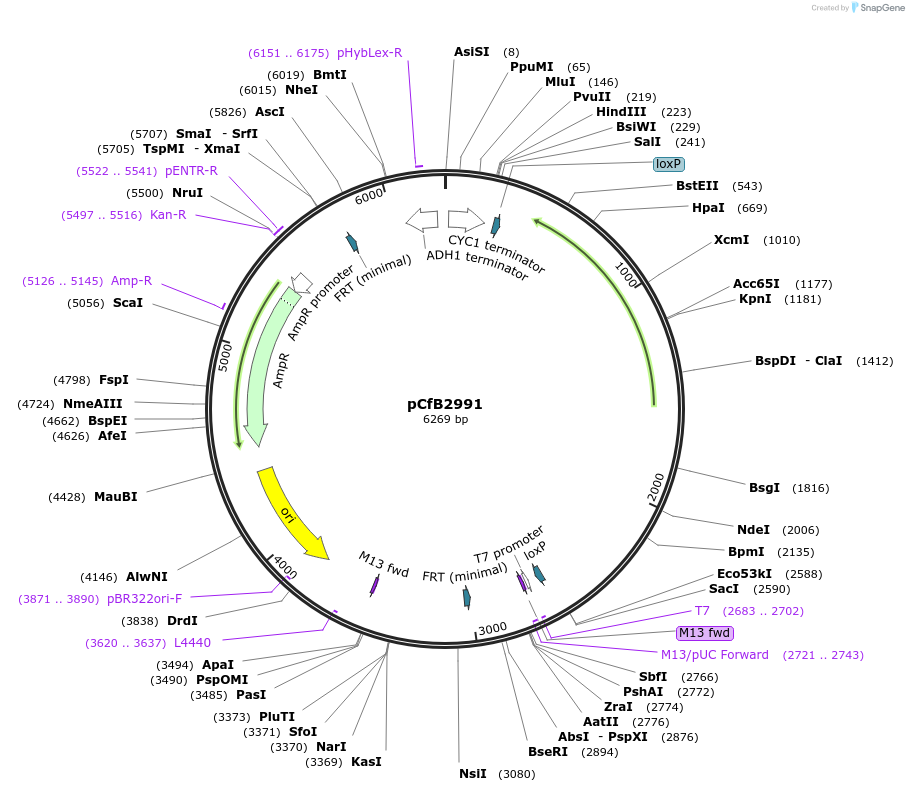
pCfB2991
Plasmid#63643PurposepCfB2991 is a vector for multiple integrations at sites sharing homology with Ty1Cons2. The selective marker is Kl.LEU2. Additionally it contains a USER cassette.DepositorTypeEmpty backboneUseCre/LoxExpressionYeastPromoternoneAvailable SinceAug. 15, 2016AvailabilityAcademic Institutions and Nonprofits only -
pSB819-PKR-hum-FFA
Plasmid#20048DepositorAvailable SinceJan. 28, 2009AvailabilityAcademic Institutions and Nonprofits only -
-
pAG416GPD-Cerulean-YPT6
Plasmid#18845DepositorAvailable SinceNov. 21, 2008AvailabilityAcademic Institutions and Nonprofits only -